Abstract
Capreomycin (CMN) belongs to the tuberactinomycin family of nonribosomal peptide antibiotics that are essential components of the drug arsenal for the treatment of multidrug-resistant tuberculosis. Members of this antibiotic family target the ribosomes of sensitive bacteria and disrupt the function of both subunits of the ribosome. Resistance to these antibiotics in Mycobacterium species arises due to mutations in the genes coding for the 16S or 23S rRNA but can also arise due to mutations in a gene coding for an rRNA-modifying enzyme, TlyA. While Mycobacterium species develop resistance due to alterations in the drug target, it has been proposed that the CMN-producing bacterium, Saccharothrix mutabilis subsp. capreolus, uses CMN modification as a mechanism for resistance rather than ribosome modification. To better understand CMN biosynthesis and resistance in S. mutabilis subsp. capreolus, we focused on the identification of the CMN biosynthetic gene cluster in this bacterium. Here, we describe the cloning and sequence analysis of the CMN biosynthetic gene cluster from S. mutabilis subsp. capreolus ATCC 23892. We provide evidence for the heterologous production of CMN in the genetically tractable bacterium Streptomyces lividans 1326. Finally, we present data supporting the existence of an additional CMN resistance gene. Initial work suggests that this resistance gene codes for an rRNA-modifying enzyme that results in the formation of CMN-resistant ribosomes that are also resistant to the aminoglycoside antibiotic kanamycin. Thus, S. mutabilis subsp. capreolus may also use ribosome modification as a mechanism for CMN resistance.
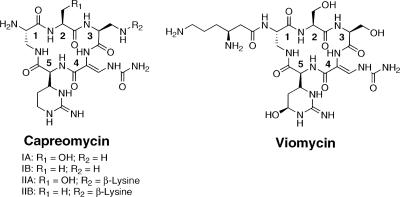
Mycobacterium tuberculosis, the causative agent of tuberculosis, has been causing disease in humans since the beginning of civilization (8). Despite more than 50 years of vaccine and antibiotic development, it has been estimated that 225 million new cases of tuberculosis will arise between the years 1998 and 2030, with 79 million tuberculosis-related deaths (25). One of the challenges in treating this disease is the widespread development of multidrug-resistant tuberculosis (MDR-TB), defined as an infection that does not respond to treatment with either of the first-line drugs isoniazid and rifampin (9). The development of MDR-TB has resulted in an increased emphasis on the use of second-line drugs to treat these infections. One of these second-line drugs is capreomycin (CMN), a collection of four structurally related peptide antibiotics (Fig. 1). For simplicity, throughout this report the individual derivatives are identified as CMN IA, IB, IIA, and IIB and are referred to collectively as CMN. The importance of CMN for the treatment of MDR-TB is reflected in this drug's being included on the World Health Organization's list of essential medicines (36). There is additional interest in CMN because of the recent finding that this drug is bactericidal for nonreplicating M. tuberculosis, suggesting the potential use of CMN to treat latent tuberculosis infections (15).
FIG. 1.
Chemical structures of the four derivatives that make up CMN (left) and the structurally related tuberactinomycin antibiotic VIO (right). Numbering of residues within structures corresponds to residue numbers mentioned in the text. R1, residue 1; R2, residue 2.
CMN and a structural analog, viomycin (VIO) (Fig. 1), disrupt the growth of Mycobacterium spp. by interfering with the function of the ribosome (22-24, 37-43). This conclusion is based on the isolation and characterization of resistant strains, along with in vitro analyses of ribosome binding and the disruption of peptide synthesis. There were two surprising results from these studies that emphasize the distinct mechanism of action of CMN and VIO. First, resistance to VIO in M. smegmatis is conferred by mutations in either the 16S or 23S rRNA, suggesting that this antibiotic interferes with the function of both ribosomal subunits (41). The second result was the recent finding that M. tuberculosis resistance to CMN and VIO can also arise from a mutation in the gene tlyA (22). Subsequent analyses determined that the TlyA enzyme likely catalyzes the methylation of both the 16S and 23S rRNAs and that the methylated regions are essential for CMN and VIO sensitivity, possibly forming part of the binding site of these antibiotics (18). Thus, resistance to CMN and VIO can arise from point mutations in the 16S or 23S rRNA or from the loss of modifications to these rRNAs.
While the CMN and VIO resistance mechanisms in Mycobacterium spp. involve mutations in the rRNA or a gene coding for an rRNA-modifying enzyme, the bacteria that naturally produce these antibiotics are proposed to have resistance mechanisms that are independent of ribosome modification (2, 28, 33). VIO resistance by Streptomyces sp. strain ATCC 11861 (previously known as Streptomyces vinaceus) occurs via antibiotic inactivation by Vph, a VIO phosphorylase (2). CMN, while commonly referred to as a single molecule, is actually a mixture of four structural derivatives (Fig. 1). The structural differences between these derivatives are particularly relevant in the context of resistance genes carried by the producing bacterium. For example, a gene coding for a homolog of Vph was isolated from the CMN producer Saccharothrix mutabilis subsp. capreolus (previously Streptomyces capreolus), but this gene confers resistance only to CMN IA and IIA, leaving CMN IB and IIB active. This selectivity is due to Cph's catalyzing the phosphorylation of the hydroxyl group of residue 2 of CMN IA and IIA that is absent from CMN IB and IIB (Fig. 1) (28, 33). A second gene, cac, was identified and confers resistance to all four derivatives of CMN (28, 33). The encoded enzyme is proposed to catalyze the acetylation of the α-amino group of residue 1 of the cyclic pentapeptide core, thereby inactivating CMN. Consistent with this proposal, Cac did not confer resistance to VIO because of the β-lysine (β-Lys) attached to the α-amino group of residue 1 of VIO.
CMN and VIO are peptide antibiotics with subtly different cyclic pentapeptide cores that can be decorated by carbamoylation, hydroxylation, or acylation with β-Lys (Fig. 1). A series of precursor labeling studies of CMN and VIO have been performed to investigate how these unusual antibiotics are assembled by the producing bacteria (4, 5, 12, 35). Our group has previously proposed a biosynthetic mechanism for the assembly of VIO using these labeling experiment results in combination with data from bioinformatic and biochemical analyses of the VIO biosynthetic gene cluster and the enzymes it encodes (19, 34). At the time, we also proposed that CMN biosynthesis would incorporate similar mechanisms with subtle differences to account for the structural differences between these antibiotics.
Our interests in deciphering how this family of antibiotics is biosynthesized are in two areas. First, we want to answer the basic biological question of how these unusual cyclic peptides, consisting of rare nonproteinogenic amino acids, are assembled. Second, we are interested in harnessing the biosynthetic machinery of CMN and VIO production to generate new structural derivatives of these antibiotics through the use of metabolic engineering. Here we describe the isolation and sequencing of the CMN biosynthetic gene cluster from S. mutabilis subsp. capreolus strain ATCC 23892. Bioinformatic analysis of this gene cluster provides a molecular blueprint for CMN biosynthesis and explains the structural differences between CMN and VIO. The integration of this biosynthetic gene cluster into the chromosome of Streptomyces lividans 1326 resulted in the heterologous production of CMN by this naturally nonproducing bacterium. This is a significant finding because S. mutabilis subsp. capreolus has proven to be intractable to genetic manipulation (27). Thus, the metabolic engineering of CMN biosynthesis in the natural producer was not possible. The results presented here circumvent this problem. Finally, while previous work suggested that S. mutabilis subsp. capreolus does not alter its ribosomes to become resistant to CMN, we present data that strongly suggest that ribosome modification is a natural CMN resistance mechanism for this bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth medium.
The strains and plasmids used in this study are listed in Table 1. S. mutabilis subsp. capreolus (ATCC 23892) was obtained from the American Type Culture Collection. S. lividans 1326 was kindly provided by Amy Gehring (Williams College). S. mutabilis subsp. capreolus and S. lividans 1326 were propagated on ISP2 medium (Difco, Becton Dickinson Microbiology Systems, Sparks, MD). S. lividans 1326 was grown in yeast extract-malt extract (YEME) medium (21) to produce mycelia for generating protoplasts. S. mutabilis subsp. capreolus was grown in YEME medium to produce mycelia for chromosomal DNA preparation. For heterologous production of CMN, S. lividans 1326 strains were grown in VIO production medium as previously described (31, 34). All Escherichia coli strains were grown in Luria-Bertani (LB) liquid medium or on LB solid medium with appropriate antibiotics. Kanamycin (50 μg/ml), apramycin (50 μg/ml), ampicillin (100 μg/ml), and chloramphenicol (30 μg/ml) were used in solid and liquid media for the propagation of plasmids or cosmids in E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Laboratory strain |
| XL1-Blue MR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| S. mutabilis subsp. capreolus | Wild type (ATCC 23892) | ATCCa |
| S. lividans 1326 | Wild type | A. Gehring |
| EAF1001 | S. lividans 1326 with pCMN-P4C8RF-436 integrated into the φC31 attB site; clone 1 | This study |
| EAF1002 | S. lividans 1326 with pCMN-P4C8RF-436 integrated into the φC31 attB site; clone 2 | This study |
| EAF1003 | S. lividans 1326 with pOJ436 inserted into the φC31 attB site; clone 1 | This study |
| EAF1004 | S. lividans 1326 with pOJ436 inserted into the φC31 attB site; clone 2 | This study |
| Plasmids | ||
| SuperCos1 | Kanr Ampr cloning cosmid | Stratagene |
| pOJ436 | Aprr oriT φC31 int φC31 attP cosmid vector | 3 |
| pCR-BluntII-TOPO | Kanr cloning vector | Invitrogen |
| pCMN-P4C8RF | S. mutabilis subsp. capreolus genomic DNA cloned into SuperCos1; contains CMN gene cluster | This study |
| pCMN-P4C8RF-436 | pCMN-P4C8RF with Aprr oriT φC31 int φC31 attP DraI fragment of pOJ436 inserted into HpaI site | This study |
| pTOPO-cmnU | cmnU cloned into pCR-BluntII-TOPO cloning vector | This study |
| pBAD33 | Arabinose-inducible expression vector | 14 |
| pET22b | Kanr cloning vector | Novagen |
| pET22b-cmnU | cmnU cloned into NdeI/HindIII site of pET22b | This study |
| pBAD33-cmnU | XbaI/HindIII fragment of pET22b-cmnU cloned into XbaI/HindIII site of pBAD33 | This study |
| pLJ102 | IPTG-inducible expression vector | 18 |
| pSJ101 | pLJ102 derivative with tlyA insert | 18 |
| pSE34 | Streptomyces sp. expression vector | 29 |
| pSE34-cmnU | pSE34 with cmnU cloned into the XbaI/HindIII site | This study |
ATCC, American Type Culture Collection.
Genomic DNA isolation and cosmid library construction.
High-molecular-weight chromosomal DNA of S. mutabilis subsp. capreolus was prepared from YEME-grown mycelia. Approximately 0.3 g (wet weight) of mycelia was washed with 500 μl of lysis buffer (10.3% [wt/vol] sucrose, 25 mM Tris-Cl, 25 mM EDTA, pH 8.0), and mycelia were collected by centrifugation and resuspended in lysis buffer containing 5 mg of lysozyme/ml. The sample was incubated at 37°C for 30 min, followed by the addition of 250 μl of 2% (wt/vol) sodium dodecyl sulfate. The sample was mixed and then added to 125 μl of phenol-chloroform-isoamyl alcohol (25:24:1). The sample was subjected to a vortex and then centrifuged (13,200 × g for 15 min). The aqueous layer was collected, and isopropanol and sodium acetate (pH 5.2) were added to precipitate the DNA. The sample was centrifuged (10,000 rpm for 10 min), the supernatant was poured off, and the DNA pellet was washed with 70% (vol/vol) ethanol. The DNA pellet was dried and resuspended in 100 μl of 10 mM Tris-Cl (pH 8.0)-1 mM EDTA (pH 8.0).
Purified DNA was digested with Sau3AI to give 30- to 50-kb fragments that were subsequently ligated into the BamHI site of SuperCos1 and packaged into lambda phage by using a Gigapack III XL packaging extract kit according to the instructions of the manufacturer (Stratagene, Cedar Creek, TX). The packaged cosmid pool was used to infect E. coli XL1-Blue MR according to the instructions of the manufacturer (Stratagene). A total of 672 cosmid-containing clones were isolated and individually frozen at −80°C in microtiter dish wells and in 25-μl pools of eight clones from each member of a microtiter dish column. Thus, first the column pools, followed by individual clones of the targeted column, could be screened for the CMN gene cluster.
Screening of cosmid library.
Column pools of the cosmid library were screened by PCR amplification of cph, one of the known CMN resistance genes (33). The primers used for this screen were as follows: Cph/For (5′-CCCACCTTGTTGACGTGGT-3′) and Cph/Rev (5′-TCAGCGGTAGGCGGTCAG-3′). Boiled cells from each cosmid pool were used as a source of template DNA for PCR amplification. Individual members of each cph-positive cosmid pool were subsequently screened by PCR amplification to identify the specific positive clone. Cosmid pCMN-P4C8RF was identified as a cph-positive cosmid. The two primers described above and two primers that were the reverse complements to these primers were used in sequencing reactions to confirm that the entire cph gene was contained on pCMN-P4C8RF.
Sequencing and annotation of pCMN-P4C8RF.
Two- to three-kilobase fragments of DNA from pCMN-P4C8RF were subcloned into pSMARTLCKan by Lucigen Corp. (Middleton, WI). The subclones were sequenced at the University of Wisconsin Biotechnology Center (fivefold coverage, twofold minimum). Contigs were assembled using the SeqMan program (Lasergene, Madison, WI). The annotations of the open reading frames (ORFs) and putative gene functions were assigned using a combination of MapDraw (Lasergene), FramePlot 3.0 (17), and blastp, PSI-BLAST, and RPS-BLAST programs (National Center for Biotechnology Information) (1).
Construction of pCMN-P4C8RF-436 and integration into the S. lividans 1326 chromosome.
The 6.7-kb DraI fragment of pOJ436 (3), containing the oriT, aac(3)IV (apramycin resistance), φC31 attP, and φC31 int genetic information, was cloned into the HpaI site of the SuperCos1 backbone of pCMN-P4C8RF. The resulting cosmid (pCMN-P4C8RF-436) was capable of integration into the φC31 attB site of the S. lividans 1326 genome, and selection for this integration was performed using apramycin. S. lividans 1326 was transformed with this cosmid by using the established protocols for protoplast formation and transformation (20). Transformants were selected by flooding transformation plates with apramycin (40 μg/ml). Transformants from each plate were streaked for isolation onto R2YE (21) and ISP2 plates supplemented with apramycin (40 μg/ml). Two integration-containing strains (EAF1001 and EAF1002) were characterized further. Two other strains (EAF1003 and EAF1004) containing the integrated pOJ436 cosmid alone were constructed as controls for analyzing the heterologous production of CMN. To confirm the integration of the CMN biosynthetic gene cluster into S. lividans 1326, all strains were screened by PCR amplification for the presence of cmnR, cmnI, and cph. These genes were used because cmnR and cph are at opposite ends of the gene cluster and cmnI is at the center of the gene cluster. The presence of all three genes in S. lividans 1326 strongly suggested that the entire CMN biosynthetic gene cluster was present in EAF1001 and EAF1002.
Analysis of EAF1001, EAF1002, EAF1003, and EAF1004 for CMN production.
Single colonies of EAF1001, EAF1002, EAF1003, and EAF1004 were used to inoculate 50 ml of YEME supplemented with apramycin (40 μg/ml). The cultures were grown at 30°C at 200 rpm until cultures were saturated (8 to 14 days). The cells were subsequently harvested by centrifugation, washed with 10.3% (wt/vol) sucrose, resuspended in 5 ml of 10.3% (wt/vol) sucrose, and frozen at −20°C until use.
To test for CMN production, 50 μl of the frozen stock of each strain was used to inoculate 100 ml of VIO production medium (31) in 1-liter unbaffled flasks. The cultures were grown at 30°C at 200 rpm for 7 days. Any potential CMN produced was purified using a previously described protocol for the purification of the structurally related antibiotic VIO (31, 34). After purification, UV-visible spectrophotometry analysis was performed (Beckman Coulter DU640) and spectra were compared to an authentic CMN standard. To identify whether one or more of the CMN derivatives were produced, the samples were analyzed by high-performance liquid chromatography (HPLC) and electrospray ionization-mass spectrometry (ESI-MS). Briefly, purified samples were run on a C18 small-pore column (Vydack) on a Beckman Gold HPLC system with a 1-ml/min flow rate. Buffer A was H2O-0.1% trifluoroacetic acid and buffer B was acetonitrile-0.1% trifluoroacetic acid. The separation profile was 5 min of isocratic development at 100% A-0% B, a 15-min linear gradient from 100% A-0% B to 50% A-50% B, a 1-min linear gradient from 50% A-50% B to 0% A-100% B, and 5 min of isocratic development at 0% A-100% B. The elution of metabolites at 268 nm, the maximum λ characteristic of tuberactinomycins (4), was monitored. For ESI-MS, samples eluting from the HPLC system were collected, the solvent was evaporated under a vacuum, and the samples were submitted to the University of Wisconsin Biotechnology Center mass spectrometry facility. Authentic CMN was obtained from MB Biomedicals Inc.
Analysis of cmnU in E. coli.
The gene cmnU was PCR amplified from pCMN-P4C8RF by using the following primers: CmnU-NdeI (5′-AAGGGCCCCCATATGCCTTCGGAAGGTCTG-3′; the NdeI site is underlined) and CmnU-HindIII (5′-GGTGTGTGTTCGAACTCACACTAACGCGCC-3′). The amplicon was cloned into pCR-BluntII-TOPO according to the instructions of the manufacturer (Invitrogen), resulting in pCR-BluntII-TOPO-cmnU. The NdeI/HindIII fragment of pCR-Blunt-TOPO-cmnU containing cmnU was subcloned into the corresponding restriction sites of pET22b (Novagen), resulting in pET22b-cmnU. The XbaI/HindIII fragment of pET22b-cmnU containing the optimized ribosome binding site and cmnU was subcloned into the corresponding sites of pBAD33 (14), resulting in pBAD33-cmnU. This construct results in the expression of cmnU under the control of the arabinose promoter of pBAD33. DH5α strains containing either pLJ102 (empty vector) or pSJ101 (tlyA inserted into pLJ102 with expression controlled by IPTG [isopropyl-β-d-thiogalactopyranoside] induction) were transformed with pBAD33 and pBAD33-cmnU (18). Each of these strains was grown in LB medium containing chloramphenicol (15 μg/ml) and ampicillin (100 μg/ml) for plasmid maintenance, IPTG (40 μg/ml) and arabinose (2% [wt/vol]) for the induced expression of tlyA and cmnU, respectively, and various concentrations of CMN (12.5 to 100 μg/ml) or kanamycin (6 to 96 μg/ml). Similar numbers of CFU of each strain were added to the media, and the optical density at 600 nm after 16 h of incubation at 37°C was determined. The value reported as the MIC is the antibiotic concentration at which the optical density at 600 nm after 16 h was <0.1.
Analysis of cmnU in S. lividans 1326.
The XbaI/HindIII fragment of pET22b-cmnU was cloned into the corresponding sites of pSE34 (29) to generate pSE34-cmnU. This vector construct results in the constitutive expression of cmnU when pSE34-cmnU is introduced into S. lividans 1326. pSE34-cmnU and pSE34 were introduced into S. lividans 1326 by transformation, and successful transformants were selected by using an overlay of thiostrepton (8 μg/ml). Transformants were streaked for isolation onto ISP2 medium containing thiostrepton. To test for CMN or kanamycin resistance, 105 spores of S. lividans 1326 containing either pSE34-cmnU or pSE34 were plated onto ISP2 plates supplemented with CMN (25 to 1,600 μg/ml) or kanamycin (3 to 1,600 μg/ml). The plates were incubated at 30°C for 2 days, and the MIC was defined as the lowest antibiotic concentration at which no growth was observed.
Nucleotide sequence accession number.
The DNA sequence from the insert of pCMN-P4C8RF has been deposited in GenBank with accession number EF472579.
RESULTS AND DISCUSSION
Isolation and sequence analysis of the CMN biosynthetic gene cluster from S. mutabilis subsp. capreolus.
To isolate the CMN biosynthetic gene cluster from S. mutabilis subsp. capreolus, a cosmid library of the organism's chromosomal DNA was constructed. Cosmids containing at least a portion of the CMN biosynthetic pathway were identified by PCR-based screening of cosmid pools and individual cosmids for the presence of cph, one of the previously identified CMN resistance genes (28, 33). The focus on cph rather than cac, the other known CMN resistance gene, was due to our prior finding that a homolog of cph in the VIO-producing bacterium Streptomyces sp. strain ATCC 11861 is associated with the VIO biosynthetic gene cluster (34). Thus, it was hypothesized that cph would be associated with the CMN biosynthetic gene cluster. By using cph-specific primers for PCR-based screening and subsequent sequencing, cosmid pCMN-P4C8RF was identified as containing cph. This cosmid was sequenced in its entirety, and 33 ORFs on the DNA inserted into pCMN-P4C8RF were identified (Fig. 2).
FIG. 2.
Schematic of the 33 S. mutabilis subsp. capreolus ORFs contained on the pCMN-P4C8RF cosmid. The proposed CMN biosynthetic gene cluster consists of ORFs highlighted in gray and black. Those ORFs coding for proteins with significant sequence identity to proteins encoded by the VIO biosynthetic gene cluster are shown in gray. The newly identified CMN resistance gene is shown in black. The remaining ORFs flanking the proposed CMN biosynthetic gene cluster are shown in white.
A comparison of these 33 ORFs with those proposed to be involved in VIO biosynthesis identified 18 ORFs encoding proteins showing significant sequence identity to proteins encoded by genes associated with the VIO biosynthetic gene cluster (Fig. 2; Table 2). Additionally, 17 of these ORFs (cmnA to cmnP and cph) were arrayed in an order identical to that of their respective homologs in the VIO biosynthetic gene cluster. The one ORF not in a location similar to that of its homolog in the VIO biosynthetic gene cluster was cmnR, coding for a putative transcriptional regulator. This gene was upstream of cmnA but separated from cmnA by an additional ORF (cmnU) that did not have a homolog in the VIO biosynthetic gene cluster. However, based on the sequence similarity of CmnU to rRNA-modifying enzymes, cmnU was hypothesized to be a newly identified CMN resistance gene. Based on these similarities with the VIO biosynthetic gene cluster and a gene coding for a putative rRNA-modifying enzyme, it was reasonable to propose that 19 ORFs were involved in CMN production. The remaining ORFs contained on pCMN-P4C8RF that surrounded the 19 proposed CMN-associated genes did not display any sequence similarity to genes that play any clear role in CMN biosynthesis (see Table S1 in the supplemental material). Of particular interest was the absence of cac, the second previously identified CMN resistance gene.
TABLE 2.
Analysis of ORF products proposed to be involved in CMN production
| Predicted ORF product (no. of amino acids) | Homolog for VIO biosynthesis (no. of amino acids) | % Identity | Predicted function(s)a |
|---|---|---|---|
| CmnR (238) | VioR (263) | 49 | Transcriptional regulator (LuxR family) |
| CmnU (240) | CMN resistance; rRNA methyltransferase | ||
| CmnA (2,642) | VioA (2,123) | 53 | NRPS (X-A-PCP-C-A-PCP-C) |
| CmnB (370) | VioB (346) | 65 | l-2,3-DAP formation |
| CmnC (339) | VioC (358) | 62 | CAM formation |
| CmnD (374) | VioD (389) | 65 | CAM formation |
| CmnE (416) | VioE (447) | 63 | CMN efflux |
| CmnF (1,057) | VioF (1,073) | 56 | NRPS (A-PCP-C) |
| CmnG (953) | VioG (1,088) | 48 | NRPS (C-A-PCP-C/) |
| CmnH (249) | VioH (262) | 52 | Type II thioesterase |
| CmnI (549) | VioI (550) | 52 | NRPS (PCP-C) |
| CmnJ (384) | VioJ (390) | 62 | l-2,3-DAP α,β-desaturase |
| CmnK (333) | VioK (360) | 62 | l-2,3-DAP formation |
| CmnL (296) | VioL (308) | 65 | Carbamoyltransferase |
| CmnM (402) | VioM (457) | 45 | NRPS (C); β-Lys attachment |
| CmnN (63) | VioN (63) | 75 | Unknown function |
| CmnO (585) | VioO (610) | 52 | NRPS (A-PCP); β-Lys attachment |
| CmnP (449) | VioP (445) | 77 | β-Lys formation |
| Cph (282) | Vph (293) | 53 | CMN resistance; CMN phosphotransferase |
Abbreviations: A, adenylation domain; PCP, peptidyl carrier protein domain; C, condensation domain; X, domain of unknown function; C/, modified condensation domain; CAM, 2S,3R-capreomycidine.
Heterologous production of CMN in S. lividans 1326.
S. mutabilis subsp. capreolus has proven to be intractable to genetic manipulation (27). This eliminated the possibility of using targeted gene disruption followed by metabolite analysis as a means to support the hypothesis that the CMN biosynthetic gene cluster had been identified. To address this issue, we investigated whether the introduction of a modified form of pCMN-P4C8RF into S. lividans 1326 would result in the heterologous production of CMN by this nonproducing bacterium. The pCMN-P4C8RF cosmid was modified to contain the genetic information from pOJ436 that enables conjugal transfer between E. coli and S. lividans 1326, integration into the φC31 site of the S. lividans 1326 genome, and apramycin resistance. S. lividans 1326 was transformed with the resulting cosmid, pCMN-P4C8RF-436, and transformants were selected using apramycin resistance. Two of the isolated transformants were characterized further. Of particular interest was the finding that the transformants were resistant not only to apramycin but also to CMN. The isolation of CMN-resistant colonies was an important finding because while the integrating cosmid contained cph, conferring resistance to CMN IA and IIA (28, 33), it did not contain the other reported resistance gene, cac, to give resistance to CMN IB and IIB (28, 33). Importantly, when pOJ436 alone was introduced into S. lividans 1326, it did not convey CMN resistance. The fact that the transformants were resistant to a mixture of all four CMN derivatives suggested that there was an additional mechanism of CMN resistance encoded by the DNA inserted into S. lividans 1326. Analysis of this additional resistance gene will be discussed in more detail below.
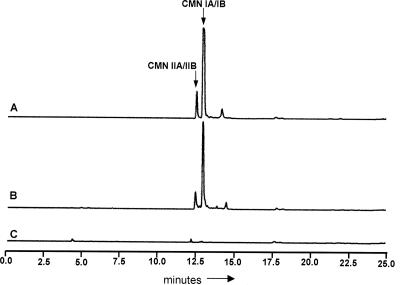
Two successful transformants from the apramycin selection were analyzed for CMN production. Each of these strains (EAF1001 and EAF1002), along with two negative control strains in which just pOJ436 was integrated (EAF1003 and EAF1004), was first grown in YEME medium containing apramycin. These cells were washed to remove the antibiotic and were subsequently used to inoculate VIO production medium lacking an antibiotic for selection. After 7 days of growth, the strains were analyzed for CMN production by purifying metabolites from the culture supernatant by an established protocol for VIO purification (31, 34) and analyzing the purified metabolites by HPLC and ESI-MS. Figure 3 shows the HPLC traces of the metabolites purified from EAF1001 and EAF1003 in comparison to that of authentic CMN. Strains EAF1002 and EAF1004 showed results similar to those of EAF1001 and EAF1003, respectively (data not shown). These data clearly showed that strains carrying integrated pCMN-R4C8RF-436 produced metabolites with the same retention times as authentic CMN IA and IB and CMN IIA and IIB but that the strains carrying integrated pOJ436 did not. To confirm that all four CMN derivatives were generated by EAF1001, the metabolites eluting from the HPLC system at 12.5 and 13 min were collected from the HPLC column, dried to completion under a vacuum, and then analyzed by ESI-MS. The results from this analysis determined that the metabolites eluting at 12.5 min had masses consistent with both CMN IIA (theoretical mass [M +H]+, 541.25; experimental mass average [M + H]+, 541.59) and CMN IIB (theoretical mass [M + H]+, 525.26; experimental mass average [M + H], 525.32). Additionally, the metabolites eluting at 13 min had masses consistent with both CMN IA (theoretical mass [M + H]+, 669.35; experimental mass average [M + H]+, 669.27) and CMN IB (theoretical mass [M + H]+, 653.35; experimental mass average [M + H]+, 653.26). Further analysis of the purified metabolites by strong-cation exchange HPLC determined that CMN IA and IB were produced at a nearly 1:1 ratio; however, the ratio of CMN IIA to CMN IIB could not be determined by this approach (data not shown). From these data, we concluded that all the necessary genetic information for the production of and resistance to CMN was contained on pCMN-P4C8RF-436. Furthermore, strain EAF1001 produced CMN at approximately 50 mg/liter based on comparisons of HPLC traces corresponding to known concentrations of authentic CMN and purified CMN from EAF1001 (data not shown).
FIG. 3.
HPLC traces of authentic CMN (A), metabolites purified from strain EAF1001 (S. lividans 1326 containing the integrated pCMN-P4C8RF-436 cosmid) (B), and metabolites purified from EAF1003 (S. lividans 1326 containing the integrated pOJ436 cosmid) (C). Metabolite elution was monitored at 268 nm. Arrows above peaks in trace A identify the peaks associated with the coelution of CMN IA and IB and the coelution of CMN IIA and IIB. Twenty milligrams of authentic CMN was injected. For EAF1001 and EAF1003 samples, 100 ml of culture was used in the purification. The purified compounds were resuspended in 500 μl of H2O. A portion of these samples was diluted 1:10 in H2O, and 20 μl of these diluted samples was injected into the HPLC column.
Analysis of the CMN biosynthetic gene cluster.
Based on bioinformatic analysis and comparisons with the previously identified VIO biosynthetic gene cluster, 19 ORFs were proposed to be involved in CMN production (Fig. 2; Table 2). A comparison of the CMN biosynthetic gene cluster with the analogous VIO biosynthetic gene cluster gave insights into the putative functions of each ORF and the molecular reasons for the structural differences between CMN and VIO (Fig. 1).
First, three proteins (VioQ, VioS, and VioT) corresponding to the VIO biosynthetic gene cluster are not encoded by the CMN biosynthetic gene cluster. It was not surprising that a VioQ homolog was missing. We previously proposed that this enzyme catalyzed the hydroxylation of the capreomycidine ring of residue 5 of the cyclic pentapeptide core, and this proposal has been confirmed by the results of a prior study (10). Since none of the CMN derivatives are hydroxylated at this position, it was not surprising that the CMN biosynthetic gene cluster did not code for a VioQ homolog. The two remaining enzymes, VioS and VioT, were proposed to be a VIO exporter and a transcriptional regulator, respectively, involved in VIO production. Neither of these proposals has been experimentally confirmed. We propose that the 19 ORFs with designated cmn nomenclature as highlighted in Fig. 2 and Table 2 code for all the proteins needed for CMN biosynthesis, transcriptional regulation of the biosynthetic genes, the export of CMN, and resistance to all four CMN derivatives.
(i) Precursor biosynthesis.
Based on prior analysis of the VIO biosynthetic gene cluster, CMN was expected to require three nonproteinogenic amino acids to be synthesized. These amino acids are l-2,3-diaminopropionate (l-2,3-DAP), 2S,3R-capreomycidine, and β-Lys. We have previously proposed (34) or biochemically established (19) how each of these precursors is formed during VIO biosynthesis, and some of our hypotheses have been independently confirmed (44, 45). Homologs of each of the precursor biosynthetic enzymes from the VIO system are encoded within the CMN biosynthetic gene cluster (Table 2), suggesting that similar mechanisms occur during CMN biosynthesis. The proposed mechanisms for the formation of these precursors are shown in Fig. S1 in the supplemental material.
(ii) Assembly of the cyclic pentapeptide core of CMN.
There are two differences in the cyclic pentapeptide cores of CMN and VIO. First, residue 2 of CMN can be either l-serine (CMN IA or IIA) or l-alanine (CMN IB or IIB) while in VIO it is only l-serine (Fig. 1). Second, residue 3 of CMN is l-2,3-DAP while the corresponding residue of VIO is l-serine. Based on these structural differences, it was anticipated that a comparison of the nonribosomal peptide synthetases (NRPSs) for these two systems would uncover variations in the enzymology that controls the incorporation of the amino acids at these positions.
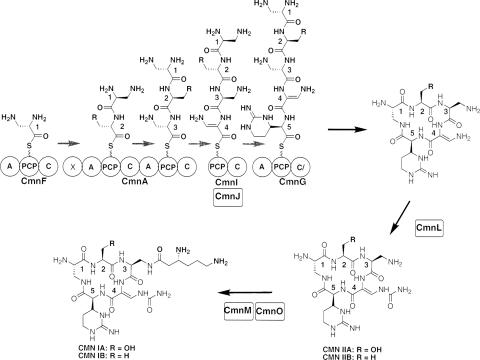
The cyclic pentapeptide core of CMN was predicted to be synthesized by an enzyme complex consisting of CmnF, CmnA, CmnI, and CmnG NRPS subunits along with CmnJ as an additional modifying enzyme that catalyzes the α,β-desaturation of residue 4 (Fig. 4). CmnF, CmnI, and CmnG all showed domain organization patterns similar to those of their homologs in the VIO NRPS, and the adenylation domain (A domain) specificity codes for CmnF and CmnG were nearly identical to those seen in the VIO components (Fig. 4; Table 3). Thus, it was reasonable to presume that they function in a manner similar to that previously proposed for VIO (34). The NRPS modules contained on CmnA and VioA control the incorporation of residues 2 and 3 into the cyclic pentapeptide core of their respective antibiotics. As expected, a comparison of CmnA with VioA revealed the key differences between the CMN and VIO NRPS systems.
FIG. 4.
Schematic for the proposed NRPS for the formation of the cyclic pentapeptide core of CMN (CmnF, CmnA, CmnF, CmnJ, and CmnG), followed by downstream carbamoylation by CmnL and the acylation of CMN IIA and IIB by the combined functions of CmnM and CmnO. The individual domains of the NRPS are noted as circles with the appropriate abbreviation to denote function. Abbreviations are as follows: A, adenylation domain; PCP, peptidyl carrier protein domain; C, condensation domain; X, domain of unknown function; and C/, modified condensation domain. The arrows above the NRPS components depict the direction of pentapeptide synthesis on the NRPS. The numbering identifies the order in which the residues are incorporated into the cyclic pentapeptide backbone of CMN. The 2,3-l-DAP tethered to the PCP domain of CmnI is proposed to be introduced by the A domain of CmnF, analogous to the mechanism proposed for the VIO NRPS (34). R, residue.
TABLE 3.
Comparison of A domain specificity codes from CMN and VIO NRPSs
| CMN or VIO NRPS componenta | Specificity codeb | Amino acid proposed to be activatedc |
|---|---|---|
| CmnF | D A Q S L A V V | l-2,3-DAP |
| VioF | D A Q S L A I V | l-2,3-DAP |
| CmnA, A1 | D V Y H F S L V | l-Ser (or l-Ala) |
| VioA, A1 | D V Y H F S L V | l-Ser |
| CmnA, A2 | D V R S L S M V | l-2,3-DAP |
| VioA, A2 | D V R H M S M V | l-Ser |
| CmnG | D P Q D I G I V | CAM |
| VioG | D P Q D V G I G | CAM |
| CmnO | D T E D V G T M | β-Lys |
| VioO | D T E D V G V G | β-Lys |
NRPS components are grouped according to homology (e.g., VioF is the homolog of CmnF). CmnA and VioA each contain more than one A domain. The first A domain is noted as A1, and the second is noted as A2.
Specificity code as described in references 6 and 30. The alignment program to identify the substrate specificity code is available at http://www.tigr.org/jravel/nrps.
This proposal is based on the results of the A domain substrate specificity code and the chemical structures of CMN and VIO. CAM, 2S,3R-capreomycidine.
The first A domain of CmnA has a substrate specificity code that was identical to that seen in VioA (Table 3), and this code is for l-serine, the amino acid found in VIO and in CMN IA and IIA (Fig. 1). However, for CMN biosynthesis, the amino acid at residue 2 of the cyclic pentapeptide core is either l-serine or l-alanine (Fig. 1). The answer to how two different amino acids can be found at this position may come from the finding of an extra enzymatic domain at the N terminus of CmnA, referred to as domain X (Fig. 4). This domain showed a low level of amino acid sequence similarity to epimerase domains of NRPSs. With this in mind, one could envision a mechanism whereby l-serine is first activated and tethered to the first peptidyl carrier protein (PCP) domain of CmnA. A catalytic base within the X domain would subsequently abstract the α-carbon proton as if it were functioning as an epimerase; however, this abstraction instead results in the dehydration of l-serine to form dehydroalanine. The conversion of dehydroalanine into l-alanine would then require hydride transfer to the desaturated C-3, likely from NADH, with the catalytic base returning the abstracted proton back to the molecule with retention of the initial stereochemistry. This process would not be fully efficient since the ratio of l-serine to l-alanine at residue 2 was observed to be approximately 1:1. This mechanism is analogous to that seen for l-serine-to-d-alanine conversion during lantibiotic biosynthesis (7). The alternative explanation that the first A domain of CmnA incorporates either l-serine or l-alanine regardless of the specificity code of the A domain cannot be eliminated. Further investigations will be needed to test these hypotheses.
The internal module of CmnA (consisting of the second conservation-adenylation-PCP set of domains) controls the incorporation of residue 3 of the cyclic pentapeptide (Fig. 4). A comparison between this region of CmnA and that of VioA showed that while the domain architectures are the same, the specificity code of the A domain in CmnA is different from that in VioA (Table 3). Most significant was the residue at position 4 of the specificity code, which in CmnA is a seryl residue but which is a histidinyl residue in VioA. Residues at this position of an A domain are proposed to be at the base of the substrate binding pocket and interact with the side chain of the bound amino acid (6, 30). One possibility is that a histidinyl residue at this position, as seen in VioA, would hinder the binding of l-2,3-DAP due to the unfavorable interactions between the β-amino group of l-2,3-DAP and the histidinyl residue. However, a change to a seryl residue would allow such a substrate to bind. This subtle difference between CmnA and VioA likely explains the different amino acids found at this position in CMN and VIO. The other steps involved in synthesizing the cyclic peptapeptide core of CMN were likely to proceed in a manner analogous to that previously proposed for VIO (Fig. 4) (34).
(iii) Modification of the cyclic pentapeptide core to generate CMN.
There are two possible modifications to the cyclic pentapeptide core once it has been synthesized. First, CmnL, a homolog of ornithine carbamoyltransferases, catalyzes the carbamoylation of the amino group of residue 4, resulting in the formation of the β-ureidodehydroalanine moiety. If the molecule is not processed any further, CMN IIA and IIB are generated (Fig. 4). The second possible modification is the addition of β-Lys to the β-amino group of residue 3 to generate CMN IA or CMN IB (Fig. 4). The activation and tethering of β-Lys to this position was proposed to be catalyzed by a monomodular NRPS. CmnO is a didomain protein consisting of an N-terminal A domain with a specificity code for β-Lys (Table 3) and a C-terminal PCP domain. These properties suggested that CmnO recognizes β-Lys and catalyzes the covalent tethering of β-Lys to the CmnO PCP domain. The β-Lys would then be transferred to CMN IIA or IIB by the action of CmnM, a homolog of condensation domains of NRPSs. Thus, CmnO and CmnM work in unison as a monomodular NRPS to acylate CMN IIA or CMN IIB, producing CMN IA or CMN IB, analogous to the way VioO and VioM are proposed to work during VIO biosynthesis (34).
(iv) Transcriptional regulation and export of CMN.
CmnR is a homolog of VioR, and both proteins show sequence similarity to the LuxR family of transcriptional regulators (11). Finally, CmnE is a homolog of VioE, and both show sequence similarity to the major facilitator superfamily MFS_1 (DUF894), involved in the efflux of various metabolites (26).
Analysis of CMN and kanamycin resistance.
The pCMN-P4C8RF-436 cosmid integrated into the S. lividans 1326 genome conferred resistance to all four CMN derivatives on the S. lividans 1326 strains. Since pCMN-P4C8RF-436 codes for only Cph, this finding suggested that there was an additional resistance gene encoded within pCMN-P4C8RF. The most likely candidate for conferring this resistance activity was the gene cmnU. CmnU is a homolog of 16S rRNA methyltransferases that are known to confer resistance to kanamycin and apramycin by modifying residue A1408 (E. coli numbering) of the 16S rRNA (16). The relevance of this finding is that mutations in the analogous residue in M. tuberculosis and M. smegmatis 16S rRNA result in VIO and kanamycin resistance (32) and a mutation in the analogous residue of Thermus thermophilus results in CMN resistance (13). Furthermore, the enzyme TlyA from M. tuberculosis that methylates both 16S and 23S rRNA to make the ribosome more sensitive to CMN methylates residue C1409 of the 16S rRNA (18). From these results, it was reasonable to hypothesize that CmnU modifies the 16S rRNA of S. mutabilis subsp. capreolus, likely at the position equivalent to A1408, resulting in CMN-resistant ribosomes. To investigate whether cmnU confers antibiotic resistance on bacteria expressing this gene, cmnU was cloned into vectors that enabled the expression of the gene in E. coli or S. lividans 1326. The strains carrying these expression constructs were analyzed for both CMN and kanamycin resistance. The latter was tested based on the amino acid similarity between CmnU and aminoglycoside methyltransferases.
E. coli does not methylate its ribosomes in a manner similar to that of M. tuberculosis or M. smegmatis because it lacks a TlyA homolog, and this results in E. coli being less sensitive to CMN (18). Therefore, we evaluated whether the expression of cmnU in E. coli resulted in increased CMN resistance in the presence and absence of tlyA from M. smegmatis. As seen previously (18), the expression of tlyA in E. coli resulted in increased sensitivity to CMN but had no effect on kanamycin resistance (Table 4). However, regardless of the methylation state of the E. coli ribosomes, the expression of cmnU in E. coli resulted in increased resistance to CMN (Table 4). The expression of cmnU in E. coli also resulted in kanamycin resistance; however, the coexpression of tlyA resulted in a decrease in the kanamycin resistance compared to that of a strain lacking tlyA (Table 4). It is not clear at this time why the expression of tlyA impairs the ability of cmnU to confer full kanamycin resistance.
TABLE 4.
Changes in CMN and kanamycin sensitivity upon expression of cmnUa
| Strain (relevant genotype)b | MIC (μg/ml) of:
|
|
|---|---|---|
| CMN | Kanamycin | |
| E. coli DH5α/pBAD33/pLJ102 (cmnU tlyA) | 50 | 12 |
| E. coli DH5α/pBAD33-cmnU/pLJ102 (cmnU+tlyA) | 100 | 96 |
| E. coli DH5α/pBAD33/pSJ101 (cmnU tlyA+) | 25 | 12 |
| E. coli DH5α/pBAD33-cmnU/pSJ101 (cmnU+tlyA+) | 50 | 24 |
| S. lividans 1326/pSE34 (cmnU) | 200 | 6 |
| S. lividans 1326/pSE34-cmnU (cmnU+) | >1,600 | >1,600 |
For E. coli strains, the MIC was defined as the lowest antibiotic concentration at which the optical density at 600 nm after 16 h of incubation at 37°C in LB medium was <0.1. For S. lividans 1326 strains, the MIC was defined as the lowest antibiotic concentration at which no growth on ISP2 plates was observed after 2 days of incubation at 30°C. MICs reported are from three separate experiments. See Materials and Methods for details.
Gene designations lacking a superscript plus sign represent genes not carried on plasmids.
When cmnU was expressed in S. lividans 1326, it resulted in increased resistance to both CMN and kanamycin (Table 4). The observed resistance to all four CMN derivatives supported the hypothesis that cmnU was a newly identified CMN resistance gene from S. mutabilis subsp. capreolus. Furthermore, the expression of cmnU also conferred resistance to the aminoglycoside kanamycin (Table 4). CmnU is a homolog of the proteins encoded by kamB and kamC, two genes isolated for their ability to confer kanamycin resistance on S. lividans 1326 (16). It has been shown previously that the expression of kamB or kamC in E. coli results in the modification of A1408 of the 16S rRNA. Based on the similarity between CmnU, KamB, and KamC, it is reasonable to hypothesize that CmnU catalyzes the modification of the 16S rRNA of the ribosome, resulting in CMN- and kanamycin-resistant ribosomes. Thus, ribosome modification in S. mutabilis subsp. capreolus may be a mechanism for resistance to the CMN produced by this bacterium.
Summary.
We have isolated and sequenced the CMN biosynthetic gene cluster from S. mutabilis subsp. capreolus. These data provide a molecular blueprint for the way CMN is biosynthesized by this organism and give some insights into the reasons for the structural differences between CMN and VIO. We also showed that the transfer of this gene cluster into S. lividans 1326 resulted in the heterologous production of the CMN antituberculosis drug, providing an important first step toward the metabolic engineering of CMN biosynthesis. Finally, we have provided evidence that ribosome modification by CmnU confers CMN resistance on E. coli and S. lividans 1326, and this is likely to be true for S. mutabilis subsp. capreolus. The similarity between CmnU and aminoglycoside resistance genes gives further support for the hypothesis that the ribosome binding sites of CMN and aminoglycosides overlap on the ribosome.
Supplementary Material
Acknowledgments
This work was supported, in part, by the American Lung Association of Wisconsin and the National Institutes of Health (RO1AI065850). E.A.F. was supported by a National Institutes of Health-sponsored Biotechnology Training Program predoctoral fellowship. H.A.C. was supported by a National Science Foundation predoctoral fellowship.
We thank Stephen Douthwaite (University of Southern Denmark) for generously providing pLJ102 and pSJ101, David Sherman (University of Michigan) for generously providing plasmid pSE34, and Amy Gehring (Williams College) for generously providing S. lividans 1326.
Footnotes
Published ahead of print on 11 May 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibb, M. J., J. M. Ward, and S. N. Cohen. 1985. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol. Gen. Genet. 199:26-36. [DOI] [PubMed] [Google Scholar]
- 3.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 4.Carter, J. H., II, R. H. Du Bus, J. R. Dyer, J. C. Floyd, K. C. Rice, and P. D. Shaw. 1974. Biosynthesis of viomycin. I. Origin of alpha, beta-diaminopropionic acid and serine. Biochemistry 13:1221-1227. [DOI] [PubMed] [Google Scholar]
- 5.Carter, J. H., II, R. H. Du Bus, J. R. Dyer, J. C. Floyd, K. C. Rice, and P. D. Shaw. 1974. Biosynthesis of viomycin. II. Origin of beta-lysine and viomycidine. Biochemistry 13:1227-1233. [DOI] [PubMed] [Google Scholar]
- 6.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, P. D., P. M. O'Connor, L. A. Draper, E. M. Lawton, L. H. Deegan, C. Hill, and R. P. Ross. 2005. Posttranslational conversion of L-serines to D-alanines is vital for optimal production and activity of the lantibiotic lacticin 3147. Proc. Natl. Acad. Sci. USA 102:18584-18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels, T. M., J. H. Bates, and K. A. Downes. 1994. History of tuberculosis, p. 13-24. In B. R. Bloom (ed.), Tuberculosis: pathogenesis, protection, and control. ASM Press, Washington, DC.
- 9.Dye, C., M. A. Espinal, C. J. Watt, C. Mbiaga, and B. G. Williams. 2002. Worldwide incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 185:1197-1202. [DOI] [PubMed] [Google Scholar]
- 10.Fei, X., X. Yin, L. Zhang, and T. M. Zabriskie. 2007. Roles of VioG and VioQ in the incorporation and modification of the capreomycidine residue in the peptide antibiotic viomycin. J. Nat. Prod. 70:618-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 12.Gould, S. J., and D. A. Minott. 1992. Biosynthesis of capreomycin. 1. Incorporation of arginine. J. Org. Chem. 57:5214-5217. [Google Scholar]
- 13.Gregory, S. T., J. F. Carr, and A. E. Dahlberg. 2005. A mutation in the decoding center of Thermus thermophilus 16S rRNA suggests a novel mechanism of streptomycin resistance. J. Bacteriol. 187:2200-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heifets, L., J. Simon, and V. Pham. 2005. Capreomycin is active against non-replicating M. tuberculosis. Ann. Clin. Microbiol. Antimicrob. 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes, D. J., D. Drocourt, G. Tiraby, and E. Cundliffe. 1991. Cloning of an aminoglycoside-resistance-encoding gene, kamC, from Saccharopolyspora hirsuta: comparison with kamB from Streptomyces tenebrarius. Gene 102:19-26. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 18.Johansen, S. K., C. E. Maus, B. B. Plikaytis, and S. Douthwaite. 2006. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell 23:173-182. [DOI] [PubMed] [Google Scholar]
- 19.Ju, J., S. G. Ozanick, B. Shen, and M. G. Thomas. 2004. Conversion of (2S)-arginine to (2S,3R)-capreomycidine by VioC and VioD from the viomycin biosynthetic pathway of Streptomyces sp. strain ATCC11861. Chembiochem 5:1281-1285. [DOI] [PubMed] [Google Scholar]
- 20.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics, p. 229-252. The John Innes Foundation, Crowes, Norwich, United Kingdom.
- 21.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics, p. 405-420. The John Innes Foundation, Crowes, Norwich, United Kingdom.
- 22.Maus, C. E., B. B. Plikaytis, and T. M. Shinnick. 2005. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuguchi, Y., K. Suga, K. Masuda, and T. Yamada. 1974. Genetic and biochemical studies on drug-resistant mutants in Mycobacterium smegmatis. Jpn. J. Microbiol. 18:457-462. [DOI] [PubMed] [Google Scholar]
- 24.Mizuguchi, Y., K. Suga, and T. Yamada. 1979. Interaction between 30 S ribosomal components in a viomycin resistant mutant of Mycobacterium smegmatis. Microbiol. Immunol. 23:595-604. [DOI] [PubMed] [Google Scholar]
- 25.Murray, C. J. L., and J. A. Salomon. 1998. Modeling the impact of global tuberculosis control strategies. Proc. Natl. Acad. Sci. USA 95:13881-13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saier, M. H., Jr., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. Heijne, S. C. Huang, D. L. Jack, P. S. Jahn, K. Lew, J. Liu, S. S. Pao, I. T. Paulsen, T. T. Tseng, and P. S. Virk. 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257-279. [PubMed] [Google Scholar]
- 27.Saugar, I., E. Sanz, M. A. Rubio, J. C. Espinosa, and A. Jimenez. 2002. Identification of a set of genes involved in the biosynthesis of the aminonucleoside moiety of antibiotic A201A from Streptomyces capreolus. Eur. J. Biochem. 269:5527-5535. [DOI] [PubMed] [Google Scholar]
- 28.Skinner, R. H., and E. Cundliffe. 1980. Resistance to the antibiotics viomycin and capreomycin in the Streptomyces species which produce them. J. Gen. Microbiol. 120:95-104. [DOI] [PubMed] [Google Scholar]
- 29.Smirnova, N., and K. A. Reynolds. 2001. Engineered fatty acid biosynthesis in Streptomyces by altered catalytic function of beta-ketoacyl-acyl carrier protein synthase III. J. Bacteriol. 183:2335-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 31.Tam, A. H.-K., and D. C. Jordan. 1972. Laboratory production and 14C-labelling of viomycin. J. Antibiot. 25:524-529. [DOI] [PubMed] [Google Scholar]
- 32.Taniguchi, H., B. Chang, C. Abe, Y. Nikaido, Y. Mizuguchi, and S. I. Yoshida. 1997. Molecular analysis of kanamycin and viomycin resistance in Mycobacterium smegmatis by use of the conjugation system. J. Bacteriol. 179:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiara, A. S., and E. Cundliffe. 1995. Analysis of two capreomycin-resistance determinants from Streptomyces capreolus and characterization of the action of their products. Gene 167:121-126. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, M. G., Y. A. Chan, and S. G. Ozanick. 2003. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob. Agents Chemother. 47:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, M., and S. J. Gould. 1993. Biosynthesis of capreomycin. 2. Incorporation of l-serine, l-alanine, and l-2,3-diaminopropionic acid. J. Org. Chem. 58:5176-5180. [Google Scholar]
- 36.World Health Organization. 2005. World Health Organization model list of essential medicines. World Health Organization, Geneva, Switzerland.
- 37.Yamada, T. 1987. The role of ribosomes in sensitivity of mycobacteria to tuberactinomycin. Microbiol. Immunol. 31:179-181. [DOI] [PubMed] [Google Scholar]
- 38.Yamada, T., and K. H. Bierhaus. 1978. Viomycin favours the formation of 70S ribosome couples. Mol. Gen. Genet. 161:261-265. [DOI] [PubMed] [Google Scholar]
- 39.Yamada, T., K. Masuda, Y. Mizuguchi, and K. Suga. 1976. Altered ribosomes in antibiotic-resistant mutants of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 9:817-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada, T., K. Masuda, K. H. Nierhaus, and H. G. Wittmann. 1972. Analysis of ribosomes from viomycin-sensitive and -resistant Mycobacterium smegmatis. J. Bacteriol. 112:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada, T., Y. Mizugichi, K. H. Nierhaus, and H. G. Wittmann. 1978. Resistance to viomycin conferred by RNA of either ribosomal subunit. Nature 275:460-461. [DOI] [PubMed] [Google Scholar]
- 42.Yamada, T., Y. Mizugichi, and K. Suga. 1976. Localization of co-resistance to streptomyces, kanamycin, capreomycin and tuberctinomycin in core particles derived from ribosomes of viomycin resistant Mycobacterium smegmatis. J. Antibiot. 29:1124-1126. [DOI] [PubMed] [Google Scholar]
- 43.Yamada, T., A. Nagata, Y. Ono, Y. Suzuki, and T. Yamanouchi. 1985. Alteration of ribosomes and RNA polymerase in drug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 27:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin, X., K. L. McPhail, K.-J. Kim, and T. M. Zabriskie. 2004. Formation of the nonproteinogenic amino acid 2S,3R-capreomycidine by VioD from the viomycin biosynthesis pathway. Chembiochem 5:1278-1281. [DOI] [PubMed] [Google Scholar]
- 45.Yin, X., and T. M. Zabriskie. 2004. VioC is a non-heme iron, α-ketoglutarate-dependent oxygenase that catalyzes the formation of 3S-hydroxy-l-arginine during viomycin biosynthesis. Chembiochem 5:1274-1277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.