Abstract
Genomes of members of the family Enterobacteriaceae contain large repertoires of putative fimbrial operons. Since many of these operons are poorly expressed in vitro, a convenient method for inducing elaboration of the encoded fimbriae would greatly facilitate their functional characterization. Here we describe a new technique for identifying fimbriated bacteria from a library of transposon mutants by screening with immunomagnetic particles for ligand expression (SIMPLE). The SIMPLE method was applied to identify the T-POP mutants of Salmonella enterica serotype Typhimurium carrying on their surfaces filaments composed of PefA, the major subunit product of a fimbrial operon (pef) that is not expressed during growth in Luria-Bertani broth. Four such mutants were identified from a library of 24,000 mutants, each of which carried a T-POP insertion within the hns gene, which encodes a global silencer of horizontally acquired genes. Our data suggest that the SIMPLE method is an effective approach for isolating fimbriated bacteria, which can be readily applied to fimbrial operons identified by whole-genome sequencing.
Comparative genomics reveals that members of the family Enterobacteriaceae possess large repertoires of fimbrial gene sequences, representing hypervariable regions within their genomes (16, 29). Many of the fimbrial operons identified by whole-genome sequencing are not expressed in vitro, which is one of the reasons why only a small fraction of the encoded surface structures has been characterized functionally.
For example, sequence analysis of the enterohemorrhagic Escherichia coli O157:H7 isolate reveals the presence of 14 fimbrial operons (11). Construction of lacZ transcriptional fusions to the genes encoding the major fimbrial subunit shows that 9 of these 14 operons are not expressed under in vitro growth conditions (16). The Salmonella enterica serotype Typhimurium genome contains 13 fimbrial operons, termed csg (agf), fim, pef, lpf, bcf, stf, saf, stb, stc, std, sth, sti, and stj (18). Laboratory-grown cultures of Salmonella serotype Typhimurium commonly elaborate only type 1 fimbriae encoded by the fim operon (6, 7) and thin curled fimbriae (also known as thin aggregative fimbriae or curli) encoded by the csg gene cluster (10, 23, 28). Expression of the remaining fimbrial operons cannot be detected readily by Western blotting (12) or flow cytometry (13) after growth of Salmonella serotype Typhimurium under standard laboratory conditions. Although expression of most fimbrial operons is suppressed in vitro, seroconversion to fimbrial antigens (12) and phenotypes of Salmonella serotype Typhimurium mutants (30) during infection of mice provide indirect evidence for in vivo expression of these surface structures. The considerable gap between the number of fimbrial structures whose function can be studied using in vitro assays (n = 2) and the number of fimbrial operons present in the genome (n = 13) has been a major obstacle for mechanistic studies of the role of adherence during Salmonella serotype Typhimurium pathogenesis.
The histone-like nucleoid-structuring (H-NS) protein is a global silencer of gene expression. In Salmonella serotype Typhimurium, H-NS silences genes with GC contents lower than that of the genomic average (i.e., genes acquired by lateral gene transfer) by restricting the access of RNA polymerase to DNA (17, 20). Mutational analysis shows that inactivation of the hns gene increases transcription of the pef operon (21). Chromatin immunoprecipitation indicates the presence of H-NS binding sites upstream of the bcfA, stfA, safA, fimA, stbA, lpfA, and sthA genes (17, 20). These data suggest that H-NS functions as a transcriptional repressor of multiple fimbrial operons in Salmonella serotype Typhimurium.
The goal of this study was to devise a screening with immunomagnetic particles for ligand expression (SIMPLE) method for identifying fimbriated bacteria within mutant libraries. This method was developed using the pef operon located on the Salmonella serotype Typhimurium virulence plasmid (8). Northern blotting analysis shows that the pef operon is not expressed after growth of Salmonella serotype Typhimurium at neutral pH, but transcription is induced under acidic growth conditions (i.e., in LB broth at pH 5.1) (21). Despite transcription of the pef operon in LB broth at pH 5.1, previous studies were not able to detect expression of PefA by Western blotting in Salmonella serotype Typhimurium grown under this condition, presumably because elaboration of fimbriae is subject to posttranscriptional control mechanisms (12). Here we describe the isolation of Salmonella serotype Typhimurium mutants expressing fimbriae encoded by the pef operon, using the SIMPLE method.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains used are listed in Table 1. Unless otherwise noted, cultures were grown from a single colony, statically, at 37°C in 0.22-μm filter-sterilized Luria-Bertani (LB) broth buffered to pH 7.0 with 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) hydrate for 48 h or on Miller LB agar plates (1.5% Difco agar) overnight. Antibiotics and other chemicals were added as needed: tetracycline (Tc), 20 mg/liter; kanamycin (Km), 100 mg/liter; carbenicillin (Carb), 100 mg/liter; naladixic acid (Nal), 50 mg/liter; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 40 mg/liter.
TABLE 1.
Bacterial strains used in this study
| Bacterial strain(s) | Genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α MCR | F′ mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 phoA supE44 λ−thi-1 gyrA96 relA1 | Gibco BRL |
| ORN172 Nalr | Nalrthr-1 leuB thi-1 Δ(argF-lac)U169 malA1 xyl-7 ara-13 mtl-2 gal-6 rpsL tonA2 supE44 ΔfimBEACDFGH::Km | 31 |
| Salmonella serotype Typhimurium | ||
| IR715 | Wild-type isolate ATCC 14028, Nalr | 27 |
| EHW2 | IR715 ΔfimAICDHF::Km | 30 |
| LT2 | Laboratory strain | 15 |
| TH3923 | LT2 pJS28(Carbr P22-9+)/F′114ts Lac+zzf-20::Tn10[tetA::MudP](Tcs) zzf-3823::Tn10dTc[del-25](T-POP)/leuA414 hsdSB Fels2− | K. Hughes |
| TH4881 | LT2 pNK2880(CarbR, Tn10 tnpA[ats-1 ats-2])/fliC5050::MudJ fljB5001::MudCm | 22 |
| SL1344(T-POP) | Wild-type isolate SL1344 fimZ2::T-POP | 5 |
| SR11 | Xylose-fermenting mutant of wild-type isolate BA2 | 24 |
| AJB3 | SR11 Nalr | 3 |
| ADH19 | SR11 NalrΔpefBACD::Km | 13 |
| SPN22 | SR11 NalrΔfimAICDHF::Km | This study |
| SPN23 | LT2 ΔfimAICDHF::Km | This study |
| SPN66 and SPN67 | SR11 NalrΔfimAICDHF::Km hns(−195)::T-POP | This study |
| SPN68 | SR11 NalrΔfimAICDHF::Km hns(+281)::T-POP | This study |
| SPN69 | SR11 NalrΔfimAICDHF::Km hns(−224)::T-POP | This study |
| SPN71 | SR11 fimZ2::T-POP | This study |
| SPN74 | SR11 Nalr ΔfimAICDHF::Km fimZ2::T-POP | This study |
| SPN81 | SR11 hns(−195)::T-POP | This study |
| SPN84 | SR11 hns(+281)::T-POP | This study |
Transduction.
P22 HT int-105 and KB1 int were used to generate generalized transducing lysates of Salmonella serotype Typhimurium LT2 as previously described (19). Salmonella serotype Typhimurium strain SR11 can serve as a recipient for P22-mediated transduction, but this strain is resistant to P22-mediated lysis. To use SR11 derivatives as donors for transduction, we used phage KB1 int, which forms plaques on this Salmonella serotype Typhimurium strain. Transductants were struck for single colonies on Evans blue uridine agar (4), and light green colonies were cross-struck against P22 H5 (for P22 HT int-105) or KB1 int (for KB1) to confirm phage sensitivity.
Strain construction.
Strains SPN22 (SR11 ΔfimAICDHF::Km) and SPN23 (LT2 ΔfimAICDHF::Km) were generated by transducing SR11 and LT2, respectively, to kanamycin resistance using a P22 lysate of EHW2 (14028 Nalr ΔfimAICDHF::Km). SPN71 (SR11 fimZ2::T-POP) and SPN74 (SR11 fimZ2::T-POP ΔfimAICDHF::Km) were generated by transducing the SR11 wild type and SR11 ΔfimAICDHF::Km (SPN22), respectively, to tetracycline resistance with a P22 lysate of SL1344(T-POP) (fimZ2::T-POP). Plasmid pNK2880 was introduced into SPN22 (ΔfimAICDHF::Km) by P22 transduction to carbenicillin resistance from a lysate of TH4881. The hns(−195)::T-POP and hns(+281)::T-POP insertions were introduced into the SR11 wild type by transduction with a KB1 lysate of SPN66 [ΔfimAICDHF::Km hns(−195)::T-POP] and SPN68 [ΔfimAICDHF::Km hns(+281)::T-POP] to generate SPN81 [SR11 hns(−195)::T-POP] and SPN84 [SR11 hns(+281)::T-POP], respectively, by selecting for tetracycline resistance.
T-POP mutant pools.
Sixteen pools of random Tn10dTc[del-25] (T-POP) (22) insertion mutants were generated using a P22 lysate grown on TH3923 to transduce into Salmonella serotype Typhimurium strain SPN22 carrying plasmid pNK2880 (22). Transductants were selected on LB plus Tc agar (approximately 1,500 mutants per plate) and pooled by flooding with 5 ml of LB broth at pH 7 and resuspending colonies.
Western blotting analysis.
Polyclonal rabbit anti-PefA serum has been previously described (13). The serum was diluted 1:5 in phosphate-buffered saline (PBS) at pH 7.4 plus 0.2% sodium azide and preabsorbed eight times (9) with ORN172 carrying plasmid pGEX-4T-2 (25) and four times with ADH19 (SR11 ΔpefBACDI::Km). Unless otherwise noted, cultures were resuspended, by measuring optical density at 600 nm (OD600), to a concentration of approximately 2 × 108 CFU/10 μl in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer and boiled for 5 min to generate cell lysates. Cell lysates were then separated by 15% SDS-PAGE, transferred to Immobilon-P (Millipore) membranes using a Trans-Blot semidry transfer cell (Bio-Rad), incubated with anti-PefA serum diluted to a 1:500 final dilution, detected with goat anti-rabbit alkaline phosphatase and an Immun-Star chemiluminescent substrate (Bio-Rad), and visualized with X-Omat Blue film (Kodak).
SIMPLE protocol.
BioMag protein G particles (QIAGEN) were resuspended by vortexing and then added in 100-μl aliquots to 1.5-ml Eppendorf tubes and magnetically separated out of solution using a magnetic particle concentrator for Eppendorf tubes (Dynal MPC-S). The storage solution was aspirated, and then the particles were washed by adding 750 μl of TN buffer (0.1 M Tris-HCl, 0.15 M NaCl [pH 7.5 final], sterilized with a 0.22-μm filter) and vortexing. This wash step was repeated two more times. The particles were then resuspended in 100 μl of preabsorbed anti-PefA serum by vortexing and incubated for 1 h at room temperature, with vortexing every 5 min. The particles were then magnetically separated, the serum was aspirated, and the particles were washed three times as described before with 750 μl of TN buffer. The particles were resuspended in 650 μl of particle-blocking (PB) buffer (TN buffer plus 1% casein, added aseptically and prepared fresh 2 h prior to use) in Eppendorf tubes and incubated at room temperature for half an hour with constant inversion on an automatic roller. Culture (100 μl; mixed by gentle swirling and at no time vortexed) containing 1 to 5 × 108 CFU (by OD600) of bacteria (either a mutant pool or mixtures of two bacterial strains) was added, and the tubes were incubated for 1 h at room temperature with constant inversion. The particles were washed three times with PB buffer, and the tubes were gently inverted 10 times to resuspend the particles. Particles were either resuspended in 1 ml of PBS (to generate serial 10-fold dilutions of the resuspended particles that were spread on agar plates containing the appropriate antibiotics) or in the appropriate growth medium (to incubate bacteria statically at 37°C for 48 h; and the resulting culture was used to repeat the above protocol). In competition experiments using mixtures of two bacterial strains, the SIMPLE protocol was performed with each mixture in triplicate.
Enrichment (n-fold) was calculated by dividing the output ratio (i.e., the mutant/wild-type ratio recovered after enrichment) by the input ratio (i.e., the mutant/wild-type ratio present in the inoculum). For statistical analysis, data were transformed logarithmically and analyzed by Student's t test.
Inverse PCR.
DNA regions adjacent to the T-POP insertion site were amplified by inverse PCR (1). In brief, genomic DNA from the mutant of interest was digested with either AluI or TaqI and ligated with T4 DNA ligase, and the divergent primers TPOP-OUT (CGCTTTTCCCGAGATCATATG [bp 2168 to 2188 of GenBank accession number AY150213]) and TPOP-AluTaq (GCACTTGTCTCCTGTTTACTCC [bp c1977 to c1956 of GenBank accession number AY150213]) were used with PCR SuperMix HiFi (Invitrogen) to amplify the region of interest from the resultant circular DNA. The reactions were then run on 1% agarose gels, and the amplified fragment was purified with a QIAEX II gel purification kit (QIAGEN), cloned into pCR2.1 with a TOPO TA cloning kit (Invitrogen), transformed into chemically competent E. coli DH5α MCR, and plated on LB plus Carb plus X-Gal. Plasmid minipreps (QIAGEN) of white colonies were digested with EcoRI to confirm insertion, and the insert was sequenced by SeqWright (Houston, TX) using the universal M13-Forward and M13-Reverse primers.
Electron microscopy.
For immune electron microscopy, bacteria were grown in a static culture, washed twice in PBS, and resuspended in electron microscopy (EM)-grade water (EM Science) at a titer of approximately 1 × 109CFU/ml. Bacteria were allowed to adhere to a formvar/carbon-coated grid (EM Science) for 2 min, and grids were incubated for 20 min in a primary antiserum (rabbit) diluted 1:250 in PBS containing 1% bovine serum albumen (BSA). Grids were washed five times for 1 min each in PBS containing 1% BSA. Grids were incubated for 20 min in goat anti-rabbit antibody-10-nm-gold conjugate (EM Science) diluted 1:20 in PBS containing 1% BSA. Grids were washed three times for 1 min each in PBS containing 1% BSA and three times for 1 min each in EM-grade water (EM Science) before they were analyzed by electron microscopy.
RESULTS
Enrichment for fimbriated bacteria by immunomagnetic separation.
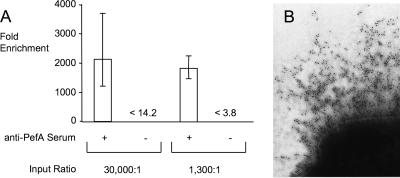
The goal of this study was to establish a genetic screen suitable for detecting mutants that elaborate fimbriae. The most direct approach to accomplishing this goal was detection of the major fimbrial subunit on the surfaces of live bacterial cells using specific antiserum. We reasoned that by incubating a bacterial culture with magnetic particles coated with antiserum raised against a fimbrial protein, it would be possible to enrich fimbriated bacteria using immunomagnetic separation. To provide a proof of concept for this approach, we chose to study expression of the pef operon because its major fimbrial subunit, PefA, can be readily detected by Western blotting in static LB broth cultures of a non-type 1-fimbriated E. coli strain (ORN172) carrying a cosmid (pFB11) bearing the Salmonella serotype Typhimurium pef operon (2). We determined whether fimbriated bacteria could be enriched by immunomagnetic separation after incubating a 30,000:1 mixture of ORN172 and ORN172(pFB11) with a slurry of anti-PefA-coated magnetic beads. Immunomagnetic separation resulted in a 2,100-fold enrichment for fimbriated bacteria [ORN172(pFB11)] (Fig. 1A). This enrichment was antibody mediated, because no fimbriated bacteria [ORN172(pFB11)] could be recovered from beads that were not coated with antiserum. Expression of PefA could be detected on the surface of ORN172(pFB11) but not on the surface of ORN172 by using immunogold labeling with rabbit anti-PefA serum (Fig. 1B), which demonstrated that the antiserum used for immunomagnetic separation specifically bound PefA assembled into fimbrial filaments. Collectively, our results show that immunomagnetic separation can be used to enrich fimbriated bacteria.
FIG. 1.
Enrichment by immunomagnetic separation of E. coli cells expressing PefA on their surface. (A) n-fold enrichment of ORN172(pFB11) was accomplished by a single round of immunomagnetic separation by particles coated with anti-PefA serum (+) or not coated with serum (−). Magnetic particles were incubated with mixtures (30,000:1 or 1,300:1) of strains ORN172 (nonfimbriated) and ORN172(pFB11) (expressing fimbriae encoded by the pef operon). Data are shown as geometric means ± standard deviations of three independent experiments. In case no colonies of ORN172(pFB11) were recovered from particles, the limit of detection is indicated (<14.2 and <3.8, respectively). (B) Detection of fimbrial filaments composed of PefA on the surfaces of E. coli ORN172(pFB11) cells by using immunogold labeling.
Type 1 fimbriae and growth conditions modulate PefA expression in Salmonella serotype Typhimurium.
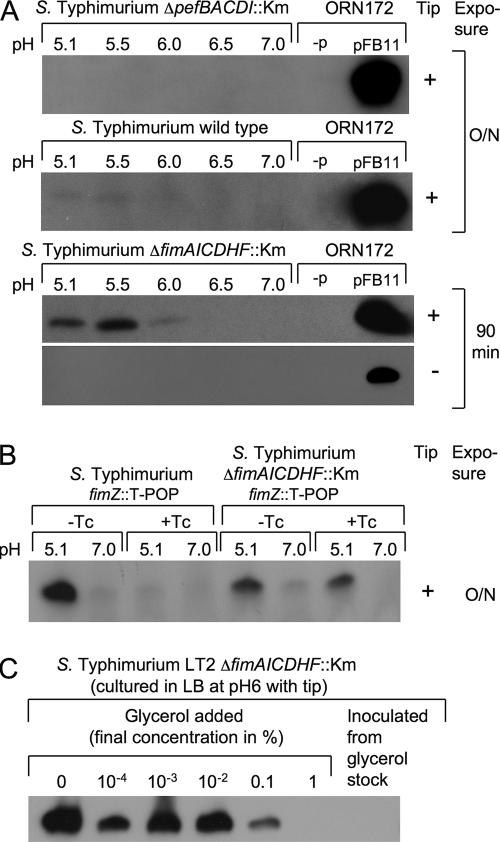
Although the cloned pef operon is well expressed in E. coli, PefA is not expressed after growth of Salmonella serotype Typhimurium in static LB broth culture (12). To test whether immunomagnetic separation could be used to isolate mutants expressing the pef operon in LB broth, we used a Salmonella serotype Typhimurium fimAICDHF deletion mutant (SPN22) to prevent the possibility that the elaboration of type 1 fimbriae would interfere with binding of bacteria to magnetic particles. PefA was not detected by Western blotting in cultures of the Salmonella serotype Typhimurium wild type (SR11) or in the fimAICDHF deletion mutant (SPN22) grown statically in LB broth at neutral pH. After growth of the Salmonella serotype Typhimurium wild type (SR11) at pH 5.1, a faint band could be detected by Western blotting after overnight exposure, suggesting poor expression of PefA under this condition. Interestingly, deletion of the biosynthesis genes for type 1 fimbriae (fimAICDHF) resulted in markedly increased PefA expression after static growth of Salmonella serotype Typhimurium in LB broth at pH 5.1 (Fig. 2A). The fimZ::T-POP insertion, which places the FimZ transcriptional activator of the fimAICDHF operon under the control of a tetracycline-inducible promoter, was introduced into the Salmonella serotype Typhimurium wild type (SR11) and the fimAICDHF deletion mutant (SPN22). PefA was expressed in the Salmonella serotype Typhimurium fimZ::T-POP mutant (SPN71) grown in LB broth at pH 5.1 only when tetracycline was absent. In contrast, the Salmonella serotype Typhimurium ΔfimAICDHF::Km fimZ::T-POP mutant (SPN74) grown in LB broth at pH 5.1 expressed PefA in both the presence and the absence of tetracycline (Fig. 2B). These data suggested that the presence of type 1 fimbrial biosynthesis genes interfered with expression of fimbriae encoded by the pef operon in Salmonella serotype Typhimurium by an unknown mechanism. Similar observations were made recently for E. coli, where expression of type 1 fimbriae affects expression of P-antigen-recognizing (P) fimbriae negatively (26).
FIG. 2.
Expression of PefA detected in Salmonella serotype Typhimurium strains by Western blotting. (A) Expression of PefA in Salmonella serotype Typhimurium strains ADH19 (ΔpefBACD::Km), SR11 (wild type), and SPN22 (ΔfimAICDHF::Km) grown statically in the presence (+) or absence (−) of a yellow pipette tip in LB broth buffered to pH 5.1, 5.5, 6.0, 6.5, or 7.0. Expression of PefA by E. coli strain ORN172 carrying a cosmid bearing the pef operon (pFB11) in LB broth at pH 7 is shown in the far right lane of each blot as a positive control. E. coli strain ORN172 without cosmid (−p) is shown as a negative control in the second lane from the right. Film was exposed for 90 min or overnight (O/N) as indicated on the right. (B) Expression of PefA in Salmonella serotype Typhimurium strains SPN71 (fimZ::T-POP) and SPN74 (ΔfimAICDHF::Km fimZ::T-POP) grown in LB broth buffered to pH 5.1 or pH 7 in the presence (+Tc) or absence (−Tc) of tetracycline. (C) Expression of PefA in Salmonella serotype Typhimurium strain SPN23 (ΔfimAICDHF::Km) grown in LB broth (pH 6) supplemented with the indicated concentrations of glycerol. Cultures were inoculated from a single colony (left lanes) or directly from glycerol stock (far right lane) and incubated overnight in the presence of a yellow pipette tip.
In the course of these studies, we noted that inoculation of LB broth cultures directly from −80°C freezer stocks prevented PefA expression in Salmonella serotype Typhimurium. A follow-up experiment suggested that this effect was due to an inhibition of PefA expression by the presence of small amounts of glycerol in the culture medium (Fig. 2C). Furthermore, expression of PefA at pH 5.1 was increased considerably in Salmonella serotype Typhimurium if the 200-μl yellow pipette tips used to inoculate static LB broth cultures were left in the culture tubes during overnight incubation (Fig. 2A). For the screen described below, bacteria were grown without inoculation with yellow tips.
Screening with immunomagnetic particles for ligand expression (SIMPLE).
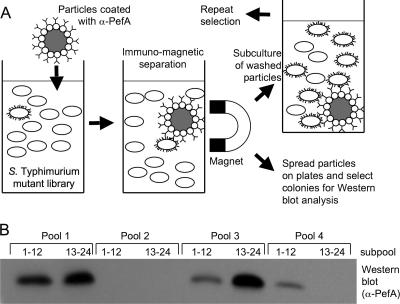
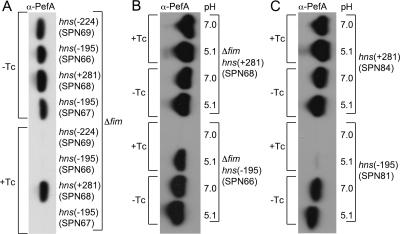
The Salmonella serotype Typhimurium fimAICDHF (SPN22) deletion mutant was used to generate a library of random T-POP insertion mutants (22). To this end, a plasmid encoding a Tn10 transposase with broad target specificity (pNK2880) was introduced into the Salmonella serotype Typhimurium fimAICDHF (SPN22) deletion mutant. Bacteriophage P22 was grown on a serotype Typhimurium strain (TH3923) carrying the T-POP transposon on an E. coli F′ plasmid, and the resulting phage lysate was used to deliver the T-POP transposon into SPN22(pNK2880). Transductants were selected on LB agar plates containing tetracycline (the resistance conferred by T-POP). Approximately 24,000 mutants generated by this method were grouped into 16 pools of approximately 1,500 mutants each. Each pool was used to inoculate a static LB broth culture buffered to pH 7, a growth condition under which PefA is not expressed in the SPN22 parental strain. Each pool was incubated with magnetic particles coated with anti-PefA serum, and particle-bound bacteria were resuspended in LB broth (pH 7) and used to inoculate a second static LB broth (pH 7) culture to repeat the enrichment procedure. After two enrichment cycles, 24 randomly selected colonies from each pool were grown statically in LB broth at pH 7, and expression of PefA was assessed for subpools of 12 mutants by Western blotting (Fig. 3). Seven of the 16 pools were positive for PefA expression. Individual bacterial colonies from each positive pool were grown statically in LB broth at pH 7 to identify T-POP mutants expressing PefA. One PefA-expressing mutant from each pool was selected, and the T-POP insertion was transduced into the Salmonella serotype Typhimurium fimAICDHF deletion mutant (SPN22) by KB1 int-mediated transduction to remove the transposase encoded by plasmid pNK2880. For three pools, we were not able to obtain transductants expressing PefA (even after repeated attempts with different colonies from each pool). In these cases, T-POP may have undergone secondary transposition events due to the continued presence of pNK2880 during the screening procedure. For the remaining four pools, transduction did yield T-POP mutants (SPN66, SPN67, SPN68, and SPN69) expressing PefA in LB broth at pH 5.1 and pH 7. Interestingly, three of the four mutants (SPN66, SPN67, and SPN69) did not express PefA after growth in LB broth at pH 7 in the presence of tetracycline (Fig. 4), suggesting that the tetA promoter of T-POP may drive expression of a negative regulator of PefA expression in these strains.
FIG. 3.
Enrichment for Salmonella serotype Typhimurium mutants expressing PefA by using the SIMPLE method. (A) Schematic illustrates the principle of the SIMPLE method. (B) Western blotting analysis of four pools, each originally consisting of approximately 1,500 independent mutants, after two rounds of enrichment by the SIMPLE method. From each pool, 24 colonies were randomly selected and grown statically in LB broth at pH 7, and a mixture of 12 mutants was loaded per lane.
FIG. 4.
Expression of PefA detected by Western blotting in Salmonella serotype Typhimurium hns mutants. Bacteria were cultured statically in LB broth at 37°C in the presence of a yellow pipette tip. (A) Salmonella serotype Typhimurium SR11 ΔfimAICDHF::Km carrying T-POP insertions in hns at position −224 (SPN69), −195 (STN66, STN67), or +281 (SPN68) was grown at pH 7 in the presence (+Tc) or absence (−Tc) of tetracycline. (B) Salmonella serotype Typhimurium SR11 ΔfimAICDHF::Km carrying T-POP insertions in hns at position −195 (SPN66) or +281 (SPN68) was grown at pH 7 or pH 5.1 in the presence (+Tc) or absence (−Tc) of tetracycline. (C) Salmonella serotype Typhimurium SR11 carrying T-POP insertions in hns at position −195 (SPN81) or +281 (SPN84) was grown at pH 7 or pH 5.1 in the presence (+Tc) or absence (−Tc) of tetracycline. α, anti.
Inactivation of hns relieves PefA suppression in Salmonella serotype Typhimurium.
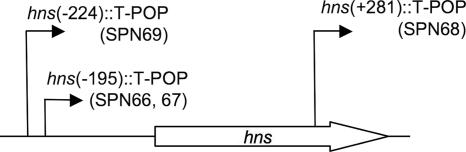
DNA regions flanking the T-POP transposon insertion sites in strains SPN66, SPN67, SPN68, and SPN69 were cloned by inverse PCR, and the respective nucleotide sequences were determined. The T-POP insertions were located either within or upstream of the hns open reading frame at base pair position −224 (SPN69), −195 (SPN66 and SPN67), or +281 (SPN68), relative to that of the start codon (Fig. 5). The fact that PefA was detected in a mutant in which the T-POP insertion had disrupted the hns open reading frame (SPN68) suggested that the expression of PefA is negatively regulated by the H-NS protein. Mutants SPN66, SPN67, and SPN69 carried T-POP insertions in an orientation in which the hns gene was located downstream of the tetA promoter. Our data suggested that the T-POP insertions in strains SPN66, SPN67, and SPN69 disrupted the hns promoter, resulting in the expression of PefA in the absence of tetracycline. Sequence analysis further suggested that growth of strains SPN66, SPN67, and SPN69 in the presence of tetracycline would result in the expression of hns from the tetA promoter, resulting in PefA repression. Predictions from the sequence analysis of T-POP insertion sites (Fig. 5) were thus in good agreement with the expression of PefA observed for the respective mutants (Fig. 4).
FIG. 5.
Positions of T-POP insertions identified by the SIMPLE method for Salmonella serotype Typhimurium mutants expressing PefA. The open arrow indicates the position and orientation of the hns open reading frame. The T-POP insertion sites in mutants SPN66, SPN67, SPN68, and SPN69 are indicated. Arrows indicate the orientation of the tetracycline-inducible tetA promoter of T-POP.
SIMPLE method validation.
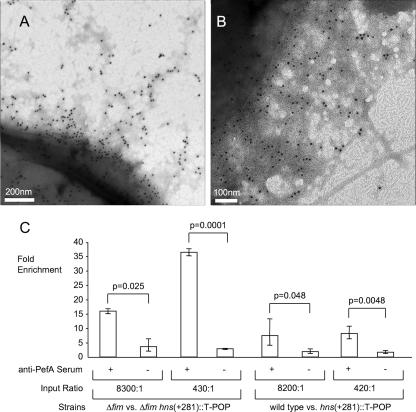
Our data suggested that the hns mutants were identified using the SIMPLE method because PefA expressed on their surfaces bound to particles coated with anti-PefA serum. This assumption was tested experimentally by detecting the expression of PefA on the surface of strain SPN68 [ΔfimAICDHF::Km hns(+281)::T-POP] cells by immunogold labeling after growth at pH 7. Expression of PefA could be detected by this method in Salmonella serotype Typhimurium ΔfimAICDHF::Km hns(+281)::T-POP (SPN68) but not in Salmonella serotype Typhimurium ΔfimAICDHF::Km (SPN22) cells (Fig. 6A).
FIG. 6.
Enrichment by immunomagnetic separation for Salmonella serotype Typhimurium hns(+281)::T-POP mutants expressing PefA on their surfaces. Detection of fimbrial filaments composed of PefA on the surface of (A) Salmonella serotype Typhimurium SR11 ΔfimAICDHF::Km hns(+281)::T-POP (SPN68) or (B) Salmonella serotype Typhimurium SR11 hns(+281)::T-POP (SPN84) by immunogold labeling. Bacteria were grown statically in LB broth at pH 7 without a yellow pipette tip. (C) n-fold enrichment is shown for SPN68 [Δfim::Km hns(+281)::T-POP; left panel] or SPN84 [hns(+281)::T-POP; right panel] accomplished by a single round of immunomagnetic separation with particles coated with anti-PefA serum (+) or not coated with serum (−). Magnetic particles were incubated with the indicated mixtures of strains SPN22 and SPN68 (left panel) or SR11 and SPN84 (right panel). Data are shown as geometric means ± standard deviations of three independent experiments. Statistical significance (P value) of differences between treatment groups (shown in brackets) is indicated above the bars.
The enrichment factor for the hns mutants, by the SIMPLE method, was quantified in a competition experiment in which magnetic beads were incubated with an 8,300:1 or a 430:1 mixture of strains SPN22 (ΔfimAICDHF::Km) and SPN68 [ΔfimAICDHF::Km hns(+281)::T-POP] grown at pH 7. Immunomagnetic separation resulted in a 16-fold or a 36-fold enrichment, respectively, for strain SPN68 [ΔfimAICDHF::Km hns(+281)::T-POP] when the experiment was performed with magnetic particles coated with anti-PefA serum (Fig. 6C). In contrast, the same experiment performed with magnetic particles that had not been coated with serum did not result in a marked enrichment for strain SPN68.
To determine whether the presence of an intact fim operon might interfere with the SIMPLE method, the hns(+281)::T-POP insertion in strain SPN68 was transduced into the Salmonella serotype Typhimurium wild type (SR11). Mixtures of strains SR11 (wild type) and SPN84 [hns(+281)::T-POP] were subjected to enrichment using magnetic particles as described above. Bacteria expressing PefA were enriched approximately 10-fold with magnetic particles coated with anti-PefA serum, while no significant enrichment was observed in the absence of anti-PefA serum. In the presence of type 1 fimbriae, enrichment for PefA-expressing bacteria by the SIMPLE method was reduced to approximately 10-fold compared to the approximately 16- to 36-fold enrichment observed in the absence of type 1 fimbrial biosynthesis genes (Fig. 6C). The degree of enrichment correlated positively with the fraction of cells expressing PefA on their cell surfaces as assessed by immunogold labeling, which was approximately 65% of cells for strain SPN68 [ΔfimAICDHF::Km hns(+281)::T-POP] and approximately 5% of cells for strain SPN84 [hns(+281)::T-POP]. These data suggested that type 1 fimbriae did not interfere directly with bacterial binding to magnetic beads but rather with PefA expression on the bacterial surface. Interestingly, similar levels of PefA expression were detected for cultures of SPN68 [ΔfimAICDHF::Km hns(+281)::T-POP] and SPN84 [hns(+281)::T-POP] by Western blotting (Fig. 4B and C), suggesting that rather than decreasing the total amount of PefA, the presence of type 1 fimbrial biosynthesis genes may interfere with the assembly of PefA on the cell surface.
In summary, our data demonstrate that SIMPLE is a powerful method for identifying mutants expressing fimbriae from a complex mutant library.
DISCUSSION
At the dawn of the genomic era, Escherichia coli and Salmonella serotypes were among the organisms best characterized genetically. Despite a large body of knowledge about these organisms’ biology, whole-genome sequencing revealed the presence of much larger repertoires of fimbrial operons than predicted by the number of fimbrial structures observed on the surfaces of these bacteria. The fact that the majority of fimbrial operons were identified only by sequence analysis reflects the tight control of their expression in vitro, a property that has prevented a thorough characterization of the encoded adhesins.
Efforts to characterize the growing number of putative fimbrial operons identified by whole-genome sequencing would greatly benefit from the development of reliable approaches for isolating fimbriated bacteria from a mutant library. Currently, such screens commonly employ transposon mutagenesis of a strain carrying a fusion between the gene of interest (in this case a fimbrial operon) and a reporter gene that, when expressed, alters the colony phenotype. Two shortcomings limit the usefulness of this approach for analysis of fimbrial operons. First, a screen for colony phenotypes requires that fimbriae be expressed on agar plates, an assumption that may not always be correct. For example, type 1 fimbriae of Salmonella serotype Typhimurium are expressed in LB broth but not on LB agar plates (13). Second, the use of transcriptional fusions cannot detect posttranscriptional control mechanisms that may affect elaboration of fimbriae on the bacterial surface. The SIMPLE method developed in this study overcomes these limitations because it allows identification of bacteria expressing a fimbrial operon by directly targeting the encoded fimbrial filaments expressed in broth culture.
The Salmonella serotype Typhimurium genome contains 4,597 open reading frames (18), of which 490 genes are essential during growth in rich medium (14). By applying the formula P = 1 − e−a/b, where P is the probability that a gene is mutated, a is the number of mutants, and b is the number of nonessential genes, it can be estimated that our library of approximately 24,000 transposon mutants was large enough to contain insertions within 99.7% of Salmonella serotype Typhimurium nonessential genes. Assuming an equal probability for insertions in each gene, a bank of 24,000 transposon mutants should, on average, contain approximately six insertions in hns. Our screen identified seven mutants expressing PefA, although three of them could not be analyzed further, presumably because T-POP insertions in these strains had undergone secondary transposition events. The remaining four mutants contained insertions in the hns gene, a putative negative regulator. The number of hns insertions identified in our study was, thus, in good agreement with theoretical predictions. Expression of H-NS-repressed genes is thought to be induced when this repressor becomes displaced by a positive regulatory element from the promoter region (17, 20). No such positive regulatory elements for pef expression were identified in this study. To identify genes encoding positive regulatory elements, the T-POP transposon has to be inserted in the correct orientation (i.e., downstream of the single tetracycline-inducible promoter present on Tn10dTc[del-25]) and in close proximity (i.e., within 200 bp or less) of a start codon, to ensure activation of its open reading frame in the presence of tetracycline. A library of approximately 24,000 T-POP mutants was expected to contain insertions in the correct orientation and in close proximity to start codons of only 38% of Salmonella serotype Typhimurium genes. The mutant bank screened in this study was thus not of sufficient size for identifying positive regulatory elements with high probability.
The presence of type 1 fimbrial biosynthesis genes reduced the fraction of Salmonella serotype Typhimurium cells expressing PefA on their surfaces. Similar observations were made recently for E. coli, where the expression of type 1 fimbriae affects the expression of P fimbriae negatively (26). The fact that our screen was performed with a strain background carrying a deletion of type 1 fimbrial biosynthesis genes resulted in increased sensitivity of the SIMPLE method (Fig. 6C). H-NS binding sites are present in promoter regions of multiple Salmonella serotype Typhimurium fimbrial operons (17, 20). The use of a hns fim mutant background may further improve analysis of Salmonella serotype Typhimurium fimbrial operons by the SIMPLE method.
Our results demonstrate that the SIMPLE method can be used to isolate mutants elaborating a fimbrial protein, PefA, whose expression is not observed in Salmonella serotype Typhimurium grown under standard laboratory conditions. One potential drawback of the SIMPLE approach is that it requires the availability of antibodies directed against fimbrial proteins. However, the fact that the majority of Salmonella serotype Typhimurium fimbrial operons are not expressed in vitro has not been a major obstacle for generating specific antibodies against major fimbrial subunits (13). The SIMPLE method should be tractable with any mutagenesis strategy and readily applicable to other bacteria and possibly to any surface structure to which antiserum can be raised.
In summation, we have demonstrated that the SIMPLE method is readily applicable to the isolation of a mutant elaborating a fimbrial structure on its surface from a complex library.
Acknowledgments
We thank Kelly Hughes and John Roth for kindly providing bacterial strains and Grete Adamson and Robert Droleskey for help with electron microscopy.
Work in A.J.B.'s laboratory was supported by USDA/NRICGP grant 2002-35204-12247 and by Public Health Service grants AI040124, AI044170, and AI065534.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Bäumler, A. J., J. G. Kusters, I. Stojiljkovic, and F. Heffron. 1994. Salmonella typhimurium loci involved in survival within macrophages. Infect. Immun. 62:1623-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäumler, A. J., R. M. Tsolis, F. Bowe, J. G. Kusters, S. Hoffmann, and F. Heffron. 1996. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäumler, A. J., R. M. Tsolis, and F. Heffron. 1996. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochner, B. R. 1984. Curing bacterial cells of lysogenic viruses by using UCB indicator plates. BioTechniques 2:234-240. [Google Scholar]
- 5.Clegg, S., and K. T. Hughes. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clegg, S., B. K. Purcell, and J. Pruckler. 1987. Characterization of genes encoding type 1 fimbriae of Klebsiella pneumoniae, Salmonella typhimurium, and Serratia marcescens. Infect. Immun. 55:281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duguid, J. P., E. S. Anderson, and I. Campbell. 1966. Fimbriae and adhesive properties in Salmonellae. J. Pathol. Bacteriol. 92:107-137. [DOI] [PubMed] [Google Scholar]
- 8.Friedrich, M. J., N. E. Kinsey, J. Vila, and R. J. Kadner. 1993. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol. Microbiol. 8:543-558. [DOI] [PubMed] [Google Scholar]
- 9.Gruber, A., and B. Zingales. 1995. Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. BioTechniques 19:28-30. [PubMed] [Google Scholar]
- 10.Grund, S., and A. Weber. 1988. A new type of fimbriae on Salmonella typhimurium. Zentbl. Vetmed. Reihe B 35:779-782. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 12.Humphries, A. D., S. DeRidder, and A. J. Bäumler. 2005. Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect. Immun. 73:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphries, A. D., M. Raffatellu, S. Winter, E. H. Weening, R. A. Kingsley, R. Droleskey, S. Zhang, J. Figueiredo, S. Khare, J. Nunes, L. G. Adams, R. M. Tsolis, and A. J. Bäumler. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357-1376. [DOI] [PubMed] [Google Scholar]
- 14.Knuth, K., H. Niesella, C. J. Hueck, and T. M. Fuchs. 2004. Large-scale identification of essential Salmonella by trapping lethal insertions. Mol. Microbiol. 51:1729-1744. [DOI] [PubMed] [Google Scholar]
- 15.Lilleengen, K. 1948. Typing of Salmonella typhimurium by means of bacteriophage. Acta Pathol. Microbiol. Scand. Suppl. 77:2-125. [Google Scholar]
- 16.Low, A. S., N. Holden, T. Rosser, A. J. Roe, C. Constantinidou, J. L. Hobman, D. G. Smith, J. C. Low, and D. L. Gally. 2006. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ. Microbiol. 8:1033-1047. [DOI] [PubMed] [Google Scholar]
- 17.Lucchini, S., G. Rowley, M. D. Goldberg, D. Hurd, M. Harrison, and J. C. Hinton. 2006. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathogens 2:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236-238. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson, B., and D. Low. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35:728-742. [DOI] [PubMed] [Google Scholar]
- 22.Rappleye, C. A., and J. R. Roth. 1997. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179:5827-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider, H. A., and N. D. Zinder. 1956. Nutrition of the host and natural resistance to infection. V. An improved assay employing genetic markers in the double strain inoculation test. J. Exp. Med. 103:207-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 26.Snyder, J. A., B. J. Haugen, C. V. Lockatell, N. Maroncle, E. C. Hagan, D. E. Johnson, R. A. Welch, and H. L. Mobley. 2005. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 73:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stolpe, H., S. Grund, and W. Schroder. 1994. Purification and partial characterization of type 3 fimbriae from Salmonella typhimurium var. copenhagen. Zentbl. Bakteriol. 281:8-15. [DOI] [PubMed] [Google Scholar]
- 29.Townsend, S. M., N. E. Kramer, R. Edwards, S. Baker, N. Hamlin, M. Simmonds, K. Stevens, S. Maloy, J. Parkhill, G. Dougan, and A. J. Bäumler. 2001. Salmonella enterica serotype Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 69:2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weening, E. H., J. D. Barker, M. C. Laarakker, A. D. Humphries, R. M. Tsolis, and A. J. Bäumler. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodall, L. A., P. W. Russell, S. L. Harris, and P. E. Orndorff. 1993. Rapid, synchronous, and stable induction of type 1 piliation in Escherichia coli by using a chromosomal lacUV5 promoter. J. Bacteriol. 175:2770-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]