Abstract
Bacteria commonly inhabit subsurface oil reservoirs, but almost nothing is known yet about microorganisms that live in naturally occurring terrestrial oil seeps and natural asphalts that are comprised of highly recalcitrant petroleum hydrocarbons. Here we report the first survey of microbial diversity in ca. 28,000-year-old samples of natural asphalts from the Rancho La Brea Tar Pits in Los Angeles, CA. Microbiological studies included analyses of 16S rRNA gene sequences and DNA encoding aromatic ring-hydroxylating dioxygenases from two tar pits differing in chemical composition. Our results revealed a wide range of phylogenetic groups within the Archaea and Bacteria domains, in which individual taxonomic clusters were comprised of sets of closely related species within novel genera and families. Fluorescent staining of asphalt-soil particles using phylogenetic probes for Archaea, Bacteria, and Pseudomonas showed coexistence of mixed microbial communities at high cell densities. Genes encoding dioxygenases included three novel clusters of enzymes. The discovery of life in the tar pits provides an avenue for further studies of the evolution of enzymes and catabolic pathways for bacteria that have been exposed to complex hydrocarbons for millennia. These bacteria also should have application for industrial microbiology and bioremediation.
Prior studies of subsurface petroleum reservoirs using culture-based methods have revealed diverse microbial communities that are able to live on complex petroleum hydrocarbon mixtures (22, 43). Nonetheless, very little is known yet about the true extent of microbial diversity in natural oil reservoirs and terrestrial petroleum deposits, such as those that occur in oil sands, shales, and natural asphalts. Initial surveys of underground reservoirs using molecular approaches so far suggest that the majority of microorganisms inhabiting these environments are new species that represent a rich pool of novel genetic diversity with potential importance for industrial and petroleum microbiology (43). Compared to underground oil reservoirs, even less is known about terrestrial habitats, where petroleum-degrading soil bacteria have come to inhabit heavy oil seeps, tar sands, and natural asphalts. With the advent of improved DNA extraction and purification methods, such bacteria and their genes may now be accessible for detailed study of their diversity and genes that encode petroleum-degrading enzymes.
The existence of bacteria in petroleum deposits at great depths suggests that many species have evolved specifically for this environment and may be carried to the surface in oil seeps. In soils that are permeated with the asphalt, bacteria may also include indigenous bacteria that survived after asphalt seeped through the soil. Selection of bacterial communities for petroleum substances occurs rapidly after even short-term exposures of soil to petroleum hydrocarbons following oil spills (41, 42). Over time spans encompassing millennia, bacteria that can tolerate this environment would be expected to undergo genetic adaptations that may lead to evolution of new ecotypes and species and enzymes for growth on petroleum hydrocarbons. During adaptation of communities, genes for petroleum hydrocarbon-degrading enzymes that are carried on plasmids or transposons may be exchanged between species. In turn, new catabolic pathways eventually may be assembled and modified for efficient regulation (27). Other cell adaptations leading to new ecotypes may include modifications of the cell envelope to tolerate solvents (28) and development of community-level interactions that facilitate cooperation within consortia.
Here we describe a survey of microbial diversity in natural asphalts at the Rancho La Brea Tar Pits in California. These natural asphalts are located in Hancock Park in downtown Los Angeles and consist of asphalt-soil mixtures formed by upwelling of heavy oil and asphaltenes in spatially separated seeps that differ in their chemical composition and age. Although the asphalt at Rancho La Brea is commonly called tar, the petroleum hydrocarbons here are correctly referred to as natural asphalt and are comprised of some of the most recalcitrant carbon compounds in nature (9). Our survey examined two excavation sites. The first site, Pit 91, has yielded thousands of plant and animal fossils and is the richest Pleistocene fossil site in the world (10, 16, 45). Carbon dating of fossils from the current depth under excavation in Pit 91 fixes their ages in a range from 10,000 to 38,000 years before the present (16). The second site, Pit 101, was excavated early last century and was closed in the 1920s, after which the pit was covered with a permanent building as part of a museum display. Microbiological studies included analysis of 16S rRNA genes from DNA extracted from the tar pits and a traditional approach employing cultivation of bacteria on agar. In conjunction with microbial diversity, we also surveyed DNA sequences for aromatic ring-hydroxylating dioxygenases that may have application for industrial microbiology and bioremediation of petroleum wastes.
MATERIALS AND METHODS
Characterization of chemical properties in Pits 91 and 101.
Ten-gram samples of the asphalt-permeated soil were physically broken into small aggregates and placed in beakers containing 10 ml deionized water or 10 mM CaCl2 buffer. The suspensions were mixed using a magnetic stir bar for 1 h, after which pH and salinity were determined. Salinity was determined in water solutions using a conductivity meter. Other samples for metal analyses were acid digested in nitric perchloric acid using digestion bombs and a microwave oven (USEPA SW-846, method 3051). Heavy metals and cations were analyzed by inductively coupled plasma mass spectrometry using an ELAN500 mass spectrometer (Perkin-Elmer-Sciex Instruments, Concord, Ontario, Canada). Carbon, nitrogen, and sulfur were analyzed using a Flash EA 1112 NC analyzer (Thermo Electron Corporation, Milan, Italy).
Sampling and DNA extraction.
Previously unexposed samples were removed from approximately 10 cm under the surfaces of Pit 91 and Pit 101 of the Rancho La Brea Tar Pits in Los Angeles in October 2004. A total of five samples were taken along a 3-m transect from the center of each pit. Samples were removed from the pits with sterile, autoclaved spatulas and were transferred into sterile 50-ml plastic tubes with screw caps and transported to the laboratory for processing. The samples from each pit were pooled prior to extraction. One of the challenges in conducting this survey was the difficulty in extracting high-quality DNA for use in cloning and sequencing. DNA was extracted from the asphalt-soil mixtures by first freezing approximately 5-g aliquots of the asphalt in liquid nitrogen. The frozen samples were transferred to a sterile ceramic mortar and were then ground under liquid nitrogen to a fine powder. Approximately 0.5-g subsamples were processed to extract DNA by bead beating using the BIO 101 Fastprep DNA extraction kit for soil following the manufacturer's protocols. Extracted DNA was concentrated using a Savant Speed Vac system (GMI Inc., Ramsey, MN), and subsamples were combined to obtain a high concentration of DNA. The DNA was purified using a QIAquick gel extraction kit (QIAGEN, Chatsworth, CA) according to the manufacturer's instructions. The purified DNA was concentrated again for use in construction of clone libraries.
Phylogenetic analysis.
16S rRNA genes were amplified by PCR and purified with a QIAquick PCR purification kit (QIAGEN). The purified PCR products from five runs were combined and used to construct clone libraries using the pGEM-T Easy vector (Promega) with selected primer sets. The Bacteria were detected using bacterium-specific primers, 27F and 1492R (21). The Archaea were detected using the domain-specific PCR primers, Ar4F and Ar958R (17). Pseudomonas spp. were identified using the Pseudomonas-selective PCR primers, Ps289F and Ps1258R (47). Dioxygenases were detected using the PCR primer set adoF to adoR for aromatic ring-hydroxylating dioxygenases (36, 37). The sequencing primers were T7 and SP6. After obtaining raw sequences using Chromas 2 (Technelysium Pty. Ltd., Tewantin, Queensland, Australia), putative chimeric sequences were identified using Bellerophon (12) and identified chimeric sequences (43 of 278 bacterial sequences) were excluded. The 16S rRNA sequences were aligned using the NAST aligner, and the aligned sequences were compared to the Lane mask (21) using the Greengenes web site (4, 5). Evolutionary distances were calculated with the Kimura 2-parameter method, and a phylogenetic tree was constructed by the neighbor-joining method (31) with MEGA3 for Windows (19). Bootstrap analyses of the neighbor-joining data were conducted based on 1,000 samples to assess the stability of the phylogenetic relationships.
Statistical analyses.
The computer program DOTUR (32) was used to calculate species richness estimates and diversity indices. A second program, LIBSHUFF (34), was used to compare the similarities of bacterial clone libraries in Pits 91 and 101. The distance matrices for both programs were obtained using an algorithm located at the Greengenes website (4, 5).
FISH.
Fluorescent in situ hybridization (FISH) was carried out as described previously (49) with minor modifications. Oligonucleotides were 5′end labeled with fluorescent dyes and included Eub338 labeled with Cy3, ARCH915 labeled with Cy5, and Pae997 with Bodipy FL. Details on these probes are available at probeBase (24). Cells were photographed with a Leica TCS/SP2 UV confocal microscope.
Cultivation.
Culturable bacteria were isolated by serial dilutions of water suspensions of asphalt-soil mixtures on agar plates containing DSMZ medium 371 amended with 20% NaCl and 10% tryptic soy agar and M9 minimum medium. The plates were incubated at 28°C for 2 to 3 weeks, after which individual isolates were transferred and processed for sequencing of 16S rRNA gene sequences. Isolates were placed in glycerol medium and transferred to a −80°C freezer for long-term preservation. Bacterial isolates were tested on agar medium with 1% asphalt as a sole carbon source.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank under accession numbers DQ001614 to DQ001623, DQ001626 to DQ001638, DQ001641, DQ001642, DQ001644, DQ001646, DQ001647, and EF157073 to EF157279 (for Bacteria), DQ192039 to DQ192061 (for isolates), DQ062817 to DQ062856 (for dioxygenase), AY860440 to AY860443 and AY939988 to AY940011 (for Archaea), and AY940013, AY940019 to AY940022, AY940024 to AY940026, AY940028 and AY940029, AY940032, and AY860446 to AY860448 (for Pseudomonas stutzeri).
RESULTS
Detailed analyses of the two tar pits revealed differences in both the chemical composition and the microbial community composition of the asphalt samples. The asphalt-permeated soil from Pit 91 contained a greater concentration of petroleum hydrocarbons, was slightly acidic, and had a relatively low salinity and metal content (Table 1). In contrast, water suspensions of asphalt-permeated soil from Pit 101 were alkaline (pH 8.4) and contained a high concentration of salts and metals. The salinity of 1:1 asphalt water suspensions of Pit 101 was 4,610 μS cm−1, which was 100 times higher than that of Pit 91. Materials from both of the tar pits are impermeable to surface water from rainfall, and bacteria in this matrix are subject to water deficits and high salinity. Water contained in the asphalt is present in stratified water pockets and pore spaces that occur throughout the pit and are likely the main sites for microbial growth. The occurrence of microbial activity in the tar pits is visually evident from the continual evolution of methane bubbles at various locations in the pits where there is still viscous liquefied asphalt.
TABLE 1.
Chemical properties of asphalt-permeated soil samples from Pit 91 and Pit 101 of Rancho La Brea Tar Pitsa
| Sample site | pH of suspension
|
EC (μS/cm) | % C | % N | % S | C/N ratio | Concn (μg/ml) of:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1:1 (CaCl2) | 1:5 (H2O) | Na | Ca | Mg | K | Al | Cd | Cr | Fe | Zn | Cu | Mn | Pb | Ni | ||||||
| Pit 91 | 6.3 | 5.36 | 46 | 22.04 | 0.3 | 0.97 | 74.16 | 13 | 2,902 | 265 | 86.1 | 356 | 2.2 | 2.65 | 484 | 33.9 | 23.4 | 13 | ND | 89.9 |
| Pit 101 | 8.44 | 7.59 | 4,610 | 7.04 | 0.18 | 0.53 | 46.74 | 178 | 3,696 | 4,245 | 1,011 | 11,395 | 1.52 | 19.9 | 14,133 | 126 | 37.1 | 178 | 58.1 | 39.4 |
EC, electrical conductivity; ND, not detected.
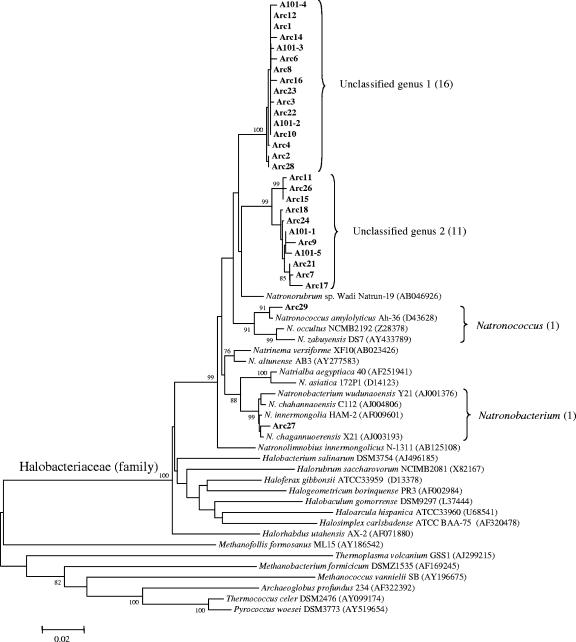
Here a total of 235 bacterial clones were sequenced to identify the predominant phylogenetic groups (see Fig. 2). The most striking difference in the microbial community composition of the two pits was the presence of halophilic Archaea in the highly saline Pit 101, which were not detected in Pit 91 (Fig. 1). Among these were 27 clones representing clusters of closely related species from two unclassified genera that were most similar to Natronococcus and Natronobacterium (Fig. 1). Of the 29 Archaea sequences that were obtained, only 2 were outside of the clusters representing the new genera.
FIG. 2.
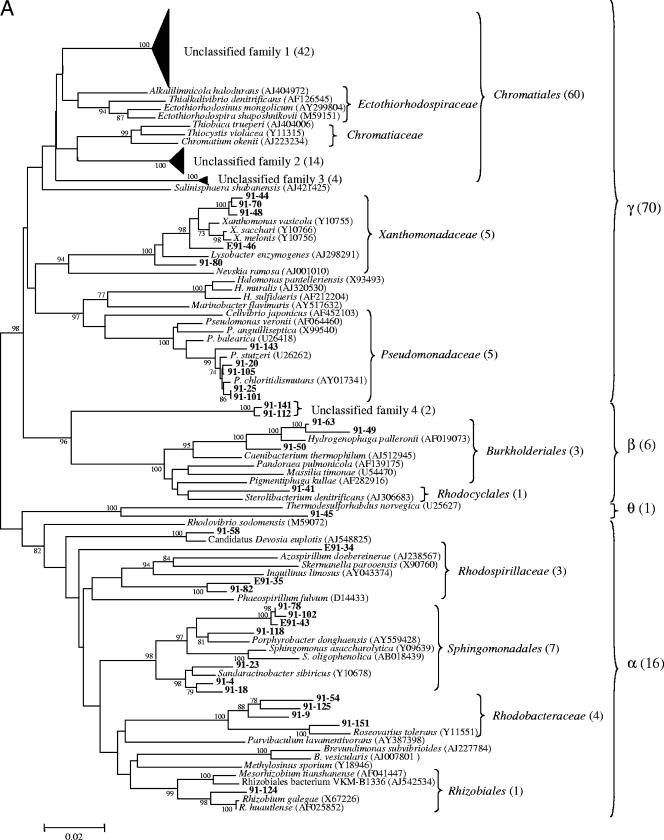
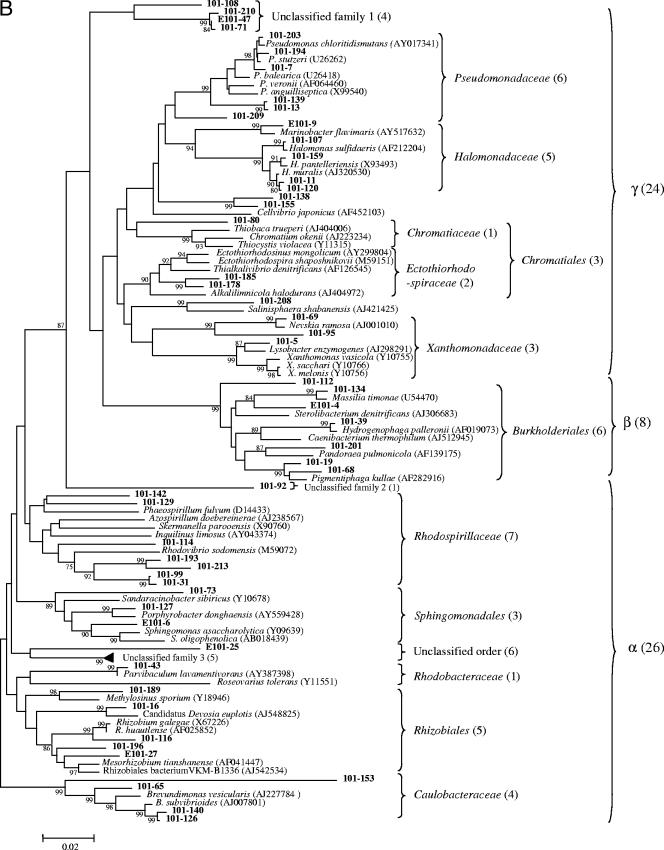
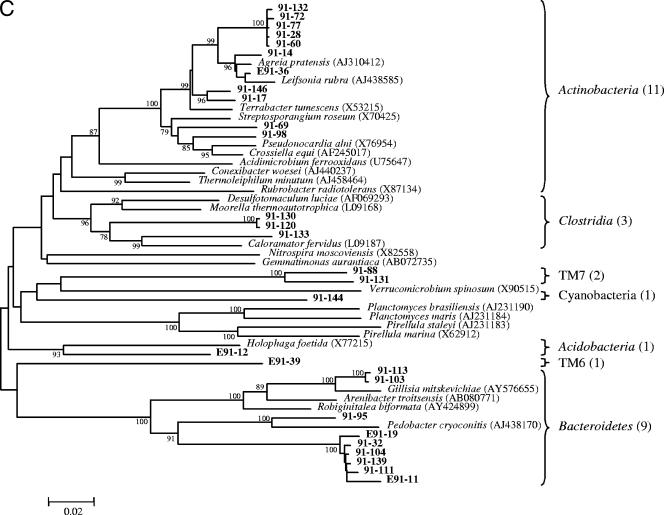
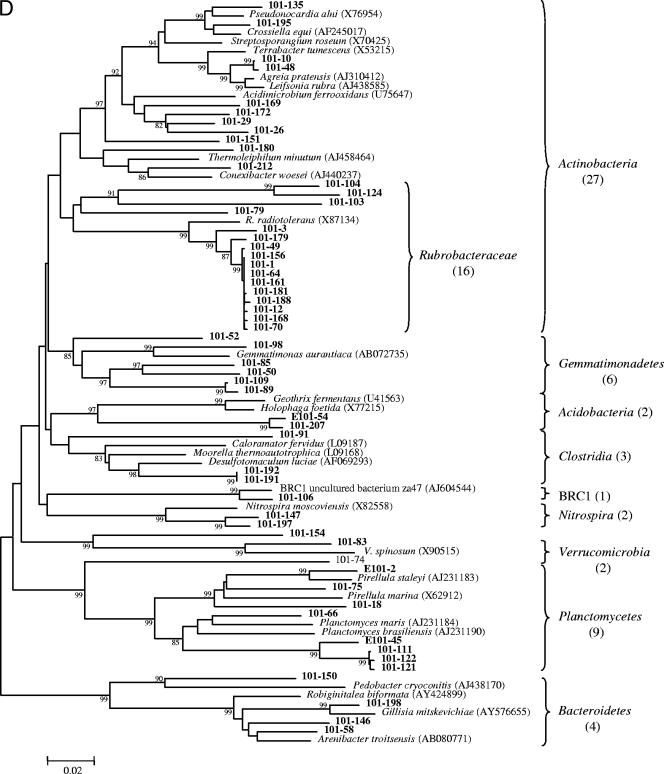
Phylogeny of the Bacteria from near-full-length 16S rRNA gene sequences identified in heavy oil from in Pits 91 and 101 of the Rancho La Brea Tar Pits. Taxa are indicated by vertical bars on the right of the tree, and values in parentheses are clone numbers. The robustness of the topology was estimated by bootstrap resampling. Bootstrap values greater than 75 are shown at branch points. Proteobacteria in Pit 91(A) and Pit 101 (B); other bacteria in Pit 91 (C) and Pit 101 (D). Accession numbers are indicated in parentheses on the right of the tree. Scale bar denotes 0.02 changes per nucleotide.
FIG. 1.
Phylogeny of halophilic Archaea identified in an asphalt soil mixture from Pit 101 of the Rancho La Brea Tar Pits. Genera are indicated by vertical bars on the right of the tree, and values in parentheses are clone numbers. The robustness of the topology was estimated by bootstrap resampling of the neighbor-joining data. Bootstrap values greater than 75 are shown at branch points. Scale bar denotes 0.02 changes per nucleotide.
In addition to Archaea, there were also differences in the distribution of bacterial phyla between the two pits. The predominant bacteria in both pits were Gammaproteobacteria (purple sulfur bacteria). Pit 91 contained 3 unclassified families (60 clones) in the order Chromatiales, none of which was found in Pit 101 (Fig. 2). Other families of the Gammaproteobacteria in Pit 91 included Xanthomonadaceae (5 clones) and Pseudomonadaceae (5 clones). (Fig. 2A and C).
Pit 101 contained a greater breadth of diversity of Gammaproteobacteria and Alphaproteobacteria than Pit 91. This included two unclassified families and one new order with two more unclassified families (Fig. 2B). In addition to 16 clones representing the family Rubrobacteraceae, other unique taxa in Pit 101 included 5 additional phyla that were represented by 9 clones from the Planctomycetes, 6 clones of Gemmatimonadetes, 1 clone of BRC1, and 2 clones each of Nitrospira and Verrucomicrobia (Fig. 2D). Taxa that were common to both Pit 91 and Pit 101 included species from Alpha-, Beta-, and Gammaproteobacteria and various representatives from the Acidobacteria, Actinobacteria, Bacteroidetes, and Clostridia (Fig. 2).
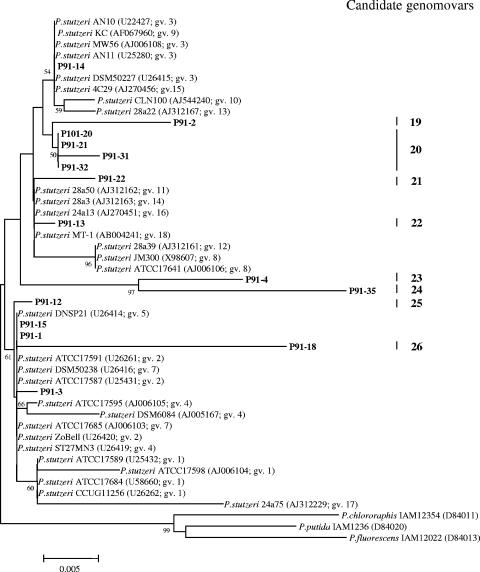
Sequences obtained using the Pseudomonas-selective primers included 14 clones of Pseudomonas stutzeri, which represented 8 new genomovars of this species (Fig. 3). All but 1 of the 14 clones was obtained from Pit 91, suggesting a differential distribution of P. stutzeri in the two tar pits (Fig. 3).
FIG. 3.
Phylogeny of Pseudomonas stutzeri. 16S rRNA gene sequences identified in asphalt from Pits 91 and 101 of the Rancho La Brea Tar Pits using PCR primers for Pseudomonas spp. Candidate genomovars (gv.) are indicated by vertical bars on the right of the tree. The robustness of the topology was estimated by bootstrap resampling of the neighbor-joining data. Bootstrap values greater than 50 are shown at branch points. Scale bar denotes 0.005 changes per nucleotide.
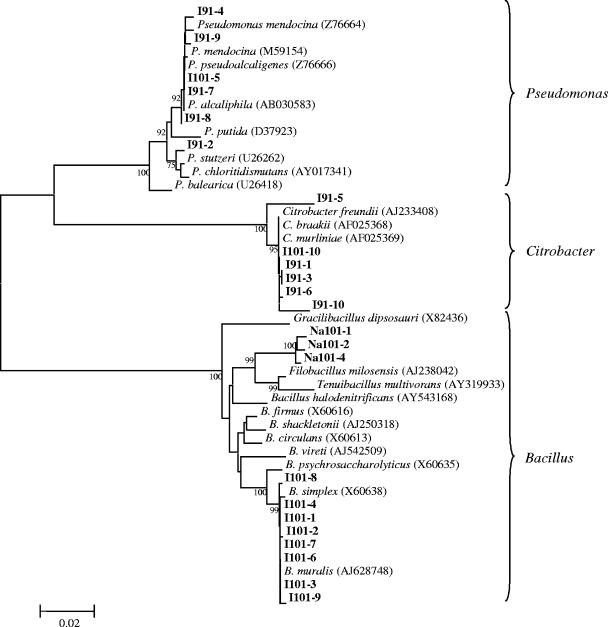
Culturable bacteria from both tar pits were represented by relatively few species. The 10% tryptic soy agar medium yielded five distinct isolates of Pseudomonas spp., eight isolates of Bacillus spp., and five isolates of Citrobacter spp. The 20% salt medium yielded three isolates of haloalkalophilic Bacillus spp. from Pit 101 (Fig. 4). All of the isolates obtained on the above media were further shown to grow on M9 medium with asphalt as the sole carbon source.
FIG. 4.
Phylogeny of cultured bacteria isolated from the Rancho La Brea Tar Pits. Bootstrap values greater than 75 are shown. Bold letters indicate isolates were obtained from Pit 91 or Pit 101. Scale bar denotes 0.02 changes per nucleotide.
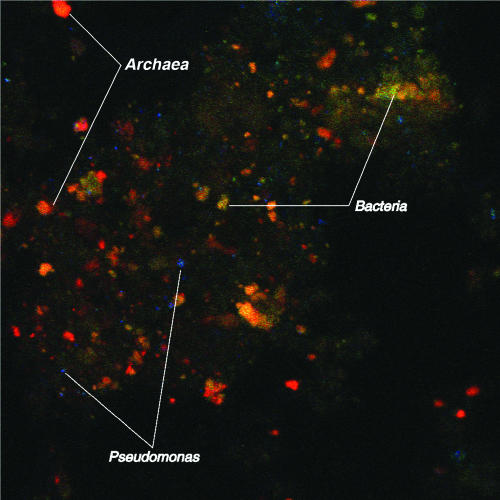
To study the physical distribution of Archaea and Bacteria in the asphalt from Pit 101, samples from the tar pit were microscopically examined using FISH with DNA probes targeted against Bacteria, Archaea, and Pseudomonas spp. (Fig. 5). Cells from each group were observed to occur in dense clusters of colonies along with randomly dispersed individual cells. The very close association of cells from different phylogenetic groups suggests that there may be consortia that cooperatively solubilize, degrade, and detoxify the complex substances contained in the asphalt. Such consortia also may be conducive for horizontal gene transfer of plasmids and DNA sequences that encode catabolic genes for use of petroleum hydrocarbons.
FIG. 5.
Microorganisms associated with heavy oil-soil as revealed by FISH using a combination of dyes that stain different organisms. Cells of the Archaea are stained red, the Bacteria are yellow, and Pseudomonas (a genus of Bacteria) is blue. The microbial colonies can be seen as clumps of different-color cells that are growing together, which suggests coexistence in diverse consortia.
Diversity analyses.
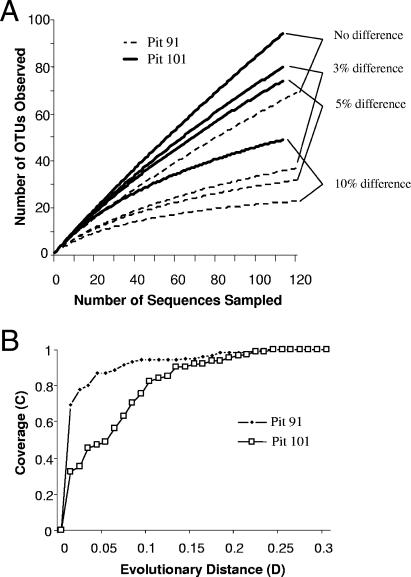
Rarefaction curves of Pit 91 and Pit 101 libraries showed significantly different numbers of operational taxonomic units (OTUs) at distance values of 0.03 corresponding to the species level, 0.05 at the genus level, and 0.1 at the family/class level (Fig. 6A). Only 37 OTUs were obtained from the Pit 91 bacterial library, while 80 OTUs were obtained from the Pit 101 bacterial library. Indices of diversity were higher for the Pit 101 library than for the Pit 91 library. The Shannon and Simpson index values were 2.8 and 0.139 for the Pit 91 library, compared with 4.2 and 0.014 for the Pit 101 library. Richness ACE (abundance-based coverage estimator) and Chao1 (13) values were 64 and 58 in Pit 91 and 268 and 242 in Pit 101 (Table 2). There was a significant difference between the rarefaction curves for the Pit 91 and Pit 101 libraries based on 0%, 3%, 5% and 10% differences (Fig. 6A). LIBSHUFF comparisons of each of the libraries indicated that two communities, Pit 91 and Pit 101, were significantly different (P < 0.001). At an evolutionary distance of 0.03, coverages of the libraries were 80% and 46% in Pit 91 and Pit 101, respectively (Fig. 6B).
FIG. 6.
Rarefaction curves of observed OTU richness using DOTUR (A) and coverage calculated using LIBSHUFF (B) from the two bacterial 16S rRNA gene libraries in Pit 91 and Pit 101 of Rancho La Brea.
TABLE 2.
Richness and diversity estimations in 16S rRNA gene libraries from Pit 91 and Pit 101 of La Brea Tar Pits
| Library | Richness
|
Diversity index value
|
Coverage | |||||
|---|---|---|---|---|---|---|---|---|
| No. of OTUs | ACE value | Boot value | Chao1 value | Jack value | Shannon | Simpson | ||
| Pit 91 | 37 | 64 | 46 | 58 | 58 | 2.8 | 0.139 | 0.8 |
| Pit 101 | 80 | 268 | 105 | 242 | 265 | 4.2 | 0.014 | 0.45 |
Bacterial dioxygenase sequences.
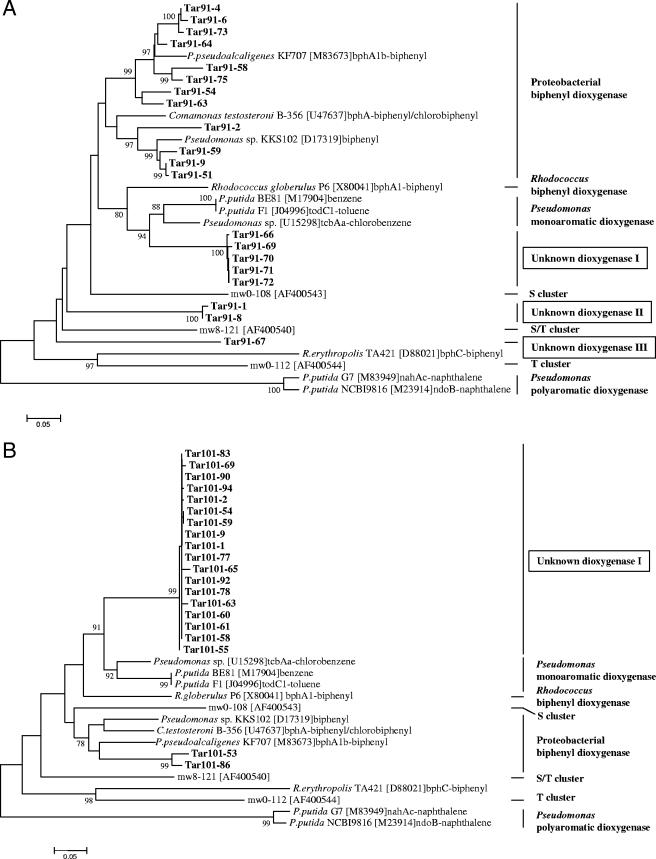
Taxonomic relationships for the dioxygenase sequences that were identified are shown in Fig. 7. The phylogenetic trees for these sequences included reference dioxygenases that were not found in the tar pits but that provide an indication of the similarities of the new sequences to those of previously described enzymes. Sequences from Pit 91 were predominantly associated with those of known proteobacterial biphenyl dioxygenases. This cluster of biphenyl dioxygenases was comprised of 12 sequences from Pit 91 and 2 sequences from Pit 101. Overall similarities to known biphenyl dioxygenases ranged from 79 to 95%. Detailed analysis revealed at least four subclusters within the biphenyl dioxygenase group. As with the microbial 16S rRNA genes, there were striking differences in the clusters of dioxygenases represented by the two sites. Ring-hydroxylating dioxygenases from Pit 101 were predominantly comprised of a new group of 18 closely related sequences that represent a novel, phylogenetically deep group of enzymes. This cluster was most closely associated with sequences encoding benzene and toluene dioxygenases but was sufficiently distant that it is not possible to infer whether these dioxygenases utilize these substances or instead transform other substrates. Two other new clusters that were discovered from sequences from Pit 91 were represented by one and two clones, respectively, and also appeared to represent deep branches of genes encoding unknown types of dioxygenases.
FIG. 7.
Phylogeny of aromatic ring hydroxylating dioxygenases identified from Rancho La Brea Tar Pits. (A) Pit 91; (B) Pit 101. Bootstrap values greater than 75 are shown. Bold letters denote clones sampled from the asphalt-soil mixtures. Sequences shown in regular typeface are known dioxygenases from GenBank used here for classification. Enzyme classes are indicated by vertical bars at the right of the trees. Scale bar indicates 0.05 changes per nucleotide.
DISCUSSION
Previous research has suggested that bacteria in deep subsurface oil reservoirs have inhabited those environments since the oil was formed (25). The origin of the bacteria in the natural asphalts at Rancho La Brea is unknown, but their presence could reflect recent entry from dust deposition on the surface, bacteria originating from the subsurface oil reservoir that seeped to the surface, or the progeny of soil bacteria that were embedded in the asphalt matrix as heavy oil seeped to the surface. Regardless of their origin, life in asphalt poses extreme conditions in which microbial growth is limited by the lack of air and water, the presence of highly recalcitrant carbon sources, and high concentrations of potentially toxic metals and chemicals. The selectivity of this environment would be expected to require specialized adaptations. Here 235 clones were described, many of which appear to comprise new genera and families of Proteobacteria (Fig. 2). An analysis of two different pits differing in their chemical properties revealed very little overlap in diversity (Fig. 6A), indicating that site-specific differences in salinity and pH strongly influence selection within the asphalt communities.
The relatively simple community structures and low complexity of Pits 91 and 101 compared to soil suggest that this environment is highly selective. The phylogenetic trees within the Proteobacteria revealed considerable breadth in taxa at the levels of family and order but also manifested branches containing discrete clusters of related sequences. The occurrence of many closely related species within the Gammaproteobacteria in both pits and the Archaea in Pit 101 suggested either an evolutionary radiation of species or an initial selection of closely related bacteria that share specific traits that enable them to survive in this environment.
Important questions arising from this research are the functional properties and adaptations of the taxa that inhabit the tar pits. It is very difficult to infer functionality at the level of phylum, and there is sparse information on many of the bacterial species that were found here. Among the predominant bacteria were 60 clones representing three unknown families from the Gammaproteobacteria in the order Chromatiales. This order has previously been characterized by two families, the Ectothiorhodospiraceae and Chromatiaceae, both of which comprise phototrophic anaerobic bacteria that produce sulfur from hydrogen sulfide gas (14, 15). The former produce granules of sulfur on the outside of their cells, while the latter produce internal sulfur granules. The three new families discovered here are not likely to derive energy from photosynthesis given that they live in complete darkness within the asphalt-soil matrix. Nonetheless, an ability to utilize electrons from hydrogen sulfide is consistent with life in the tar pits, where hydrogen sulfide and methane are produced during anaerobic metabolism of hydrocarbons contained in the asphalt.
Another important cluster in Pit 101 was classified within the Rubrobacteraceae. There were 16 closely related sequences from this family. The Rubrobacteraceae are in the phylum Actinobacteria and have been previously reported to occur in high-ionizing-radiation environments and in Australian desert soils (11). Rubrobacter strains have received considerable attention as being among the bacteria most resistant to ionizing radiation (7). Whether the bacteria from Pit 101 are radiation resistant is unknown, but it can be speculated that this trait could be of importance as an adaptation to protection from DNA damage in mixtures of heterocyclic aromatic hydrocarbons which are potent mutagens (48).
The occurrence of Pseudomonas stutzeri sequences, which included eight new genomovars, is consistent with prior reports on the distribution of this species. P. stutzeri is a well-known petroleum hydrocarbon degrader (8) and appears to be a cosmopolitan species that is readily isolated from various petroleum-contaminated environments (20, 29). Previously 17 genomovars have been described (20); this research has added another 8 candidate genomovars. As noted for several taxa in the communities from the tar pits, the existence of closely related sequences of P. stutzeri could be explained either by selection for bacteria that shared essential characteristics that allow them to survive in the asphalt or as an evolutionary radiation of ecotypes. With the exception of Pseudomonas spp., all of the identified taxa were dissimilar to those reported earlier for two studies investigating a high-temperature oil reservoir, which are the only other studies in the literature reporting a bacterial survey of a natural petroleum habitat using culture-independent methods (3, 26).
The discovery of many halophilic Archaea sequences in Pit 101 is particularly intriguing and provides an opportunity for future studies on the role of Archaea in petroleum hydrocarbon degradation. Based on studies conducted with enrichment cultures of bacterial petroleum degraders, biodegradation typically involves consortia in which the species composition of the degrader community is strongly influenced by salinity (18, 44). Whether this selection also occurs with Archaea is not known. The Archaea have been reported to occur in crude oil sludge but not in crude oil samples in oil stockpiles in Japan (40) or in petroleum reservoirs in California (26). Various Halobacteria isolated from soil and sediments that are capable of degrading hydrocarbons under saline conditions have been described. These include species tentatively identified as Halobacterium (2), Haloferax (51), and Haloarcula (40). A prior report on Archaea that can degrade aromatic hydrocarbons under anaerobic conditions using Fe(III) as an electron acceptor was published in 2001 (39). Although not yet cultivable, genes from these bacteria potentially could be used for improving culturable hydrocarbon degraders used for bioaugmentation or could serve as a source of catabolic genes that could be seeded into the environment on plasmids to facilitate adaptation of indigenous strains for degradation of recalcitrant hydrocarbons (38). It will also be important to determine whether the large unknown cluster of ring-hydroxylating dioxygenases identified in Pit 101 are carried by the Archaea, which is suggested by their cooccurrence in this particular site.
As expected from prior experience with environmental samples, relatively few isolates were obtained using culture-based methods. The culturable bacteria included strains of Pseudomonas spp., Citrobacter spp., and Bacillus spp. that were similar to known strains from oil-contaminated environmental samples (Table 2). The ability to culture these strains, especially isolates of Pseudomonas, provides opportunities for full genome analysis and examination of genetic exchange that may have occurred within this group of bacteria, as well as determination of plasmid-borne genes or catabolic pathways.
From an applied perspective, the most practical aspect of this research may be the confirmation of heavy oil seeps and natural asphalts as sources of novel genes for biodegradation of petroleum hydrocarbons. Here we focused on genes encoding aromatic-ring-hydroxylating dioxygenases that are important for degradation of BTEX and aromatic chemicals, such as polychlorinated biphenyls (PCBs) and polycyclic aromatic hydrocarbons, that are common environmental pollutants. The maintenance of genes encoding dioxygenases by bacteria that have undergone selection for life in an anaerobic system is somewhat of a paradox but is common for known oil-degrading bacteria. Previously described toluene-degrading bacteria placed under anaerobic conditions have been shown to first use dioxygenases to degrade toluene until all of the oxygen is consumed, after which the cells switch to a benzylsuccinate pathway that is coupled to denitrification (33). Among the sequences that were obtained were a large number that were similar to those encoding known biphenyl dioxygenases that function for degradation of PCBs. Subtle variations in key regions of these genes can lead to large differences in substrate range and specificity for different PCB congeners. In the future, enrichment culture methods may be used to identify still other enzymes that can target specific substrates.
Relatively little is known yet about anaerobic petroleum hydrocarbon degradation, although there has been steady progress in this field (1, 23, 30, 50). Tentative mechanisms that function for anaerobic degradation of alkylbenzenes and nonaromatic hydrocarbons are proposed to involve hydrolases and carboxylases and are coupled to sulfate or nitrate reduction (35, 46). In our survey, anaerobic bacteria that were identified included members of Gammaproteobacteria (60 clones of purple sulfur bacteria), Bacteroidetes, Clostridia, and Acidobacteria, none of which have been studied with respect to their possible contributions to anaerobic degradation of petroleum hydrocarbons. Homologous genes for the benzylsuccinate synthase (bss), which is the key enzyme for anaerobic toluene degradation, have been cloned (33) and may provide an entry point for future studies on the relevance of this pathway in the tar pit bacteria. In addition to direct catabolism of hydrocarbons, anaerobic bacteria may also contribute to hydrocarbon degradation by syntrophy, in which metabolically linked consortia function to consume fatty acids and degradation products of hydrocarbons to generate methane (46).
Looking toward future research on the Rancho La Brea microorganisms, the discovery of closely related bacterial clusters and genes encoding new dioxygenases is of particular interest for understanding the evolutionary biology of bacteria during adaptation to the extreme environment posed by life in asphalt. Detailed studies on efficient regulator-promoter pairs are now being conducted to understand and design improved operons for xenobiotic degrading bacteria (6). New approaches using high-throughput DNA sequencing will be the next step for obtaining insight into the function and diversity of oil-inhabiting bacteria and the catabolic pathways for degradation of petroleum hydrocarbons.
Acknowledgments
We gratefully acknowledge the assistance and review comments of John Harris and Christopher Shaw and the cooperation of the George C. Page Museum at Hancock Park, Rancho La Brea Tar Pits.
This project was supported in part by National Research Initiative Competitive Grant 2004-35107-15021 from the USDA Cooperative State Research, Education, and Extension Service.
Footnotes
Published ahead of print on 6 April 2007.
REFERENCES
- 1.Aitken, C. M., D. M. Jones, and S. R. Larter. 2004. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 431:291-294. [DOI] [PubMed] [Google Scholar]
- 2.Bertrand, J. C., M. Almallah, M. Acquaviva, and G. Mille. 1990. Biodegradation of hydrocarbons by an extremely halophilic archaebacterium. Lett. Appl. Microbiol. 11:260-263. [Google Scholar]
- 3.Bonch-Osmolovskaya, E. A., M. L. Miroshnichenko, A. V. Lebedinsky, N. A. Chernyh, T. N. Nazina, V. S. Ivoilov, S. S. Belyaev, E. S. Boulygina, Y. P. Lysov, A. N. Perov, A. D. Mirzabekov, H. Hippe, E. Stackebrandt, S. L'Haridon, and C. Jeanthon. 2003. Radioisotopic, culture-based, and oligonucleotide microchip analyses of thermophilic microbial communities in a continental high-temperature petroleum reservoir. Appl. Environ. Microbiol. 69:6143-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeSantis, T. Z., Jr., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz, E., and M. A. Prieto. 2000. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr. Opin. Biotechnol. 11:467-475. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira, A. C., M. F. Nobre, E. Moore, F. A. Rainey, J. R. Battista, and M. S. da Costa. 1999. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles 3:235-238. [DOI] [PubMed] [Google Scholar]
- 8.Ferrero, M., E. Llobet-Brossa, J. Lalucat, E. Garcia-Valdes, R. Rossello-Mora, and R. Bosch. 2002. Coexistence of two distinct copies of naphthalene degradation genes in Pseudomonas strains isolated from the western Mediterranean region. Appl. Environ. Microbiol. 68:957-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freemantle, M. 1999. What's that staff? Asphalt Chem. Eng. News 77:81. [Google Scholar]
- 10.Harris, J. M. 2001. Rancho La Brea: death trap and treasure trove. (Natural History Museum of Los Angeles County, Los Angeles). Terra 38:1-56. [Google Scholar]
- 11.Holmes, A. J., J. Bowyer, M. P. Holley, M. O'Donoghue, M. Montgomery, and M. R. Gillings. 2000. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 33:111-120. [DOI] [PubMed] [Google Scholar]
- 12.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imhoff, J. F. 2006. The Chromatiaceae, p. 846-873. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 6. Springer, New York, NY. [Google Scholar]
- 15.Imhoff, J. F. 2006. The family Ectothiorhodospiraceae, p. 874-886. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 6. Springer, New York, NY. [Google Scholar]
- 16.Janczewski, D. N., N. Yuhki, D. A. Gilbert, G. T. Jerfferson, and S. J. O'Brien. 1992. Molecular phylogenetic inference from saber toothed cat fossils of Rancho La Brea. Proc. Natl. Acad. Sci. USA 89:9769-9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurgens, G., F. O. Glockner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Munster. 2000. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 18.Kleinsteuber, S., V. Riis, I. Fetzer, H. Harms, and S. Muller. 2006. Population dynamics within a microbial consortium during growth on diesel fuel in saline environments. Appl. Environ. Microbiol. 72:3531-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 20.Lalucat, J., A. Bennasar, R. Bosch, E. Garcia-Valdes, and N. J. Palleroni. 2006. Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 70:510-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 22.L'Haridon, S., A. L. Reysenbacht, P. Glénat, D. Prieur, and C. Jeanthon. 1995. Hot subterranean biosphere in a continental oil reservoir. Nature 377:223-224. [PubMed] [Google Scholar]
- 23.Lovley, D. R. 2000. Anaerobic benzene degradation. Biodegradation 11:107-116. [DOI] [PubMed] [Google Scholar]
- 24.Loy, A., M. Horn, and M. Wagner. 2003. probeBase—an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magot, M., B. Ollivier, and B. K. C. Patel. 2000. Microbiology of petroleum reservoirs. Antonie Leeuwenhoek 77:103-116. [DOI] [PubMed] [Google Scholar]
- 26.Orphan, V. J., L. T. Taylor, D. Hafenbradl, and E. F. Delong. 2000. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66:700-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27-36. [DOI] [PubMed] [Google Scholar]
- 28.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godyoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 29.Roling, W. F. M., M. G. Milner, D. M. Jones, F. Fratepietro, R. P. J. Swannell, F. Daniel, and I. M. Head. 2004. Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl. Environ. Microbiol. 70:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rueter, P., R. Rabus, H. Wilkes, F. Aeckersberg, F. A. Rainey, H. W. Jannasch, and F. Widdel. 1994. Anaerobic oxidation of hydrocarbons in crude-oil by new types of sulfate-reducing bacteria. Nature 372:455-458. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinoda, Y., Y. Sakai, H. Uenishi, Y. Uchihashi, A. Hiraishi, H. Yukawa, H. Yurimoto, and N. Kato. 2004. Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1. Appl. Environ. Microbiol. 70:1385-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spormann, A. M., and F. Widdel. 2000. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 11:85-105. [DOI] [PubMed] [Google Scholar]
- 36.Taylor, P. M., and P. H. Janssen. 2005. Variations in the abundance and identity of class II aromatic ring-hydroxylating dioxygenase genes in groundwater at an aromatic hydrocarbon-contaminated site. Environ. Microbiol. 7:140-146. [DOI] [PubMed] [Google Scholar]
- 37.Taylor, P. M., J. M. Medd, L. Schoenborn, B. Hodgson, and P. H. Janssen. 2002. Detection of known and novel genes encoding aromatic ring-hydroxylating dioxygenases in soils and in aromatic hydrocarbon-degrading bacteria. FEMS Microbiol. Lett. 216:61-66. [DOI] [PubMed] [Google Scholar]
- 38.Top, E. M., D. Springael, and N. Boon. 2002. Catabolic mobile genetic elements and their potential use in bioaugmentation of polluted soils and waters. FEMS Microbiol. Ecol. 42:199-208. [DOI] [PubMed] [Google Scholar]
- 39.Tor, J. M., and D. R. Lovley. 2001. Anaerobic degradation of aromatic compounds coupled to Fe(III) reduction by Ferroglobus placidus. Environ. Microbiol. 3:281-287. [DOI] [PubMed] [Google Scholar]
- 40.Usami, R., T. Fukushima, T. Mizuki, Y. Yoshida, A. Inoue, and K. Horikoshi. 2005. Organic solvent tolerance of halophilic archaea, Haloarcula strains: effects of NaCl concentration on the tolerance and polar lipid composition. J. Biosci. Bioeng. 99:169-174. [DOI] [PubMed] [Google Scholar]
- 41.van der Meer, J. R. 1994. Genetic adaptation of bacteria to chlorinated aromatic-compounds. FEMS Microbiol. Rev. 15:239-249. [DOI] [PubMed] [Google Scholar]
- 42.van der Meer, J. R., W. M. Devos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Hamme, J. D., A. Singh, and O. P. Ward. 2003. Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 67:503-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward, D. M., and T. D. Brock. 1978. Hydrocarbon biodegradation in hypersaline environments. Appl. Environ. Microbiol. 35:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward, J. K., J. M. Harris, T. E. Cerling, A. Wiedenhoeft, M. J. Lott, M. D. Dearing, J. B. Coltrain, and J. R. Ehleringer. 2005. Carbon starvation in glacial trees recovered from the La Brea tar pits, southern California. Proc. Natl. Acad. Sci. USA 102:690-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259-276. [DOI] [PubMed] [Google Scholar]
- 47.Widmer, F., R. J. Seidler, P. M. Gillevet, L. S. Watrud, and G. D. Di Giovanni. 1998. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl. Environ. Microbiol. 64:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue, W. L., and D. Warshawsky. 2005. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol. Appl. Pharmacol. 206:73-93. [DOI] [PubMed] [Google Scholar]
- 49.Zarda, B., D. Hahn, A. Chatzinotas, W. Schonhuber, A. Neef, R. I. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185-192. [Google Scholar]
- 50.Zhang, C. L., and G. N. Bennett. 2005. Biodegradation of xenobiotics by anaerobic bacteria. Appl. Microbiol. Biotechnol. 67:600-618. [DOI] [PubMed] [Google Scholar]
- 51.Zvyagintseva, I. S., S. S. Belyaev, I. A. Borzenkov, N. A. Kostrikina, E. I. Milekhina, and M. V. Ivanov. 1995. Halophilic archaebacteria from the Kalamkass oil field. Microbiology 64:67-71. [PubMed] [Google Scholar]