Abstract
Bacterioplankton of the marine Roseobacter clade have genomes that reflect a dynamic environment and diverse interactions with marine plankton. Comparative genome sequence analysis of three cultured representatives suggests that cellular requirements for nitrogen are largely provided by regenerated ammonium and organic compounds (polyamines, allophanate, and urea), while typical sources of carbon include amino acids, glyoxylate, and aromatic metabolites. An unexpectedly large number of genes are predicted to encode proteins involved in the production, degradation, and efflux of toxins and metabolites. A mechanism likely involved in cell-to-cell DNA or protein transfer was also discovered: vir-related genes encoding a type IV secretion system typical of bacterial pathogens. These suggest a potential for interacting with neighboring cells and impacting the routing of organic matter into the microbial loop. Genes shared among the three roseobacters and also common in nine draft Roseobacter genomes include those for carbon monoxide oxidation, dimethylsulfoniopropionate demethylation, and aromatic compound degradation. Genes shared with other cultured marine bacteria include those for utilizing sodium gradients, transport and metabolism of sulfate, and osmoregulation.
In surface waters of the open ocean, 1 in 10 bacterial cells is a member of the Roseobacter group (17). In coastal waters, the number of Roseobacter cells increases to 1 in 5 (11, 19). Despite their obvious ecological success, however, roseobacters do not fit the stereotype of a small, metabolically conservative, “oligotrophic” bacterium (8, 18). Instead, they are large (0.08 μm3) (38), easily cultured (19), and respond readily to increased substrate availability (7). Analysis of the first Roseobacter genome sequence, that of Silicibacter pomeroyi, revealed a fairly large genome (4.5 Mb) housing abundant and diverse transporters, complex regulatory systems, and multiple pathways for acquiring carbon and energy in seawater. Roseobacters thus appear to be quite versatile from metabolic and ecological standpoints (43), with an assortment of strategies for obtaining carbon and nutrients and, directly or indirectly, affecting the biogeochemical status of seawater.
The availability of two additional closed genome sequences of cultured roseobacters provides the opportunity for an ecologically based analysis of the genetic capabilities of this bacterial taxon. The three organisms are assumed to have different niches in the surface ocean based on the conditions of their isolation: S. pomeroyi is a free-living heterotrophic bacterioplankter obtained from coastal seawater (43), congener Silicibacter sp. strain TM1040 (96% 16S rRNA sequence identity to S. pomeroyi [Fig. 1]) is an associate of the marine dinoflagellate Pfiesteria piscicida (1, 40), and Jannaschia sp. strain CCS1 (with 94% 16S rRNA sequence identity to the two Silicibacter species) represents a recently discovered class of marine aerobic bacteriochlorophyll a-based phototrophs (4). Our comparative analysis of the three Roseobacter genomes centered on three questions. (i) What physiological and ecological traits of roseobacters can be inferred from the genome sequences (focusing on genes that define ecological strategies and biogeochemical roles)? (ii) What makes a roseobacter a roseobacter (focusing on genes shared among the species)? (iii) What makes a roseobacter marine (focusing on genes shared with other cultured marine bacteria from phylogenetically distant taxa)?
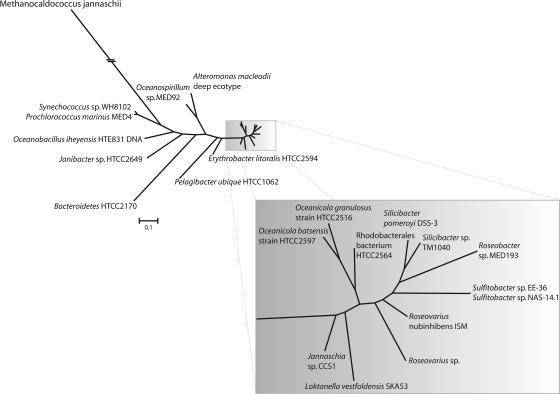
FIG. 1.
Phylogenetic tree of 16S rRNA gene sequences from 12 roseobacters and other selected marine bacterioplankton for which a genome sequence is available. The tree is based on positions 21 to 1490 of the 16S rRNA gene (E. coli numbering system). The tree was constructed with the PAUP* package (53), version 4.10b, using the maximum likelihood method. The bar corresponds to the number of changes per nucleotide for the main tree.
MATERIALS AND METHODS
Organisms.
Silicibacter pomeroyi strain DSS-3 was isolated from Atlantic coastal seawater (Georgia; salinity = 31) in 1999 on a minimal seawater medium enriched with 10 μM dimethylsulfoniopropionate (DMSP). Silicibacter sp. strain TM1040 was isolated from a marine dinoflagellate culture (Pfiesteria piscicida) in 2000 on 0.5× Marine agar 2216. Jannaschia sp. strain CCS1 was isolated from Pacific coastal seawater (Bodega Head, CA; salinity = 33) in 2003 on 1/10-strength YTSS agar (full strength YTSS is 0.4% yeast extract plus 0.25% tryptone made with 20 g liter−1 sea salts).
Genomic sequencing and annotation.
Silicibacter sp. strain TM1040 and Jannaschia sp. strain CCS1 were sequenced by the random shotgun method (see www.jgi.doe.gov for details of library construction and sequencing). Large, medium, and small insert random libraries (38 kb, 9.5 kb, and 3 kb for Silicibacter sp. strain TM1040; 38 kb, 13 kb, and 5.6 kb for Jannaschia sp. strain CCS1) were sequenced with an average success rate of 96% and an average high-quality read length of 685 nucleotides. Reads were assembled with parallel Phrap (High Performance Software, LLC), and possible misassemblies were corrected with Dupfinisher (22) or a transposon bomb of bridging clones (Epicentre Biotechnologies, Madison, WI). Gaps between contigs were closed by editing, custom primer walking, or PCR amplification. The completed genome sequence of Silicibacter sp. strain TM1040 contains 59,153 reads, achieving an average of 13-fold sequence coverage per base. The completed genome sequence of Jannaschia sp. strain CCS1 contains 67,683 reads, achieving an average of 14-fold sequence coverage per base. The error rate was <1 in 100,000. Sequencing and annotation of S. pomeroyi DSS-3 are described by Moran et al. (43).
Sequence analysis and annotation.
The Silicibacter sp. strain TM1040 and Jannaschia sp. strain CCS1 genomes were annotated with the GenDB system (39).
Synteny analysis.
Comparative genetic arrangements were analyzed by the Artemis Comparison Tool (50). Files were prepared using separate one-way tBLASTx analysis of chromosomes, megaplasmids, and plasmids with cutoffs of >45% identity and scores of >35.
Signal transduction analysis.
Signal transduction proteins were identified by searching against the Pfam (3) and SMART (35) domain databases and filtering for proteins that contained domains implicated in signal transduction (55). Transmembrane regions were predicted using Phobius (27). Cultured marine bacteria used in comparisons were Cellulophaga sp. strain MED134, Croceibacter atlanticus HTCC2559, Flavobacterium sp. strain MED217, Polaribacter irgensii 23-P, Robiginitalea biformata HTCC2501, Tenacibaculum sp. strain MED152, Cytophaga hutchinsonii, Crocosphaera watsonii WH8501, Synechococcus sp. strain WH8102, Prochlorococcus marinus MIT9313, Bacillus sp. strain NRRL B-14911, Oceanobacillus iheyensis HTE831, Rhodopirellula sp. strain 1, Pelagibacter ubique HTCC1062, Erythrobacter litoralis HTCC2594, Erythrobacter sp. strain NAP1, Nitrobacter sp. strain Nb-311A, Oceanicaulis alexandrii HTCC2633, Parvularcula bermudensis HTCC2503, Sphingopyxis alaskensis RB2256, Alteromonas macleodii, Colwellia psychrerythraea 34H, Idiomarina baltica OS145, Idiomarina loihiensis L2TR, Marinobacter aquaeolei VT8, Marinomonas sp. strain MED121, Nitrosococcus oceani ATCC 19707, Oceanospirillum sp. strain MED92, Photobacterium profundum SS9, Photobacterium sp. strain SKA34, Pseudoalteromonas atlantica T6c, Pseudoalteromonas haloplanktis TAC125, Shewanella baltica OS155, Shewanella frigidimarina NCIMB 400, Vibrio fischeri ES114, Vibrio splendidus 12B01, and Magnetococcus sp. strain MC-1. Alphaproteobacteria used in comparisons were Agrobacterium tumefaciens C58 UWash, Caulobacter crescentus CB15, Mesorhizobium loti MAFF303099, Mesorhizobium sp. strain BNC1, Paracoccus denitrificans PD1222, Rhodobacter sphaeroides 2.4.1, Rhodopseudomonas palustris BisB18, and Sinorhizobium meliloti 1021.
Transporter profiles and ANOSIM analysis.
Complete predicted protein sequences were searched against a curated set of proteins with family assignment for similarity to known or putative transporter proteins and against a nonredundant general protein database using a semiautomated pipeline (48). Manual annotation for final assignments was based on the number of hits to the transporter database, maximum, minimum, and average BLAST E values, the description of top hits to the general protein database, and cluster of orthologous group (COG) assignments. The roseobacters were compared to a selected group of bacteria for which similar transporter analysis was available, consisting of six closely related nonmarine alphaproteobacteria (A. tumefaciens C58 Uwash, C. crescentus CB15, R. palustris CGA009, M. loti MAFF303099, R. sphaeroides 2.4.1, and S. meliloti 1021) and seven marine heterotrophic bacteria (Vibrio vulnificus CMCP6, Desulfotalea psychrophila LSv54, Oceanobacillus iheyensis HTE831, Rhodopirellula sp. strain 1, Colwellia psychrerythraea 34H, Photobacterium profundum SS9, and Idiomarina loihiensis L2TR). Using the 70 transporter families that had ≥2 occurrences in at least one genome, a Bray-Curtis similarity matrix was created. A one-way analysis of similarity (ANOSIM) (15) was used to test for significant differences in transporter composition among the three groups defined a priori. Differences among groups were visualized using nonmetric multidimensional scaling of the similarity matrix (PRIMER 5 software) (15).
Identification of shared and unique genes.
Genes shared among the three Roseobacter strains were identified by three-way reciprocal best hit (RBH) analysis using BLAST thresholds of E < 10−5 and amino acid identity of >30%. Core genes in the 12 total Roseobacter genome sequences were identified by sequential two-way RBH analysis, beginning with the S. pomeroyi genome and adding the other 11 genomes one at a time. Core genes were defined as those found in all 12 genomes based on RBH to S. pomeroyi genes. As each genome was sequentially added to the analysis, the total gene count was incremented if a gene had no RBH in any previous genome. Both the core gene count and total gene count are likely to be underestimated with this approach because nine of the genomes are draft. A second approach, the in silico genomic subtraction (ISGS) method (16), was also used because the RBH method misses orthologs if open reading frames (ORFs) are overlooked or miscalled. This method is based on sequential subtractions of ORFs based on one-way BLASTp and tBLASTn analysis. S. pomeroyi genes were blasted sequentially against the genomes of nonmarine alphaproteobacterial relatives R. sphaeroides 2.4.2, P. denitrificans PD1222, S. meliloti 1021, and M. loti MAFF303099, and orthologous genes were removed from the data set. The remaining ORFs were compared by tBLASTn to the genome sequences of Silicibacter sp. strain TM1040 and Jannaschia sp. strain CCS1 to obtain a final ISGS unique gene set consisting of 121 ORFs shared by all three roseobacters but missing from the four nonmarine alphaproteobacterial genomes. The final composition of this gene set is affected by the bioinformatic decision of which alphaproteobacterial relatives to use in the subtraction but consists largely of genes more typical of roseobacters than their alphaproteobacterial relatives.
Identification of genes with a marine signal.
We determined whether orthologs of Roseobacter genes were found more commonly in marine than in nonmarine genomes. Twenty pairs of non-Roseobacter genomes were assembled, each consisting of the genome of a marine isolate (most from surface seawater) paired with its closest nonmarine relative at the time of the analysis: Synechococcus sp. strain WH8102 and Synechococcus elongatus PCC 7942, Rhodopirellula baltica SH 1 and Moorella thermoacetica ATCC 39073, P. ubique HTCC1062 and Wolbachia sp. strain TRS, P. marinus MIT9313 and Gloeobacter violaceus PCC 7421, Pseudoalteromonas haloplanktis TAC125 and Legionella pneumophila, Shewanella frigidimarina NCIMB 400 and Shewanella putrefaciens CN-32, Magnetococcus sp. strain MC-1 and Rhodospirillum rubrum, Sphingomonas alaskensis and Zymomonas mobilis ZM4, Erythrobacter litoralis HTCC2594 and Novosphingobium aromaticivorans F199, Flavobacterium sp. strain MED217 and Bacteroides fragilis ATCC 25285, C. atlanticus HTCC2559 and Porphyromonas gingivalis W83, V. splendidus 12B01 and Escherichia coli K-12, I. baltica OS145 and Pseudomonas putida KT2440, Janibacter sp. strain HTCC2649 and Arthrobacter sp. strain FB24, Parvularcula bermudensis HTCC2503 and Brucella abortus 9-941, Bacillus sp. strain NRRL B-14911 and Bacillus halodurans C-125, Marinomonas sp. strain MED121 and Pseudomonas syringae B728a, Nitrosococcus oceani ATCC 19707 and Pseudomonas fluorescens Pf0-1, Oceanicaulis alexandrii HTCC2633 and R. sphaeroides 2.4.2, and Nitrobacter sp. strain Nb311A and R. palustris BisB18. Orthologs to Roseobacter genes were identified in these genomes with BLASTp analysis and using the criteria of RBH, an E value of ≤10−5, and a percent identity of ≥30%. Whether the observed gene distribution was different from expected (the null hypothesis being that the gene occurred in an equal number of marine and nonmarine genomes) was estimated by computing a chi-square statistic and associated P value.
Nucleotide sequence accession numbers.
The complete Silicibacter sp. strain TM1040 and Jannaschia sp. strain CCS1 genome sequences are available under GenBank accession no. NC_008043 and NC_007802. The complete Silicibacter pomeroyi DSS-3 sequence was previously deposited under GenBank accession number NC_003911. Nine draft Roseobacter genomes available for comparison at the time of analysis were Loktanella vestfoldensis SKA53 (AAMS00000000), Roseovarius sp. strain 217 (AAMV00000000), Roseobacter sp. strain MED193 (AANB00000000), Sulfitobacter sp. strain EE-36 (AALV00000000), Roseovarius nubinhibens ISM (AALY00000000), Sulfitobacter sp. strain NAS-14.1 (AALZ00000000), Oceanicola batsensis HTCC2597 (AAMO00000000), Oceanicola granulosus HTCC2516 (AAOT00000000), and Rhodobacterales bacterium HTCC2654 (AAMT00000000).
RESULTS AND DISCUSSION
General genome organization and content.
All three Roseobacter genomes contain ∼4,000 genes (Table 1), but there is considerable variability in genome organization and content (see Fig. S1 in the supplemental material). S. pomeroyi DSS-3 has a 491-kb megaplasmid (pSPD1), Jannaschia sp. strain CCS1 has an 80-kb plasmid (pJCS1), and Silicibacter sp. strain TM1040 has both an 823-kb megaplasmid (pSTM1) and a 131-kb plasmid (pSTM2). Neither the S. pomeroyi nor Jannaschia sp. strain CCS1 plasmids have rRNA operons, but the two plasmids of Silicibacter sp. strain TM1040 together harbor four of its five rRNA operons (Table 1). Designation of pSTM1 and pSTM2 as plasmids rather than chromosomes is based on the finding that none of 11 single-copy genes essential for bacterial growth (51) is found on either and that each contains a plasmid-type replication origin (rep and parAB) (12). The relatively small plasmids of Silicibacter sp. strain TM1040 and Jannaschia sp. strain CCS1 (pSTM2 and pJCS1) do not appear closely related, and only nine genes are similar (including rep, parAB, several genes involved in sugar or polysaccharide metabolism, and a gene coding for a single ABC transporter-like protein). A plasmid from an uncharacterized Roseobacter isolate (149-kb pSD25) (57) likewise has only 15 genes related to those of pSTM2 and three related to those of pJCS1, suggesting the presence of a pool of diverse Roseobacter genes harbored on small (∼100- to 150-kb) plasmids (see Fig. S1 in the supplemental material).
TABLE 1.
Features of three Roseobacter genomes
| Feature | Result fora:
|
||
|---|---|---|---|
| S. pomeroyi DSS-3 | Silicibacter sp. strain TM1040 | Jannaschia sp. strain CCS1 | |
| Total no. of coding sequences | 4,283 | 3,863 | 4,283 |
| G+C content (%) | 64.0 | 60.0 | 62.2 |
| No. of rRNA operons | 3 | 5 | 1 |
| No. of replicons | 2 | 3 | 2 |
| Molecule length by replicon (bp) | |||
| Main chromosome | 4,109,442 | 3,201,640 | 4,317,977 |
| Megaplasmid | 491,611 | 823,032 | NA |
| Plasmid | NA | 131,885 | 86,072 |
| No. of coding sequences by replicon | |||
| Main chromosome | 3,838 | 3,013 | 4,212 |
| Megaplasmid | 445 | 747 | NA |
| Plasmid | NA | 103 | 71 |
| G+C content by replicon (%) | |||
| Main chromosome | 64.2 | 60.4 | 62.3 |
| Megaplasmid | 62.8 | 59.3 | NA |
| Plasmid | NA | 55.3 | 57.8 |
| No. of rRNA operons by replicon | |||
| Main chromosome | 3 | 1 | 1 |
| Megaplasmid | 0 | 3 | NA |
| Plasmid | NA | 1 | 0 |
NA, not applicable.
Each genome contains a number of genes that have no orthologs in the other two; these account for 31% of the S. pomeroyi DSS-3 genome, 27% of the Silicibacter sp. strain TM1040 genome, and 39% of the Jannaschia sp. strain CCS1 genome (Table 2). No prophages are found in the S. pomeroyi or Jannaschia sp. strain CCS1 genomes, but four are present in the Silicibacter sp. strain TM1040 genome (see Table S1 in the supplemental material). Three of these can be experimentally induced by mitomycin C treatment (9), and the fourth lacks required structural proteins and may be defective. Variable and dynamic genome organization seems to be the rule for the Roseobacter lineage (46).
TABLE 2.
Numbers and percentages of shared and unique genes in three Roseobacter genomes
| Method | No. (%) of genes that are:
|
|||
|---|---|---|---|---|
| Shared 3 waysa | Unique to S. pomeroyi DSS-3 | Unique to Silicibacter sp. strain TM1040 | Unique to Jannaschia sp. strain CCS1 | |
| RBH analysis | 1,939 | 1,336 (31) | 1,027 (27) | 1,685 (39) |
| ISGS analysis | 121 | 1,260 (29) | 983 (25) | 1,600 (37) |
Genes shared three ways are those with orthologs in all three genomes. RBH analysis used the criterion of RBH between pairs of genomes with a corresponding BLAST E value of <10−5 and amino acid identity of >30%. ISGS analysis used one-way BLASTp and tBLASTn analysis to identify genes shared by the three roseobacters but missing from four nonmarine alphaproteobacterial relatives (R. sphaeroides, P. denitrificans, S. meliloti, and M. loti).
Inferred ecology and physiology.
Because of the numerical and ecological importance of marine roseobacters in surface ocean bacterioplankton communities, our annotation efforts paid greatest attention to genes of biogeochemical interest.
(i) Interfacing with the environment.
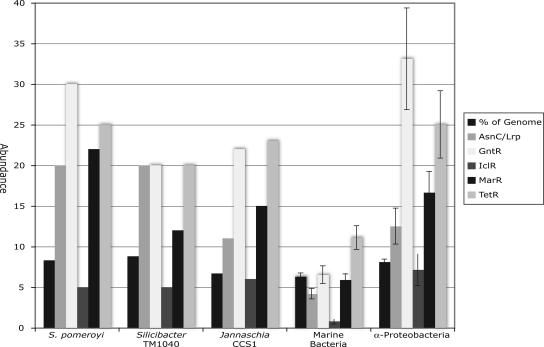
Genes encoding signal transduction proteins comprise approximately 7% of the Roseobacter genomes, similar to the average for a marine comparison group (6.3% ± 2.7%) and a nonmarine alphaproteobacterial comparison group (8.1% ± 0.9% [Fig. 2]). Two-component systems constitute a low percentage (<20%) of the overall signal transduction machinery in these Roseobacter species. Consequently, the cells respond to environmental stimuli predominantly via one-component systems that account for 86% (302 out of 353) of the signaling proteins in the S. pomeroyi genome and slightly lower percentages in the Silicibacter sp. strain TM1040 (70%) and Jannaschia sp. strain CCS1 (80%) genomes. The Roseobacter genomes contain a particularly high number of AsnC/Lrp regulatory proteins, at >3 times the average for the marine comparison group (for which genome sizes are comparable [Fig. 2]). This suggests that amino acid metabolism is particularly important and closely regulated in roseobacters; indeed several of the AsnC/Lrp regulators are located adjacent to amino acid transporters or amino acid metabolism-related genes. The roseobacters also have a larger number of other one-component systems relative to other cultured marine bacteria, including GntR, IclR, MarR, and TetR-like repressors. Generally, the Roseobacter signal transduction profiles are more similar to those of their closest (and nonmarine) alphaproteobacterial relatives than to marine bacteria (Fig. 2).
FIG. 2.
Signal transduction proteins in three Roseobacter genomes and two comparison groups consisting of marine bacterioplankton or closely related nonmarine Alphaproteobacteria. The abundance of genes encoding signal transduction proteins are shown as percentages of total coding sequences (% of genome) or as numbers of the one-component systems AsnC/Lrp (likely involved in amino acid metabolism), GntR (repression of gluconate utilization), IclR (repression of the acetate operon), MarR (repression of antibiotic resistance or stress response), and TetR (repression of tetracycline resistance). The marine comparison group consists of 37 genomes with an average size of 4.0 Mb, while the nonmarine alphaproteobacterial comparison group consists of 8 genomes with an average size of 5.5 Mb.
One-component signal transduction systems with BLUF domains sense blue light based on a conformational change following photon absorption (26). Homologs are found in the genomes of Silicibacter sp. strain TM1040 (TM1040_2025 and TM1040_2027) and Jannaschia sp. strain CCS1 (Jann_2030, Jann_2780, and Jann_3321), but not S. pomeroyi. A two-component bacterial phytochrome system that may be involved in detection of red and far-red light is also found in Silicibacter sp. strain TM1040 (TM1040_2684). The presence of systems for sensing ambient light is noteworthy in a taxon largely restricted to surface waters and may allow light sensing for phototrophy (in Jannaschia sp. strain CCS1), protective pigment formation (47), or synchronization of activity with that of photosynthetic plankton. Genomes of many other marine heterotrophs representing diverse taxa (but few nonmarine heterotrophs) likewise were found to contain proteins with BLUF domains, including other roseobacters (Sulfitobacter sp. strain NAS-14.1, Sulfitobacter sp. strain EE-36, Roseobacter sp. strain MED193, and Roseovarius sp. strain 217), other alphaproteobacteria (Sphingopyxis alaskensis SKA58), gammaproteobacteria (Alteromonas macleodii “deep ecotype,” Pseudoalteromonas tunicata, Marinomonas sp. strain MED121, Oceanospirillum sp. strain MED92, Reinekea sp. strain MED297, Vibrio sp. strain MED222, and V. splendidus 12B01), and bacteroidetes (Psychroflexus torquis and Tenacibaculum sp. strain MED152).
Swimming motility and chemotaxis behavior determine the ability of Roseobacter cells to position themselves in a heterogeneous environment. Motility has been demonstrated experimentally in the three strains, and flagellar structural genes were indeed found for all (see Table S2 in the supplemental material). Silicibacter sp. strain TM1040 has six copies of the flagellar filament gene fliC compared to just one for S. pomeroyi and Jannaschia sp. strain CCS1. Chemotaxis receptors (methyl-accepting chemotaxis proteins [MCPs]) and signal transducers (che gene orthologs) are abundant in Silicibacter sp. strain TM1040 and Jannaschia sp. strain CCS1 but surprisingly are completely lacking in S. pomeroyi (43) (see Table S3 in the supplemental material). Twenty MCPs are available to bind chemical attractants or repellants in the membrane or cytoplasm of Silicibacter sp. strain TM1040, and 14 cytoplasmic chemotaxis signal transducers receive signals from these MCPs to control changes in swimming direction. Jannaschia sp. strain CCS1 has four MCPs and eight signal transducers. Initial clues about which compounds stimulate chemotaxis in roseobacters have emerged from experimental studies of Silicibacter sp. strain TM1040: positive chemotactic responses are induced by amino acids, dinoflagellate cell homogenates, the algal osmolyte DMSP, and DMSP breakdown products (41).
ABC-type transporters average 95 per genome for the three roseobacters compared to 150 for an alphaproteobacterial comparison group and 66 for a marine heterotroph group; major facilitator superfamily (MFS) transporters average 22 per genome for roseobacters compared to 39 for alphaproteobacteria and 19 for marine heterotrophs. The set of Roseobacter transporter genes is not statistically different from either comparison group, although alphaproteobacteria and marine heterotrophs differ from each other (ANOSIM; P = 0.008; and see Fig. S2 in the supplemental material). Two particular transporter families are more abundant in roseobacters than in either comparison group, however: tripartite ATP-independent periplasmic (TRAP) transporters often associated with uptake of dicarboxylic acids and sugars (averaging 22 genes per genome compared to only 7 for alphaproteobacteria and 6 for marine heterotrophs), and the drug/metabolite (DMT) superfamily involved in cellular export of toxins and metabolites (averaging 30 genes per genome compared to 16 for alphaproteobacteria and 14 for marine heterotrophs). Both of these families use ion gradients instead of ATP to drive substrate uptake, which may be beneficial given the external salt concentrations roseobacters experience. If so, not all marine heterotrophs utilize this transport strategy.
(ii) Acquiring carbon and energy.
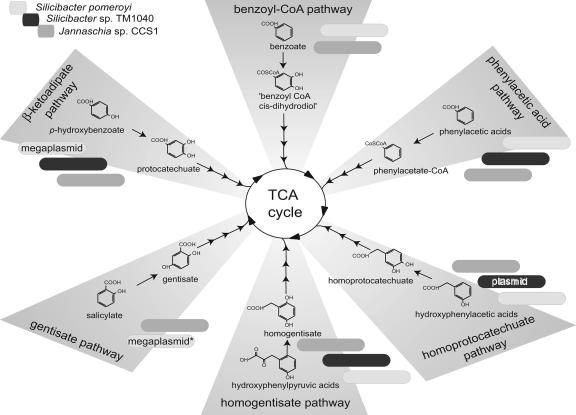
The Roseobacter genomes reveal a surprising number of pathways for catabolism of structurally diverse aromatic substrates. A pathway for the degradation of protocatechuate was identified previously in members of the Roseobacter clade (6). Now, five other distinct pathways representing both oxygenase-dependent and -independent ring-cleaving mechanisms have been found. These encode the aerobic degradation of the aromatic intermediates gentisate, homoprotocatechuate, homogentisate, benzoate, and phenylacetate. S. pomeroyi DSS-3 and Jannaschia sp. strain CCS1 possess genes for all six pathways; Silicibacter sp. strain TM1040 possesses genes for four (Fig. 3). Possible substrates for this wealth of catabolic pathways more typical of soil bacteria include phenolic metabolites (e.g., antioxidants, toxins, sunscreens, and predator deterrents) from marine plankton (13, 14), as well as lignin derivatives from coastal marshes (42).
FIG. 3.
Six distinct ring-cleaving pathways present in three Roseobacter genomes. Shaded tabs indicate the presence of a pathway (light gray, S. pomeroyi; dark gray, Silicibacter sp. strain TM1040; medium gray, Jannaschia sp. strain CCS1). The gene(s) encoding the ring-cleaving enzymes is chromosomally located unless otherwise indicated within the tab. *, S. pomeroyi contains two copies of the gentisate pathway: one on the chromosome and one on megaplasmid pSPD. The arrowheads leading into the TCA cycle indicate multiple steps. CoA, coenzyme A.
All three Roseobacter genomes contain a gene for a demethylase that mediates the first step in DMSP degradation (dmdA) (24). Half of the cultured roseobacters for which a genome sequence is available have this gene, and the DMSP demethylating phenotype has been shown experimentally for S. pomeroyi and Silicibacter sp. strain TM1040 (24, 40) (Table 3). DMSP can be a major source of both carbon and sulfur for marine bacterioplankton in ocean surface waters (30).
TABLE 3.
Survey of biogeochemically relevant genes in three complete and nine draft Roseobacter genomesa
| Genome or parameter | Result for:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AAnP genes | cox genes | pcaGH | boxC | soxB | nirS/K | nasA | phn genes | vir genes | dmdA | |
| Genomes | ||||||||||
| Complete | ||||||||||
| Silicibacter pomeroyi DSS-3 | + | + | + | + | + | + | + | |||
| Silicibacter sp. strain TM1040 | + | + | + | + | + | |||||
| Jannaschia sp. strain CCS1 | + | + | + | + | + | + | ||||
| Draft | ||||||||||
| Sulfitobacter sp. strain EE-36 | + | + | + | + | + | |||||
| Sulfitobacter sp. strain NAS-14.1 | + | + | + | + | + | + | ||||
| Roseovarius nubinhibens ISM | + | + | + | + | + | |||||
| Roseovarius sp. strain 217 | + | + | + | + | + | + | + | + | + | |
| Oceanicola batsensis HTCC2597 | + | + | + | + | ||||||
| Oceanicola granulosus HTCC2516 | + | |||||||||
| Rhodobacterales bacterium HTCC2654 | + | + | + | + | + | + | + | |||
| Loktanella vestfoldensis SKA53 | + | + | + | + | ||||||
| Roseobacter sp. strain MED193 | + | + | + | + | + | + | + | |||
| Parameters | ||||||||||
| % of Roseobacter genomes | 25 | 92 | 83 | 25 | 58 | 25 | 50 | 92 | 50 | 50 |
| Experimental evidence (reference) | ND | Yes (43) | Yes (5) | ND | Yes (21) | Yes (Fig. S7)b | Yes (Fig. S4)b | Yes (Fig. S5)b | ND | Yes (24) |
| Ortholog in P. ubique HTCC1062 | No | No | No | No | No | No | No | No | No | Yes |
The nine draft Roseobacter genomes include those for aerobic anoxygenic phototrophy (AAnP genes), carbon monoxide oxidation (cox genes), aromatic compound degradation (pcaGH and boxC), sulfur oxidation (soxB), denitrification (nirS/K), nitrate assimilation (nasA), phosphonate use (phn genes), type IV secretion (vir genes), and DMSP demethylase (dmdA). + indicates the presence of a homolog with an E value of ≤10−40 and amino acid percent sequence identity of ≥40%. Phenotypes that have been demonstrated experimentally for a Roseobacter strain are indicated by “yes.” ND, not determined. The presence of an ortholog in the genome of SAR11 member P. ubique is indicated.
See the supplemental material.
S. pomeroyi harbors two cox operons that have been shown experimentally to mediate carbon monoxide oxidation at the low concentrations typical of ocean surface waters (≤5 nM) (43). The other two Roseobacter genomes likewise contain cox clusters: two in Jannaschia sp. strain CCS1 (Jann_1763, Jann_1765, and Jann_1766 and Jann_2095, Jann_2096, and Jann_2097) and one in Silicibacter sp. strain TM1040 (TM1040_1764, TM1040_1765, and TM1040_1766). Without any apparent mechanism for CO2 fixation, this pathway may be used by the roseobacters as an energy supplement, potentially resulting in higher heterotrophic growth yields in ocean surface waters where organic matter is limiting but CO is ubiquitous (albeit at low concentrations). Including the nine available draft sequences, all but 1 of 12 Roseobacter genomes contain at least one cox operon (Table 3).
Members of the Roseobacter clade have been found to contribute to aerobic anoxygenic phototrophy (AAnP) in ocean surface waters (4, 46). The genome sequence of Jannaschia sp. strain CCS1, along with the recently published genome of Roseobacter denitrificans (52), provides insight into the genes of a marine AAnP. With no recognizable carbon fixation pathways, phototrophy may be used by Jannaschia sp. strain CCS1 to obtain energy and thereby increase efficiency of heterotrophic growth, as hypothesized for carbon monoxide oxidation. Forty-three genes form a 45.8-kb photosynthetic cluster (Jann_0412-0184) that includes genes for carotenoid biosynthesis (crt), bacteriochlorophyll synthesis (bch), reaction center and light-harvesting complex I (puf), and regulation (ppaA and ppsR). A smaller group of genes located 146 kb downstream of the main cluster contains genes for the light-harvesting complex II (pucBACD; Jann_1581 to Jann_1584). Overall, there is considerable congruence in the photosynthetic gene complements of Jannaschia sp. strain CCS1, the marine aerobic anoxygenic phototroph Erythrobacter sp. strain NAP1 (which is not a roseobacter), and several anaerobic anoxygenic phototrophs (see Table S4 and Fig. S3 in the supplemental material). Neither the S. pomeroyi nor Silicibacter sp. strain TM1040 genomes contain phototrophy genes. However, searches of nine additional Roseobacter genomes available in draft form (Fig. 1) showed that 25% of these cultured roseobacters are predicted to be phototrophs (Table 3). None of the Roseobacter genomes contains evidence of proteorhodopsin-based phototrophy.
All three Roseobacter genomes encode a complete tricarboxylic acid (TCA) cycle as well as peripheral pathways such as phosphoenolpyruvate (PEP) carboxylase and (except for Silicibacter sp. strain TM1040) the glyoxylate shunt, permitting growth on a variety of organic acids (e.g., acetate and pyruvate). All three strains also harbor genes for the Embden-Meyerhof, Entner-Doudoroff, and pentose phosphate pathways.
(iii) Acquiring nitrogen and phosphorus.
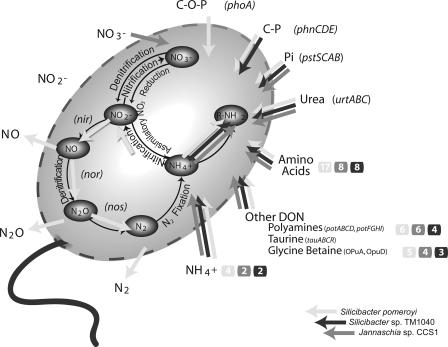
Organic nitrogen and ammonium appear to serve as the primary sources of cellular nitrogen for marine roseobacters. All three genomes contain genes for the assimilation of urea, amino acids, other nitrogen-rich organic compounds often associated with eukaryotic plankton (polyamines and the osmolytes taurine and glycine betaine), and ammonium (Fig. 4). This reliance on organic nitrogen and ammonium is in keeping with both the observed vertical distribution of roseobacters (highest abundance in surface waters where regenerated rather than “new” nitrogen dominates) and frequent physical associations with organic N-rich phytoplankton cells (25). Neither nitrate nor nitrite transporters are found in the two Silicibacter genomes, and experimental evidence confirms that S. pomeroyi cannot assimilate nitrate (see Fig. S4 in the supplemental material). However, Jannaschia sp. strain CCS1 has genes for uptake and assimilatory reduction of nitrate (nrtA, -B, and -D; nirB and -A; and nasA) (Fig. 4).
FIG. 4.
Genes for acquisition of nitrogen and phosphorus in three Roseobacter genomes. Shaded arrows indicate the presence of a pathway (light gray, S. pomeroyi; dark gray, Silicibacter sp. strain TM1040; medium gray, Jannaschia sp. strain CCS1). Numbers in colored circles indicate the number of ORFs if ≥2 copies. Amino acid transporters and branched-chain amino acid transporters are summed. C-O-P, phosphoesters; C-P, phosphonates; Pi, phosphate.
Inorganic P appears to be acquired by all three species through the high-affinity phosphate transport system (pstSCAB). Both Silicibacter species also have genes for transport (phnCDE) and cleavage (phnGHIJKLN) of organic P in the form of phosphonates (C-P-bonded phosphorus), one of two major organic P reservoirs in seawater (see Fig. S5 in the supplemental material). Only the Jannaschia sp. strain CCS1 genome and one other draft Roseobacter genome lack phosphonate transport genes (Table 3). S. pomeroyi but not Silicibacter sp. strain TM1040 can also obtain organic phosphorus from the other major organic P reservoir, phosphoesters (C-O-P bonded phosphorus; mediated by phoA) (Fig. 4). While Jannaschia sp. strain CCS1 has neither of the organic phosphorus uptake systems, the presence of genes for inorganic P storage as polyphosphate (ppk) (which is missing in the other two genomes) may provide an alternative strategy for maintaining a reliable supply of phosphorus.
(iv) Interfacing with other organisms.
Roseobacters harbor a number of genes that may be involved in information, metabolite, and DNA exchange with Roseobacter relatives, other bacteria, and marine eukaryotes.
Nonribosomal peptide synthetases (NRPSs) catalyze the synthesis of a wide array of bacterial antibiotics, toxins, siderophores, and other bioactive compounds from common or modified amino acids. S. pomeroyi and Silicibacter sp. strain TM1040, but not Jannaschia sp. strain CSS1, harbor a very similar 10-gene cluster containing two adjacent NRPSs, an NRPS loading enzyme (phosphopantetheinyl transferase family), and four glycosyl transferases. Close homologs of these genes are not found outside the Roseobacter group, suggesting they may encode a novel glycosylated peptide that plays a role in defense, signaling, or host-microbe interactions with marine plankton (7).
Roseobacter genes may encode an alternative synthesis pathway for the plant auxin indoleacetic acid (IAA) via indole-3-acetonitrile, including indoleacetamide hydrolase and nitrile hydratase (32) in S. pomeroyi and Silicibacter sp. strain TM1040 (SPO2938, -1314, and -1315 and TM1040_1578, TM1040_1971, and TM1040_1972) and arylacetone-specific nitrilase (33) in Jannaschia sp. strain CCS1 (Jann_3735). If this is the case, the potential benefits of harboring IAA-related genes would include initiation of cell wall loosening and sugar release from phytoplankton cells, as has been found for vascular plants (36).
Twelve vir-related ORFs on the pSTM2 plasmid of Silicibacter sp. strain TM1040 indicate the presence of a type IV secretion system for translocating DNA or proteins into other cells. These genes have homology to virD2 and virD4, which code for the relaxase and coupling proteins (providing the energetics for export of DNA), and virB1 to -11 (excluding virB7), which encode the inner membrane channel and pilus structure of the transfer machinery (10) (see Table S5 in the supplemental material). In Agrobacterium tumefaciens, homologous vir genes move the transferred DNA (T-DNA) into plant cells to initiate gall formation. The association between Silicibacter sp. strain TM1040 and dinoflagellate P. piscicida (1, 40, 41) tempts speculation that Vir proteins enable roseobacters to transfer DNA or protein directly to phytoplankton cells. The S. pomeroyi and Jannaschia sp. strain CCS1 genomes do not have vir orthologs (see Table S5 in the supplemental material), although half of the 12 complete and draft Roseobacter genome sequences do (Table 3).
Overall, the Roseobacter gene collection suggests members of this clade have multiple mechanisms for sensing and reacting to their environment while acquiring diverse substrates and nutrients for growth. This is a distinctly different ecological tactic from the “genome streamlining” strategy of other successful marine heterotrophs (18).
Which genes make a roseobacter a roseobacter?
Whether or not microbial biogeochemical functions can be predicted from the taxonomic composition of natural communities has been debated among microbial ecologists for more than a decade. While the answer may turn out to be different for each taxon, it hinges on whether functions encoded by a set of unique and predictable genes can be ascribed to most members of a bacterial clade. The Roseobacter lineage is a coherent group based on 16S rRNA phylogeny (7), yet the diversity in physiology and habitat within the taxon is considerable (56). Thus, we asked whether shared genes can be identified that form the basis of a predictable ecological role for marine roseobacters.
The three roseobacters share 1,939 genes, accounting for ∼50% of each genome (Table 2). To focus specifically on unique genes, we first narrowed down the three-way ortholog gene set by excluding genes that were also found in nonmarine alphaproteobacterial relatives. We used both protein (BLASTp) and nucleic acid (tBLASTn) homology searches to condense the 1,939 three-way-shared genes to 121 unique Roseobacter genes based on their presence in all three Roseobacter genomes but their absence in the Rhodobacter sphaeroides 2.4.2, Paracoccus denitrificans, Sinorhizobium meliloti 1021, and Mesorhizobium loti MAFF303099 genomes (see Table S6 in the supplemental material).
The 121 Roseobacter unique genes are clearly dominated by transport proteins, which account for 37 (31%) of the gene set. Since transporters average 9% of ORFs in Bacteria (48) and 6% in the roseobacters, this disproportionately high percentage argues for a central role for transporters in defining the taxon. The compounds predicted to move across Roseobacter membranes by the unique transporters include glyoxylate (a product of algal photorespiration and organic matter photoxidation), acetate, allophanate (a breakdown product of urea in chlorophytes and bacteria) (28), arginine, branched-chain amino acids, ammonia, and secondary metabolites. The presence of three sodium-based transporters (one symporter, one antiporter, and one voltage-gated ion channel) is consistent with adaptations to a high-salt environment, as is the abundance of TRAP transporters that likely depend on sodium ions for activity (29). Other gene categories well represented in the unique genes are involved in aromatic compound degradation (four genes), sulfur transport and metabolism (four genes), and methyl group transfer (two genes). There were also genes for extracellular peptide-mediated signaling (rhomboid protein homolog), extracellular degradation of the storage compound polyhydroxybutyrate (poly-3-hydroxybutyrate depolymerase), and resistance to peroxides (hydroperoxide resistance protein) (see Table S6 in the supplemental material). Fifty-one (42%) of the 121 unique genes are in at least 10 of the 12 Roseobacter genomes (see Table S6 in the supplemental material). Megaplasmids pSPD1 and pSTM1 of S. pomeroyi and Silicibacter sp. strain TM1040, respectively, contain unique genes in an unbiased proportion relative to the total gene distribution (e.g., pSPD1 contains 10.4% of the total genes and 12.4% of the unique genes). No unique genes were found on the small Silicibacter sp. strain TM1040 or Jannaschia sp. strain CCS1 plasmids (pJCS1 and pSTM2), consistent with the idea that the smaller Roseobacter plasmids harbor a diverse and nonessential gene pool.
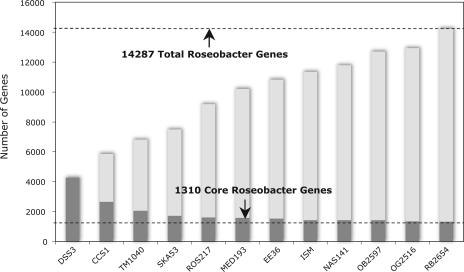
We also analyzed Roseobacter genes using a more standard concept of “core” genes, defined here as those present in the three complete plus all nine draft Roseobacter genomes using an RBH criterion. Of the 14,287 different genes harbored within the 12 Roseobacter genomes, 1,310 are core (Fig. 5). The core gene set includes those for cellular metabolism, energy generation, and ribosomal protein synthesis, among other critical processes (see Table S7 in the supplemental material). The gene set does not include many of the biogeochemically relevant pathways ascribed to roseobacters, such as carbon monoxide oxidation (cox genes), sulfur oxidation (sox genes), DMSP demethylation (dmdA), denitrification (nirK, nirS, nor genes, and nos genes), and phosphonate utilzation (phn genes). Thus, while the total Roseobacter gene repertoire consists of a number of ecologically relevant metabolic pathways, only a subset of these are present in any single genome (Table 3). This “mix-and-match” genome arrangement may allow for adaptation to a wide diversity of ecological niches, as has been observed for this group.
FIG. 5.
Roseobacter core genes (dark) and total genes (dark plus light) as a function of increasing genome number. Core genes are those with an RBH to the S. pomeroyi DSS-3 genome for the indicated genome plus all preceding genomes. Total genes are distinct genes in the Roseobacter group, incremented for each genome by the number of genes without an RBH in any of the preceding genomes.
Which genes make a roseobacter marine?
Classic studies on the existence of distinctly marine bacteria in the 1960s concluded that metabolic pathways of cultured marine bacteria are quite similar to those of nonmarine bacteria and that transport systems based on sodium ions is the main difference between the two groups (37). We took a genomic approach to this question of “marine-biased” functions by identifying Roseobacter genes more likely to be found in genomes of other cultured marine taxa than in nonmarine taxa. Using BLASTp, S. pomeroyi ORFs were compared against a set of 20 genome pairs, each consisting of a cultured marine bacterioplankter outside the Roseobacter clade and its closest nonmarine relative. Forty S. pomeroyi genes were identified as having a distribution statistically biased toward the marine bacteria (see Table S8 in the supplemental material [chi square, P ≤ 0.05]). Three functional gene categories dominated this set: genes involved in sodium transport or utilizing sodium gradients, genes involved in sulfate transport and metabolism, and genes involved in osmoregulation. Sodium-related genes include two sodium symporters that cotransport sodium with other compounds (likely amino acids; SPO1810 and SPO2370), and a respiratory sodium pump (SPOA0030; a component of a multigene NADH:ubiquinone oxidoreductase) that establishes a sodium gradient for ATP synthesis. The sulfur-related genes include two sulfate permeases (SPO1956 and SPO3058) that transport sulfate across the cytoplasmic membrane, a predicted glyoxalase with carbon-sulfur lyase activity (SPO0721), a methionine-γ-lyase (SPOA0318) potentially involved in metabolism of organic sulfur compounds, and an antioxidant that may be specific to sulfur-containing radicals (AhpC/Tsa family protein; SPO2103). Osmoregulation-related genes include osmotically inducible protein osmC (SPO2301) and a homolog of the glycine betaine synthesis gene betA (SPO0190) (see Table S8 in the supplemental material). The 44 genes in the Silicibacter sp. strain TM1040 genome and 36 in the Jannaschia sp. strain CCS1 genome, also found significantly more often in other cultured marine than in nonmarine genomes, similarly have sodium-, sulfate-, and osmoregulation-related functions well represented (see Table S8 in the supplemental material). In general, most of the Roseobacter genes on the marine-biased list are indeed those that permit the organisms to benefit from or manage the plentiful supply of ions in seawater and few appear to mediate metabolic processes that are unique to or characteristic of marine bacteria. A recent culture-independent study of marine operons also found that genes involved in osmolyte transport systems and sodium-based respiratory pumps were significantly enriched in marine bacterial communities (54).
Marine bacteria in the SAR11 group are very abundant in ocean surface waters and commonly co-occur with Roseobacter cells (17). A comparison of the Roseobacter genomes to that of SAR11 member P. ubique HTCC1062 showed that the P. ubique genome contains 547 ORFs (out of 1,354 total) that are not shared with the Roseobacter genomes; over a quarter of these are for hypothetical proteins or proteins of unknown function. We looked in the P. ubique genome for selected biogeochemically relevant genes identified in Roseobacter genomes. Except for the presence of genes for DMSP catabolism (dmdA), there was no overlap, suggesting quite distinct ecological roles for these two abundant taxa (Table 3).
Hypothesis generation.
Genome sequences of cultured marine bacterioplankton can serve to generate hypotheses about ecological activities and biogeochemical roles of their uncultured relatives. Aspects of the Roseobacter genomes suggest that they live in nutrient-replete plankton aggregates within the bulk oligotrophic ocean. Typical components of eukaryotic cytosols can be transported by the roseobacters, such as polyamines, taurine, phosphoesters, phosphonates, aromatic metabolites, glyoxylate, allophanate, acetate, glycine betaine, DMSP, branched-chain amino acids, and organic acids. Many of these compounds are produced by phytoplankton or zooplankton (23, 30, 34, 44) and may become available to roseobacters through exudation, diffusion-driven loss, or trophodynamic interactions including grazing and viral lysis. Regular observations of associations between roseobacters and marine phytoplankton in algal blooms (45, 49) and phycospheres of cultured dinoflagellates and diatoms (1, 25) are consistent with this idea. Members of the Roseobacter clade appear to exhibit a gradient in their reliance on “hot spots” in ocean waters, however. For example, Jannaschia sp. strain CCS1 differs from the other two roseobacters in that its genome indicates it is phototrophic, transports and reduces nitrate, only assimilates inorganic forms of P, and has only one rRNA operon with which to gear up metabolic activity in response to transient nutrient availability (compared to three in S. pomeroyi and five in Silicibacter sp. strain TM1040) (31).
The repertoire of Roseobacter genes also generates the hypothesis that they frequently interact with neighboring cells, possibly to increase their own access to resources. Abundant toxin and metabolite (DMT) transporters, NRPS and possibly IAA synthesis pathways, extracellular polyhydroxybutyrate degradation, β-lactamase-related proteins, extracellular peptide signaling genes, and RND-MFP (resistance nodulation division-membrane fusion protein) family proteins support this idea. Although speculative, it is possible that roseobacters directly capture organic matter from eukaryotic cells through Vir proteins, IAA production, or hemolysin-type proteins. Such activity would influence the amount of carbon and nutrients entering the microbial food web (2) and affect their availability to metazoans.
At least two successful ecological strategies for marine bacterioplankton thus appear to operate in ocean surface waters. One tactic, typified by the roseobacters, is based on metabolically versatile cells that can compete well with other organisms for labile substrates within plankton-dense microzones. Another tactic, typified by SAR11 clade members, is based on metabolically conservative, free-living cells that scavenge dilute organic matter dissolved in seawater (18). Members of the Roseobacter and SAR11 clades represent a significant fraction of marine bacterioplankton communities, together accounting for ≥30% of the bacterioplankton in most ocean surface waters (17, 19, 20). As more genomic information emerges for these and other key bacterioplankton clades, including those yet to be cultured, insights into the biochemical capabilities of individual cells will enrich understanding of bacterial ecology and ocean-scale biogeochemical processes.
Supplementary Material
Acknowledgments
We thank M. Joye for unpublished data. The Department of Energy provided genome sequences of Silicibacter sp. strain TM1040 and Jannaschia sp. strain CCS1.
This work was supported by grants from the National Science Foundation (MCB-0531476 to M.A.M., MCB-0446001 to R.B., and MCB-0534203 to A.B.) and the Gordon and Betty Moore Foundation.
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alavi, M., T. Miller, K. Erlandson, R. Schneide, and R. Belas. 2001. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3:380-396. [DOI] [PubMed] [Google Scholar]
- 2.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Béjà, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, et al. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 5.Buchan, A., L. S. Collier, E. L. Neidle, and M. A. Moran. 2000. Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage. Appl. Environ. Microbiol. 66:4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchan, A., E. L. Neidle, and M. A. Moran. 2004. Diverse organization of genes of the β-ketoadipate pathway in members of the marine Roseobacter lineage. Appl. Environ. Microbiol. 70:1658-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchan, A., J. M. González, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Button, D. K. 1991. Biochemical basis for whole-cell uptake kinetics: specific affinity, oligotrophic capacity, and the meaning of the Michaelis constant. Appl. Environ. Microbiol. 57:2033-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, F., K. Wang, J. Stewart, and R. Belas. 2006. Induction of multiple prophages from a marine bacterium: a genomic approach. Appl. Environ. Microbiol. 72:4995-5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. F. 2005. Microbial community genomics in the ocean. Nat. Rev. Microbiol. 3:459-469. [DOI] [PubMed] [Google Scholar]
- 12.Dolowy, P., J. Mondzelewski, R. Zawadzka, J. Baj, and D. Bartosik. 2005. Cloning and characterization of a region responsible for the maintenance of megaplasmid pTAV3 of Paracoccus versutus UW1. Plasmid 53:239-250. [DOI] [PubMed] [Google Scholar]
- 13.Duval, B., K. Shetty, and W. H. J. Thomas. 1999. Phenolic compounds and antioxidant properties in the snow alga Chlamydomonas nivalis after exposure to UV light. Appl. Phycol. 11:559-566. [Google Scholar]
- 14.Faulkner, D. J. 2000. Marine natural products. Nat. Prod. Rep. 17:7-55. [DOI] [PubMed] [Google Scholar]
- 15.Field, J. G., K. R. Clarke, and R. M. Warwick. 1982. A practical strategy for analysing multispecies distribution patterns. Mar. Ecol. Prog. Ser. 8:37-52. [Google Scholar]
- 16.Gabriel, D. W., C. Allen, M. Schell, T. P. Denny, J. T. Greenberg, et al. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant-Microbe Inter. 19:69-79. [DOI] [PubMed] [Google Scholar]
- 17.Giovannoni, S. J., and M. S. Rappé. 2000. The uncultured microbial majority, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. John Wiley & Sons, New York, NY.
- 18.Giovannoni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin, et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242-1245. [DOI] [PubMed] [Google Scholar]
- 19.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González, J. M., J. S. Covert, W. B. Whitman, J. Henricksen, F. Mayer, et al. 2003. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., DMSP demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 53:1261-1269. [DOI] [PubMed] [Google Scholar]
- 22.Han, C. S., and P. Chain. 2006. Finishing repeat regions automatically with Dupfinisher, p. 141-146. In H. R. Arabnia and H. Valafar (ed.), Proceedings of the 2006 International Conference on Bioinformatics and Computational Biology. CSREA Press, Las Vegas, NV.
- 23.Hellebust, J. A. 1965. Excretion of some organic compounds by marine phytoplankton. Limnol. Oceanogr. 10:192-206. [Google Scholar]
- 24.Howard, E. C., J. R. Henriksen, A. Buchan, C. R. Reisch, H. Bürgmann, et al. 2006. Bacterial taxa limiting sulfur flux from the ocean. Science 314:649-652. [DOI] [PubMed] [Google Scholar]
- 25.Jasti, S., M. E. Sieracki, N. J. Poulton, M. W. Giewat, and J. N. Rooney-Varga. 2005. Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 71:3483-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung, A., T. Domratcheva, M. Tarutina, Q. Wu, W. H. Ko, et al. 2005. Structure of a bacterial BLUF photoreceptor: insights into blue light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 102:12350-12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Käll, L., A. Krogh, and E. L. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 28.Kanamori, T., N. Kanou, H. Atomi, and T. Imanaka. 2004. Enzymatic characterization of a prokaryotic urea carboxylase. J. Bacteriol. 186:2532-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly, D. J., and G. H. Thomas. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25:405-424. [DOI] [PubMed] [Google Scholar]
- 30.Kiene, R. P., L. J. Linn, and J. A. Bruton. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43:209-224. [Google Scholar]
- 31.Klappenbach, J. A., J. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi, M., H. Izui, T. Nagasawa, and H. Yamada. 1993. Nitrilase in biosynthesis of the plant hormone indole-3-acetic acid from indole-3-acetonitrile: cloning of the Alcaligenes gene and site-directed mutagenesis of cysteine residues Proc. Natl. Acad. Sci. USA 90:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi, M., T. Suzuki, T. Fujita, M. Masuda, and S. Shimizu. 1995. Occurrence of enzymes involved in biosynthesis of indole-3-acetic acid from indole-3-acetonitrile in plant-associated bacteria, Agrobacterium and Rhizobium. Proc. Natl. Acad. Sci. USA 92:714-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, C., and N. O. G. Jorgensen. 1995. Seasonal cycling of putrescine and amino-acids in relation to biological production in a stratified coastal salt pond. Biogeochemistry 29:131-157. [Google Scholar]
- 35.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, et al. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32:D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacLeod, R. A. 1965. The question of the existence of specific marine bacteria. Bacteriol. Rev. 29:9-23. [PMC free article] [PubMed] [Google Scholar]
- 38.Malmstrom, R. R., R. P. Kiene, and D. L. Kirchman. 2004. Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49:597-606. [Google Scholar]
- 39.Meyer, F., A. Goesmann, A. C. McHardy, D. Bartels, T. Bekel, et al. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, T. R., and R. Belas. 2004. Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp. Appl. Environ. Microbiol. 70:3383-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, T. R., and R. Belas. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8:1648-1659. [DOI] [PubMed] [Google Scholar]
- 42.Moran, M. A., and R. E. Hodson. 1994. Dissolved humic substances of vascular plant origin in a coastal marine environment. Limnol. Oceanogr. 39:762-771. [Google Scholar]
- 43.Moran, M. A., A. Buchan, J. M. González, J. F. Heidelberg, W. B. Whitman, et al. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 44.Nishibori, N., A. Yuasa, M. Sakai, S. Fujihara, and S. Nishio. 2001. Free polyamine concentrations in coastal seawater during phytoplankton bloom. Fish. Sci. 67:79-83. [Google Scholar]
- 45.Pinhassi, J., R. Simó, J. M. González, M. Vila, L. Alonso-Sáez, et al. 2005. Dimethylsulfoniopropionate turnover is linked to the composition and dynamics of the bacterioplankton assemblage during a microcosm phytoplankton bloom. Appl. Environ. Microbiol. 71:7650-7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pradella, S., M. Allgaier, C. Hoch, O. Päuker, E. Stackebrandt, et al. 2004. Genome organization and localization of the pufLM genes of the photosynthesis reaction center in phylogenetically diverse marine Alphaproteobacteria. Appl. Environ. Microbiol. 70:3360-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quest, B., and W. Gärtner. 2004. Chromophore selectivity in bacterial phytochromes. Eur. J. Biochem. 271:1117-1126. [DOI] [PubMed] [Google Scholar]
- 48.Ren, Q., and I. T. Paulsen. 2005. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 1:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 51.Santos, S. R., and H. Ochman. 2004. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 6:754-759. [DOI] [PubMed] [Google Scholar]
- 52.Swingley, W. D., S. Sadekar, S. D. Mastrian, H. J. Matthies, J. Hao, et al. 2007. The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J. Bacteriol. 189:683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swofford, D. L. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (and other methods) 4.0. Illinois Natural History Survey, University of Illinois, Champaign.
- 54.Tringe, S. G., C. von Mering, A. Kobayashi, A. A. Salamov, K. Chen, et al. 2005. Comparative metagenomics of microbial communities. Science 308:554-557. [DOI] [PubMed] [Google Scholar]
- 55.Ulrich, L. E., E. V. Koonin, and I. B. Zhulin. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 13:52-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner-Döbler, I., and H. Biebl. 2006. Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60:255-280. [DOI] [PubMed] [Google Scholar]
- 57.Zhong, Z., R. Caspi, D. Helinski, V. Knauf, S. Sykes, et al. 2003. Nucleotide sequence based characterizations of two cryptic plasmids from the marine bacterium Ruegeria isolate PR1b. Plasmid 49:233-252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.