Abstract
To elucidate the potential importance of deep-water viruses in controlling the meso- and bathypelagic picoplankton community, the abundance, decay rate, and diversity of the virioplankton community were determined in the meso- and bathypelagic water masses of the eastern part of the subtropical North Atlantic. Viral abundance averaged 1.4 × 106 ml−1 at around 100 m of depth and decreased only by a factor of 2 at 3,000 to 4,000 m of depth. In contrast, picoplankton abundance decreased by 1 order of magnitude to the Lower Deep Water (LDW; 3,500- to 5,000-m depth). The virus-to-picoplankton ratio increased from 9 at about 100 m of depth to 110 in the LDW. Mean viral decay rates were 3.5 × 10−3 h−1 between 900 m and 2,750 m and 1.1 × 10−3 h−1 at 4,000 m of depth, corresponding to viral turnover times of 11 and 39 days, respectively. Pulsed-field gel electrophoresis fingerprints obtained from the viral community between 2,400 m and 4,000 m of depth revealed a maximum of only four bands from 4,000 m of depth. Based on the high viral abundance and the low picoplankton production determined via leucine incorporation, we conclude that the viral production calculated from the viral decay is insufficient to maintain the high viral abundance in the deep North Atlantic. Rather, we propose that substantial allochthonous viral input or lysogenic or pseudolysogenic production is required to maintain the high viral abundance detected in the meso- and bathypelagic North Atlantic. Consequently, deep-water prokaryotes are apparently far less controlled in their abundance and taxon richness by lytic prokaryotic phages than the high viral abundance and the virus-to-picoplankton ratio would suggest.
The ecological role of viruses in marine surface waters has been studied intensively over the past 2 decades (11, 52). Bacteriophages, i.e., viruses infecting bacteria, not only may influence the biogeochemical cycling of organic matter and trace metals (13, 57) but might also regulate prokaryotic plankton diversity (49, 62). One of the preconditions for a viral control of prokaryotic plankton diversity is that prokaryotic viruses show some host specificity (3, 23). Typically, phages do not trespass the genus barrier. The general scarcity of isolated virus-host systems studied in this respect thus far, however, precludes any firm statements on the host specificity of marine viruses. Moreover, experiments with viral communities obtained from terrestrial and limnetic systems indicate that a substantial number of these viruses are capable of infecting marine coastal prokaryotic (essentially Bacteria) plankton communities (41). This finding is rather surprising in light of the generally assumed virus-host specificity and casts doubts on the common view of the host specificity of aquatic viruses.
Prokaryotic phage production depends on several factors, such as contact rate between virus and host, contact success (i.e., infection rate), the growth rate of the host, and the burst size (number of viruses liberated per infected cell) (26, 34, 52, 60). Generally, the grand average ratio between the abundance of viruses and prokaryotes is around 10 in surface waters, although major deviations have been reported as well (10, 52). Viral production is counterbalanced by viral losses via grazing by flagellates (12) and viral decay. In sunlit surface waters, virioplankton decay is commonly in the range of 1 to 2 days (17, 55, 58). The main factors determining viral decay in surface waters are UV radiation, adsorption to particles, temperature, capsid thickness, and the density of the packaged genome (33) and overall genome size.
There is considerable information available on the role of viruses on the microbiota in coastal systems and on the production and decay rates of these viruses. Much less is known for the oceanic euphotic layer, and our knowledge on the ecology of viruses in the meso- and bathypelagic realms of the ocean is rather limited (17, 31). In a study performed in Antarctic waters (15), viral abundance at 800 m of depth was around 1 × 106 ml−1. The same authors reported viral abundances of 1 × 107 ml−1 and 6 × 106 ml−1 at 70 and 200 m of depth, respectively, for the western Mediterranean Sea (14). Hara et al. (16) report a viral abundance of 4 × 105 ml−1 and a virus-to-picoplankton ratio (VPR) of about 5 for the bathypelagic Pacific. Similar VPRs have been reported for waters influenced by hydrothermal vents and the overlying water column (32, 64).
While picoplankton abundance decreases with depth from the euphotic to the bathypelagic layer by about 1 or 2 orders of magnitude to 104 cells ml−1, prokaryotic production decreases by 2 or 3 orders of magnitude from the open ocean's euphotic zone to about 1 μmol C m−3 day−1 in the bathypelagic realm (29, 39). The reported picoplankton generation time of 30 to 55 days for the northern part of the bathypelagic North Atlantic (from 65°N to 35°N) (39) is therefore substantially longer than the decay time of viruses of 1 to 2 days reported for surface oceanic waters. Hara et al. (16) concluded that bathypelagic picoplankton abundance is too low to sustain the detected viral abundance in deep waters and that the deep-water viral community might originate, at least partly, from allochthonous sources such as sedimenting particles. One uncertainty in the few reported viral and prokaryotic abundance data for the deep ocean is the decay of viruses and prokaryotes in glutaraldehyde- or formaldehyde-fixed samples. Ortmann and Suttle (32) reported picoplankton and viral abundances in nonhydrothermal regions of the deep Pacific 10-fold higher than those found in other deep-sea studies (16). These authors attributed the difference between viral and prokaryotic abundances found in their study and those found in other studies to differences between sample storage using fixatives and instant filtration of unfixed samples. Thus, at present there is uncertainty pertaining to the actual viral abundance in the deep ocean, the extent to which viruses in the bathypelagic waters are autochthonously produced, and whether viruses in the deep ocean play a role in controlling prokaryotic abundance similar to that shown for surface waters (10).
Given the at least 1-order-of-magnitude-lower picoplankton abundance, the approximately 2-orders-of-magnitude-lower picoplankton production (PPP) in the bathypelagic layers compared to surface waters, and the simultaneous reduction in prokaryotic taxon richness in the bathypelagic realm to only about 30 to 50% of that of the surface waters (19, 27), the contact rates of viruses with their potential hosts and their production should be greatly reduced in the dark ocean. It has been shown that under low host abundance, the lysogenic cycle of viruses prevails over the lytic cycle (51).
The aim of this study was to determine the viral abundance, decay rates, and diversity in the mesopelagic (200- to 1,000-m depth) and bathypelagic (1,000- to 5,000-m depth) waters of the North Atlantic. In concert with basic picoplankton parameters, they allow elucidating specific aspects of the ecology of viruses in the deep North Atlantic. Particularly, we aimed at answering the question of whether viruses in the deep ocean play a role in controlling prokaryotic biomass similar to that shown for viruses in the euphotic layer (61). We hypothesized that the picoplankton abundance and production are too low in the deep ocean to support a significant autochthonously produced viral biomass by lytic infection.
MATERIALS AND METHODS
Study site and sampling.
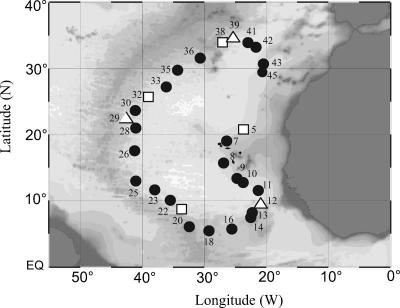
During the ARCHIMEDES-1 cruise (November to December 2005), 45 stations were occupied by R/V Pelagia in the eastern basin of the subtropical North Atlantic; at 30 stations viral parameters were measured (Fig. 1). Meso- and bathypelagic samples were taken from the main water masses down to 5,000 m of depth with 22 10-liter NOEX bottles mounted in a frame also holding sensors for conductivity, temperature, depth, chlorophyll fluorescence, and optical backscattering. Additionally, one sample per cast was taken from the lower euphotic layer at around 100 m of depth, termed hereafter the “surface-water” sample. Surface waters consisted of the more saline subtropical and the fresher tropical surface waters. The waters below the thermocline (at around 250 m of depth) were formed by the North Atlantic Central Water (NACW) and the South Atlantic Central Water (SACW) (Table 1). The intermediate waters with cores between 900 m and 1,100 m of depth included the more saline Mediterranean Sea Outflow Water (MSOW) and the less saline Antarctic Intermediate Water (AAIW) (Table 1). Deeper in the water column, Northeast Atlantic Deep Water (NEADW) is flowing southwards, and below the NEADW, Antarctic Bottom Water (AABW) is entering the eastern Atlantic basin via the Romanche Fracture Zone, forming the Lower Deep Water (LDW) together with the NEADW and the Labrador Sea Water (LSW). The specific water masses were identified based on their distinct temperature-salinity characteristics and inorganic nutrient signatures (Table 1). A detailed overview on the current pattern of the major water masses of the North Atlantic can be found elsewhere (50).
FIG. 1.
Map of stations occupied during the ARCHIMEDES-1 cruise in the subtropical North Atlantic Ocean where viral parameters were determined. Stations where additional samples were taken for viral decay and viral diversity are indicated by open squares and triangles, respectively. EQ, equator.
TABLE 1.
Physical and chemical characteristics of the core water masses sampled during ARCHIMEDES-1a
| Water sample | Depth (m) | Temp (°C) | Salinity | Oxygen (μmol/kg) | NO3 (μmol/liter) | PO4 (μmol/liter) | SiO4 (μmol/liter) |
|---|---|---|---|---|---|---|---|
| Surfaceb | 100 | 20.25 (2.02) | 36.806 (0.281) | 198.8 (13.6) | 1.9 (3.1) | 0.12 (0.19) | 1.11 (0.62) |
| SACW | 250-500 | 11.27 (0.54) | 35.138 (0.076) | 96.5 (21.0) | 27.7 (2.1) | 1.72 (0.10) | 11.45 (0.77) |
| NACW | 250-500 | 16.11 (0.95) | 36.246 (0.183) | 176.5 (27.8) | 9.1 (5.2) | 0.50 (0.32) | 3.04 (1.79) |
| MSOW | 700-1,500 | 8.84 (0.60) | 35.439 (0.132) | 161.1 (14.9) | 22.9 (4.0) | 1.42 (0.28) | 13.58 (3.12) |
| AAIW | 700-1,500 | 5.60 (0.45) | 34.739 (0.084) | 109.7 (9.2) | 34.8 (1.0) | 2.28 (0.08) | 26.20 (1.84) |
| NEADW | 2,500-3,500 | 2.73 (0.05) | 34.952 (0.008) | 245.7 (3.0) | 19.7 (0.4) | 1.03 (0.03) | 26.04 (1.16) |
| LSW | 1,500-3,000 | 3.38 (0.05) | 35.016 (0.041) | 254.4 (6.5) | 18.8 (0.5) | 1.21 (0.03) | 19.07 (0.41) |
| LDW | 3,500-5,000 | 2.03 (0.02) | 34.893 (0.003) | 232.4 (1.6) | 22.3 (0.6) | 1.47 (0.02) | 42.10 (1.35) |
| AABW | 5,000 | 1.87 (0.00) | 34.878 (0.000) | 232.7 (0.0) | 23.3 (0.0) | 1.55 (0.00) | 50.70 (0.00) |
The mean values for the physical and chemical parameters for each water mass are given (SD in parentheses). The mixed water masses are not given.
“Surface” denotes the bottom of the euphotic layer at around 100 m of depth.
Water from the NOEX bottles was collected in acid-rinsed (0.1 N HCl) polycarbonate flasks rinsed three times with sample water prior to the collection of samples for picoplankton and viral abundance and PPP. At selected stations, water was also collected for viral decay experiments and viral diversity (Table 2). At each station and depth, samples were also collected for additional parameters such as dissolved organic carbon, dissolved organic nitrogen, dissolved organic phosphorus, and inorganic nutrients (unpublished data). The samples were either immediately processed for specific measurements aboard the ship or stored for analyses back in the home lab (as described below).
TABLE 2.
Location and characteristics of the sampling stations in the North Atlantic where water was collected for viral decay experiments and viral diversitya
| Sampling purpose and station no. | Latitude (°N) | Longitude (°W) | Depth (m) | Water mass | PA (105 cells ml−1) | VA (105 ml−1) | PPP (cells ml−1 h−1) |
|---|---|---|---|---|---|---|---|
| Viral decay | |||||||
| 5 | 20.00 | 22.99 | 2,750 | NEADW/LSW | 0.2 | 11.9 (2.0) | 0.4 |
| 20 | 8.45 | 32.35 | 4,000 | LDW/AABW | 0.2 | 16.1 (7.3) | 0.8 |
| 32 | 25.66 | 38.38 | 900 | AAIW/MSOW | 0.5 | 13.3 (5.0) | 3.5 |
| 38 | 34.09 | 26.76 | 2,400 | NEADW/LSW | 0.2 | 12.1 (3.1) | 0.7 |
| Viral diversity | |||||||
| 12 | 10.90 | 20.64 | 4,000 | LDW/AABW | ND | ND | ND |
| 29 | 20.29 | 40.98 | 2,750 | NEADW/LSW | 0.1 | 8.4 (0.3) | ND |
| 39 | 34.51 | 26.28 | 2,400 | NEADW/LSW | 0.1 | 14.7 (4.0) | ND |
The picoplankton abundance (PA), viral abundance (VA), and picoplankton (PPP) are given. ND, not determined. Mean values are given for VA with SD in parentheses.
Viral decay experiments.
At specific stations (stations 5, 20, 32, and 38; Fig. 1), water was collected from different depths in the meso- and bathypelagic zones to determine deep-water viral decay rates (Table 2). Sample water was twice filtered through 0.2-μm polycarbonate filters (47-mm filter diameter; Millipore) into acid-rinsed suction flasks. Prior to the collection of this filtrate, the suction flasks were rinsed three times with 0.2-μm-filtered sample water. Incubations (each with 1 liter of water) were established in 1.2-liter screw-cap glass bottles in triplicate in the dark within 2 h after the water was collected from the rosette sampler. All the incubations were performed at in situ temperature in temperature-controlled incubators. Subsamples from these bottles were taken for viral abundance assessment and for checking the presence of picoplankton at variable time intervals (5 to 24 h) for up to 7 days. Picoplankton were essentially absent during the course of the incubations, as indicated by the cytograms obtained by flow cytometry (see below). From the decrease in viral abundance over time, the viral decay constant k was calculated by fitting a linear regression to the log of the viral abundance versus time (17). The slope of the line is the decay constant k h−1, and the reciprocal is the viral turnover time.
Picoplankton and viral abundance determined by flow cytometry.
Picoplankton and viruses collected from the different depth layers of the water column and from the viral decay experiments were enumerated using flow cytometry. Samples (2 ml) were fixed with glutaraldehyde (0.5% final concentration), shock frozen in liquid nitrogen for 5 min, and stored at −80°C. Picoplankton cells and viral particles were stained with SYBR green I and enumerated with a FACSCalibur flow cytometer (Becton Dickinson) as described previously within 3 months (7, 24). Briefly, the samples for enumeration of viruses were thawed immediately before analysis, diluted 10- to 100-fold in TE buffer (10 mM Tris, 1 mM EDTA, pH 8), and stained with SYBR green I (Molecular Probes Inc., Eugene, OR) added at a 200-fold dilution of the commercial stock at 80°C in the dark for 10 min. The thawed picoplankton samples were diluted 5- to 10-fold in TE buffer and stained with SYBR green I at room temperature in the dark for 15 min. Fluorescent microspheres (Molecular Probes Inc.) with a diameter of 1 μm were added to all samples as an internal standard. The discriminator was set on green fluorescence, and the samples were analyzed at a viral event rate of between 100 s−1 and 1,000 s−1 for 1 min.
PPP determined by [3H]leucine incorporation.
Bulk PPP was measured by incubating 10- to 40-ml samples in duplicate and one formaldehyde-killed blank (2% final concentration) with 10 nM [3H]leucine (final concentration, 157 Ci mmol−1 specific activity; Amersham) in the dark at in situ temperature for 4 h (42). Thereafter, the incubation was terminated by adding formaldehyde (2% final concentration) to the duplicate samples and filtering the samples through 0.2-μm polycarbonate filters (25-mm filter diameter; Millipore) supported by a Millipore HAWP filter. Subsequently, the filters were rinsed with 5% ice-cold trichloroacetic acid, dried, and placed in scintillation vials. Scintillation cocktail (8 ml Canberra-Packard Filter Count) was added, and after 18 h counting took place in a liquid scintillation counter (LKB Wallac model 1212). The disintegrations per minute (dpm) of the formaldehyde-fixed blank was subtracted from the mean dpm of the respective samples, and the resulting dpm converted into leucine incorporation rates. The leucine incorporation was converted into cells produced per mol leucine incorporated by use of the conversion factor of 0.07 × 1018 cells produced per mol leucine incorporated (40).
Preparation of viral concentrates and assessment of viral diversity by pulsed-field gel electrophoresis (PFGE).
Eighty liters of seawater were collected at three stations (Table 2) and filtered first through 0.8-μm-pore-size polycarbonate filters (Isopore ATTP, 142-mm diameter; Millipore) by use of a stainless steel filter holder (Sartorius) and an air pressure pump. Subsequently, the filtrate was concentrated to a final volume of approximately 600 ml using tangential-flow filtration (Vivaflow 200 cartridge, 0.22-μm pore size; Vivascience, Lincoln, United Kingdom). The obtained 0.22-μm filtrate was further processed using a 30-kDa ultrafiltration cartridge (Vivascience, Lincoln, United Kingdom) to obtain a virus-free ultrafiltrate and a virus concentrate (60). Both filtration devices were operated using peristaltic pumps (Masterflex) at a maximum pressure of 2 bars. The entire filtration procedure was performed at in situ temperature within 1 h after sample collection. The viral concentrate (retentate of the 30-kDa filtration) was kept at −20°C until further processing in the home lab.
Upon return to the home lab, the virus concentrate was pelleted by ultracentrifugation at 141,000 × g in a fixed-angle rotor (TFT 55.38 tubes, Centrikon T-1080 rotor; Kontron Instruments) at 8°C for 2 h. Pellets were resuspended and incubated overnight at 4°C in SM buffer (0.1 M NaCl, 8 mM MgSO4·7H2O, 50 mM Tris-HCl, and 0.005% [wt/vol] glycerol) (63). Equal volumes of the viral concentrate were mixed with melted (50°C) 1.5% InCert agarose (Cambrex Bioscience, Rockland, ME) and loaded into plugs. The plugs were digested overnight at 30°C in a lysis buffer (250 mM EDTA, 1% sodium dodecyl sulfate [vol/vol], 1 mg ml−1 proteinase K; Sigma-Aldrich). Then, the plugs were washed three times in TE 10:1 buffer (10 mM Tris-1 mM EDTA, pH 8.0) for 30 min and stored in TE 20:50 buffer (20 mM Tris-50 mM EDTA, pH 8.0) at 4°C until loading onto the gel. The molecular weight markers, a lambda ladder, and a 5-kb ladder (Bio-Rad, Richmond, CA) were also loaded into plugs.
Plugged samples and markers were placed into wells of a 1% SeaKem GTG agarose gel (Cambrex Bioscience, Rockland, ME) prepared in 1× TBE gel buffer (90 mM Tris-borate, 1 mM EDTA, pH 8.0) with an overlay of melted 1% agarose. PFGE was performed with a contour-clamped homogenous electric field DR-II cell (Bio-Rad, Richmond, CA) at a pulse ramp of 1 to 6 s and 6 V/cm set at 14°C for 20 h. After electrophoresis, the gels were stained with SYBR green I (Molecular Probes, Eugene, OR) for 60 min and destained in Milli-Q water for 10 min (gradient A10; Millipore). Then, the gel was digitally scanned for fluorescence using a FluorS imager (Bio-Rad).
Each of the PFGE fingerprints of the virioplankton community was normalized against the background fluorescence to quantify the signal intensity of each band. By comparing the bands with the banding pattern of a size standard with a known amount of DNA, the relative abundance of each of the bands with a different genome size was calculated. The staining intensity and genome size were related to the number of viruses loaded into each plug (35).
Statistical analysis.
Analysis of variance (Kruskal-Wallis ANOVA on ranks) in combination with Dunn's method was used to evaluate differences within and between water masses or depths. The Mann-Whitney rank sum test and the Wilcoxon rank sum test were used for testing differences between two independent and paired samples, respectively. For comparing more than two dependent samples, the Kruskal-Wallis test was applied. The Pearson correlation coefficient was calculated to test relationships between parameters. Correlation coefficients with P values of <0.05 were assumed to be statistically significant.
RESULTS
Abundance of picoplankton and viruses and picoplankton activity in the meso- and bathypelagic water masses of the North Atlantic.
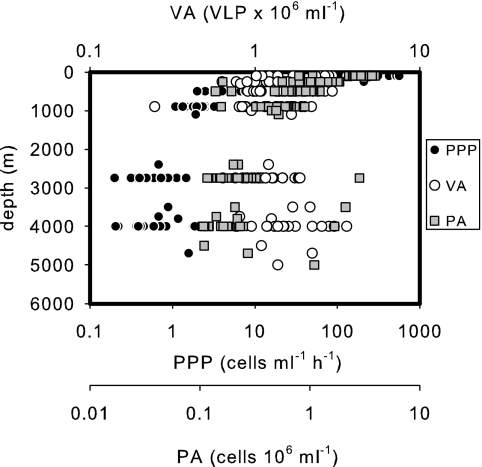
Generally, picoplankton abundance decreased exponentially with depth from 2.9 × 105 cells ml−1 at about 100 m of depth to 0.2 × 105 cells ml−1 in the LDW (Table 3), as described by the exponential function PA = 0.14−0.0006d (R2 = 0.70; n = 161), where PA is picoplankton abundance (×105 ml−1) and d is depth in m. The levels of abundance of picoplankton were significantly different between the different depth layers (Kruskal-Wallis ANOVA on ranks; P = 0.001), except for the two deepest water masses (below 2,750 m of depth) (Table 3). In contrast to picoplankton abundance, levels of viral abundance were not significantly different between the different depth layers and water masses (Table 3) (Kruskal-Wallis ANOVA on ranks; P = 0.056). Among all the microbial parameters measured, PPP exhibited the most pronounced decrease with depth (Table 3) (Kruskal-Wallis ANOVA on ranks, P < 0.001), following the function PPP = 0.0003−0.0011d (R2 = 0.58; n = 161), where PPP is in ml−1 h−1 and d is depth in m. This more pronounced decrease of PPP with depth than of abundance is reflected by the increase in picoplankton turnover time (PA/PPP) ranging from 98 days at the 100-m depth horizon to 4.4 years in the NEADW/LSW (Table 3). Since the decrease in picoplankton abundance with depth was more pronounced than that of viral abundance, the VPR increased from the 100-m layer to the LDW from 9 to 110 (Table 3).
TABLE 3.
Average abundances of picoplankton and viruses, VPR, PPP, and picoplankton turnover time in the different water massesa
| Water sample | PA (105 ml−1)b | VA (105 ml−1)c | VPRd | PPP (cells ml−1 h−1)e | PPTT (days)f |
|---|---|---|---|---|---|
| Surface | 2.9 (0.5) | 26.0 (7.4) | 9 (1) | 123.6 (73.0) | 98 |
| NACW/SACW | 1.5 (0.5) | 18.4 (5.0) | 12 (5) | 33.2 (21.0) | 272 |
| AAIW/MSOW | 0.5 (0.2) | 13.3 (5.0) | 27 (12) | 3.4 (3.2) | 604 |
| NEADW/LSW | 0.2 (0.0) | 13.4 (4.0) | 67 (22) | 0.5 (0.2) | 1620 |
| LDW | 0.2 (0.0) | 16.1 (7.3) | 110 (77) | 0.8 (0.6) | 1091 |
SD shown in parentheses.
PA, picoplankton abundance.
VA, viral abundance.
VPR, virus-to-picoplankton ratio.
PPP, picoplankton production.
PPTT, picoplankton turnover time.
Lateral variability in picoplankton and viral abundance and picoplankton activity.
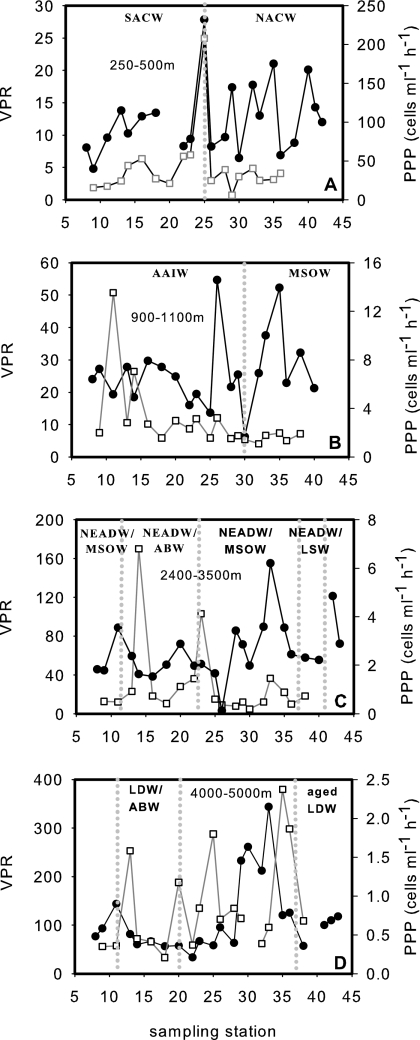
Different water masses were encountered in a given depth layer, depending on the area of sampling (northern versus southern stations, eastern versus western stations; Fig. 1 and Table 1). The upper mesopelagic zone (250- to 500-m depth) was dominated at the eastern stations by SACW and at the western stations by NACW, as indicated by their characteristic physicochemical variables (Table 1). The PPPs averaged 38.3 ± 19.2 cells ml−1 h−1 and 28.1 ± 10.9 cells ml−1 h−1 in the SACW and NACW, respectively, excluding the single data point at the boundary of both water masses (Fig. 2A). The VPRs in the SACW and NACW were on average 11 ± 3 and 13 ± 6, respectively. Neither PPPs nor VPRs was significantly different between these two water masses. The highest values for both variables were obtained in the mixing zone of the two water masses at station 25 (Fig. 2A).
FIG. 2.
Lateral variation in the VPRs (full circles) and PPPs (open squares) in different water masses following isopycnals. The dotted vertical lines separate the different water masses encountered. (A) 250- to 500-m depth layer; (B) 900- to 1,100-m layer; (C) 2,400- to 3,500-m layer; (D) 4,000- to 5,000-m layer. For main water mass hydrological characteristics, see Table 1. The absence of an indicated water mass represents a mixture between water masses not clearly identifiable.
The lower part of the mesopelagic zone was composed of AAIW at the eastern stations and modified MSOW at the western stations (Table 1). The mean PPP was significantly higher (Mann-Whitney test; P = 0.015) in the AAIW (3.5 ± 3.2 cells ml−1 h−1) than in the MSOW (1.6 ± 0.3 cells ml−1 h−1) (Fig. 2B). In contrast, VPRs were not significantly different between AAIW and MSOW, averaging 25 ± 11 and 28 ± 11, respectively (Fig. 2B).
In the depth layer between 2,400 and 3,500 m, NEADW was the main water mass; however, it was mixed with other water masses (Fig. 2C). Neither PPPs nor VPRs were significantly different between the different mixed water masses encountered. The mean PPP and VPR were 1.8 ± 2.5 cells ml−1 h−1 and 58 ± 19 in the NEADW/AABW and 1.06 ± 0.24 cells ml−1h−1 and 66 ± 42 in the NEADW/MSOW, respectively (Fig. 2C). LSW mixed with NEADW was found at 2,400 to 3,500 m of depth from stations 38 to 41 (Fig. 2C). The PPP in the NEADW/LSW was 0.7 cells ml−1 h−1, and VPR averaged 77 ± 32.
In the water column underneath the NEADW-dominated region, LDW was mixed with AABW. No significant difference was found in PPP between LDW/AABW (0.7 ± 0.5 cells ml−1 h−1) and aged LDW (Fig. 2D). The VPR was lower in LDW/AABW (78 ± 33) than in aged LDW (96 ± 27), albeit not significantly (Fig. 2D).
Pooling all the data from the different water masses, only picoplankton abundance and production were significantly related (R2 = 0.51), while no direct relations could be established for the other parameters (data not shown).
Viral decay in the meso- and bathypelagic North Atlantic.
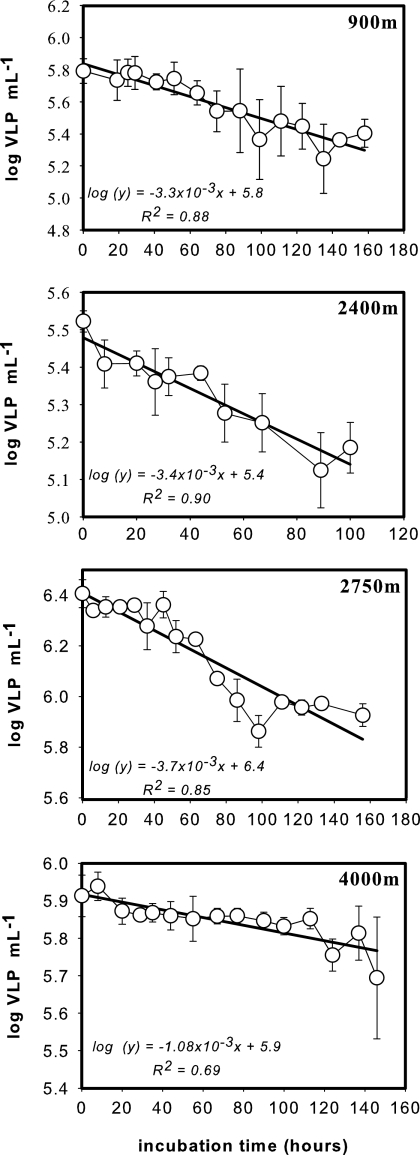
Viral decay in the dark realm of the North Atlantic was determined at four stations in different water masses representing a depth range between 900 m and 4,000 m (Table 2). The exponential decrease in viral abundance over time for the four different depth layers is shown in Fig. 3. The decay rates k ranged from 3.3 × 10−3 to 3.7 × 10−3 h−1 between 900 m and 2,750 m of depth, and k was 1.1 × 10−3 h−1 at 4,000 m of depth (LDW/AABW) (Fig. 3). As indicated by slope comparison, the obtained k values were significantly different between 2,400-m, 2,750-m, and 4,000-m depths (t test; P = 0.0029) but not significantly different between 900 m and 2,400 m of depth (t test; P = 0.65).
FIG. 3.
Decrease in the abundance of virus-like particles (VLP, log transformed) over time in 0.2-μm-filtered seawater collected at four stations and depths (see Table 2 for locations of stations and basic characteristics of collected water). Symbols represent the means of the viral abundance of triplicate incubations, and vertical lines give the standard deviations (SD). Decay rates (k) were calculated from the slope of the regression line.
When a constant viral abundance was assumed and the obtained decay rates for the different depth layers were used, the turnover time (1/k) of the viral community at 4,000 m of depth was about 930 h (39 days) and ranged from 270 to 295 h for the other deep-water masses.
Virioplankton diversity determined by PFGE.
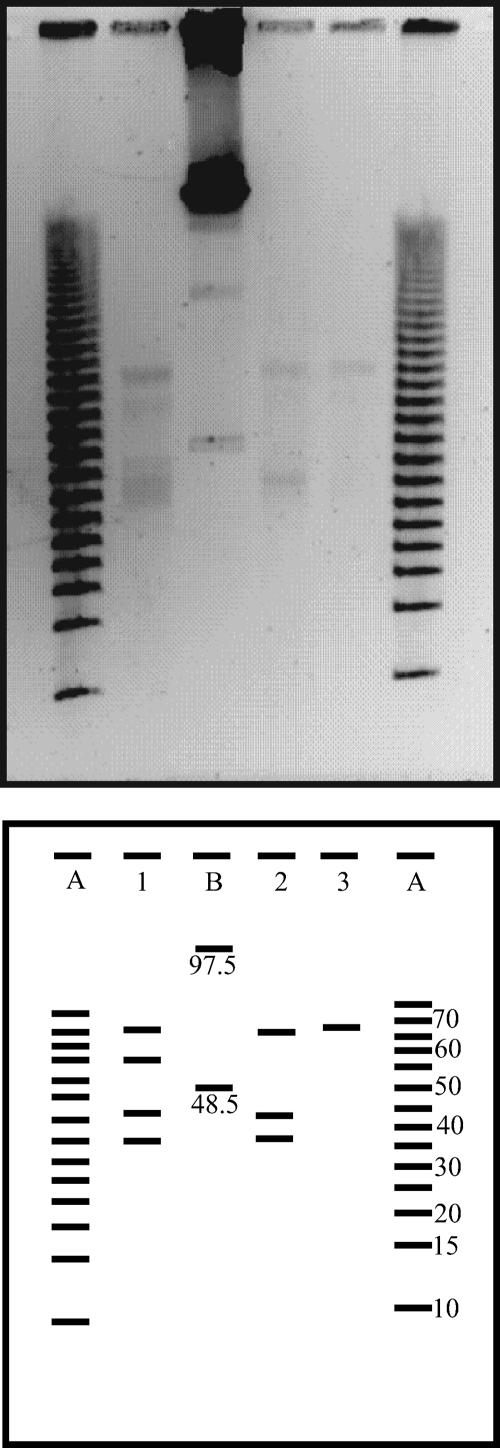
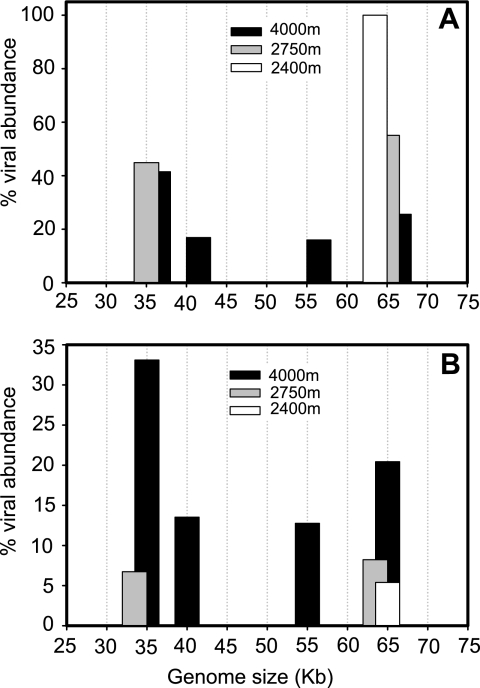
PFGE fingerprints of the virioplankton community of the deep waters of the North Atlantic were obtained at three stations from depths between 2,400 m and 4,000 m (Table 2). The resulting bands of the viral genome size ranged from about 35 kb to 65 kb (Fig. 4). The highest number of bands was detected at 4,000 m of depth in the water mass consisting of LDW/AABW. The viral genome of 65 kb was present in all three depth layers.
FIG. 4.
PFGE fingerprints of the virioplankton community collected from the bathypelagic waters of the subtropical North Atlantic and the corresponding computer-generated banding patterns. Lanes A and B are molecular size markers (sizes are shown in kb). Lane 1 denotes the sample from 4,000 m, lane 2 from 2,750 m, and lane 3 from 2,400 m of depth. See Table 2 for sampling locations and basic characteristics of collected water.
At 4,000 m of depth, the most abundant viral genome was 35 kb, comprising 41% of the total viral genome abundance (Fig. 5A) and 33% of the total viral genome abundance of all deep-water samples pooled (Fig. 5B). The second most abundant viral genome at 4,000 m of depth was 65 kb, representing 26% of the total viral genome abundance in this sample (Fig. 5A) and 20% of the total viral genome abundance of all deep-water samples pooled (Fig. 5B). The viral genome sizes of 40 kb and 55 kb were found only at 4,000 m of depth. At 2,750 m of depth, the 35-kb and 65-kb genome sizes comprised 45% and 55% of the total viral genome abundance in this sample, respectively (Fig. 5A), and 7% and 8%, respectively, of the total viral genome abundance in all deep-water samples pooled (Fig. 5B). At 2,400 m of depth, the unique band of 65 kb comprised 5% of the total viral genome abundance of all samples pooled (Fig. 5B).
FIG. 5.
Distribution of the relative abundances of viral genome sizes in bathypelagic waters collected at three different sampling locations and depths in the subtropical North Atlantic (see Table 2 for locations of stations and basic characteristics of collected waters). Percents distribution per station (A) and among all the bands detected from all the three sites (B).
DISCUSSION
Depth distribution of viruses and picoplankton.
The surprisingly high viral abundance found in the meso- and bathypelagic marine waters of the North Atlantic confirms previous findings of high viral abundance in other parts of the meso- and bathypelagic global ocean (16, 30, 32, 45, 51, 64). While picoplankton abundance and productivity decreased by at least 1 order of magnitude with depth, viral abundance varied by only a factor of 2 with depth (Table 3).
Hara et al. (16) found high viral abundance in the meso- and bathypelagic subarctic Pacific Ocean at 500-m and 2,000-m depths. These authors report a VPR of 1.6 at 5,008 m of depth in the subtropical Pacific, while the VPR in the deep North Atlantic increased with depth from around 100 m of depth to the LDW (3,500- to 5,000-m depth) from 9 to 110 (Table 3). On average, the viral abundance we measured in the deep North Atlantic is about 1 order of magnitude higher than that reported for the subtropical Pacific (16); however, it is in the range reported for the anoxic Cariaço basin (45). Guixa-Boixereu et al. (15) reported 1.1 × 106 to 2.8 × 106 viruses ml−1 at 800 m of depth in Antarctic waters and VPRs ranging from 11 to 25. The deep-water viral turnover times calculated from the viral decay rates (Fig. 3) range between 11 and 39 days (see Table 5) and are therefore substantially lower than the picoplankton turnover times, which range from about 600 to 1,090 days over the same depth range (Table 3). It is noteworthy that picoplankton leucine incorporation and picoplankton turnover times in the meso- and bathypelagic waters at our study site in the southern region of the eastern North Atlantic are 1 or 2 orders of magnitude lower than those in waters between 65°N and 35°N as reported by Reinthaler et al. (39), reflecting the aging of the main water masses from north to the southern part of the North Atlantic.
TABLE 5.
Estimates of viral production, contact rate, and burst size for the different water masses at indicated cell viability levels
| Water mass with indicated % PA VCa | VA (105 ml−1)b | VC (105 ml−1) | VP (103 VLP ml−1 h−1)c | PPP (cells ml−1 h−1) | VTT (days)d | CR (102 collisions ml−1 h−1)e | BS (viruses cell−1)f |
|---|---|---|---|---|---|---|---|
| 100% PA VC | |||||||
| AAIW/MSOW | 13.3 | 0.53 | 4.5 | 3.45 | 12 | 30 | 2 |
| NEADW/LSW | 12.1 | 0.22 | 4.1 | 0.68 | 12 | 11 | 4 |
| NEADW/LSW | 11.9 | 0.20 | 4.4 | 0.40 | 11 | 10 | 4 |
| LDW | 16.1 | 0.16 | 1.7 | 0.84 | 39 | 11 | 2 |
| 16% PA VC | |||||||
| AAIW/MSOW | 13.3 | 0.08 | 4.5 | 3.45 | 12 | 5 | 9 |
| NEADW/LSW | 12.1 | 0.03 | 4.1 | 0.68 | 12 | 2 | 22 |
| NEADW/LSW | 11.9 | 0.03 | 4.4 | 0.40 | 11 | 2 | 27 |
| LDW | 16.1 | 0.02 | 1.7 | 0.84 | 39 | 2 | 10 |
For details on water masses at indicated viable cell (VC) levels in picoplankton abundance (PA), see text.
VA, viral abundance.
VP, viral production; VLP, virus-like particles. VP was calculated by dividing VA by VTT.
VTT, viral turnover time. VTT was calculated as the reciprocal of the viral decay constant according to the method of Suttle and Chen (44).
CR, contact rate.
BS, burst size. BS was calculated by dividing VP by CR (43).
Decay rates, diversity, and source of bathypelagic virioplankton.
Viruses are thought to be generally more resistant to pressure, temperature, and osmotic stress than most prokaryotes (33). The decay rates reported for surface waters vary over 3 orders of magnitude, and the different approaches used for calculation make direct comparison between different studies difficult (Table 4). No deep-water viral decay rates have been reported prior to this study (Table 4). In deep waters, viral decay is apparently substantially slower than in surface waters (Table 4). Wall adsorption of viruses most likely did not bias our experiments, since surveys showed that this factor is negligible (6). It is known that the decay rate of viruses increases with temperature (8). Thus, the low temperature of the deep waters (2 to 8°C) should result in decay rates lower than those seen for warmer surface waters. The temperature difference between the meso- and bathypelagic waters in our study area is around 6°C, and the decay rate in mesopelagic waters is 3.3 × 10−3 h−1, and that at a 4,000-m depth is 1.08 × 10−3 h−1, potentially reflecting the temperature difference between the two depth horizons (Fig. 3). This decrease in viral decay rates with depth might contribute to the remarkable constant viral abundance below 2,000 m of depth (Fig. 6).
TABLE 4.
Compilation of viral decay rates and viral turnover times obtained in different aquatic environments
| Environment | Depth (m) | Avg viral decay (k × 10−3) (h−1) | VTTa (days) | R2 | No. of time points | Incubation time (days) | In situ temp (°C) | Source or reference | Treatmentb |
|---|---|---|---|---|---|---|---|---|---|
| North Atlantic | 900 | 3.3 (0.82) | 12.3 | 0.88 | 14 | 6 | 7.7 | This study | Log linear function; dark incubated; 0.02-μm FSW |
| North Atlantic | 2,400 | 3.4 (0.98) | 12.3 | 0.9 | 10 | 4 | 3.2 | This study | |
| North Atlantic | 2,750 | 3.7 (0.17) | 11.3 | 0.85 | 16 | 7 | 3.0 | This study | |
| North Atlantic | 4,000 | 1.1 (0.19) | 38.7 | 0.69 | 15 | 6 | 2.4 | This study | |
| Raunefjorden, Norway | Surface | 280 (40-500) | 0.15 | 35 | 14.0 | 5 | Log linear function; dark incubated; USW | ||
| Raunefjorden, Norway | Surface | 330 (20-890) | 0.13 | >5 | 19 | 15.0 | 4 | Log linear function; dark incubated; USW | |
| Bergen Harbor | Surface | 300 (10) | 0.14 | 4 | 1 | 12.0 | 17 | Log linear function; KCN | |
| Bergen | 1.5 | 1,100 (200) | 0.04 | 3 | 1 | 13.0 | 17 | Log linear function; KCN | |
| Coastal waters, Texas | Surface | 8-23 | 1.8-5.2 | 0.9-1 | 4-5 | 4 | 44 | Dark incubated; USW | |
| Coastal waters, Texas | Surface | 0-11 | 3.80 | 44 | Dark incubated; 0.02-μm FSW | ||||
| Coastal, Santa Monica | Surface | 13-18 | 2.3-3.2 | 12 | <1 | 31 | Dark incubated; USW | ||
| North Sea | Surface | 3-17 | 2.4-13.8 | 0.6-1 | 6 | 1 | 60 | Dark incubated; USW | |
| North Adriatic | Surface | 110-510 | 0.08-0.38 | 4 | 0.5 | 2 | Dark incubated; 0.02-μm FSW | ||
| North Adriatic | Surface | 450 | 0.09 | 4 | 0.5 | 2 | Dark incubated; 0.02-μm FSW | ||
| North Adriatic | Surface | 4.2-96 | 0.4-9.9 | 53 | Plaque assay | ||||
| North Adriatic | Surface | 300-1,100 | 0.04-0.14 | 54 | Direct counting method | ||||
| Bay of Århus | Surface | 1,100 | 0.04 | 6 | Log linear function; KCN | ||||
| Bacterial strain | Surface | 4.2 | 9.90 | 65 | Plaque assay | ||||
| Gulf of Mexico and North Sea | Surface | 500-8,300 | 0.01-0.08 | 43 | Light incubations, USW or 0.02-μm FSW plaque assay | ||||
| Gulf of Mexico | Surface | 0.7-0.85 | 0.05-0.06 | 58 | Dark-light incubations; USW or 0.02-μm FSW | ||||
| Mediterranean Sea | Surface | 20 | 0.002 | 9 | 2 | 14 | Dark-light incubations; KCN | ||
| Antarctic waters | Surface | 2-300 | 0.13-20 | 0.7-1 | 5 | 2 | 15 | 0.8- or 50-μm FSW; KCN | |
| Seawater, coliphages | Surface | 4-96 | <0.01-0.01 | 20 | |||||
| Danube backwater system | Surface | 20-150 | 0.27-2 | 10 | 1 | 5-25 | 25 | Log function | |
| Oxbow lake, Alte Danube | Surface | 0-130 | 0.32 | 18 | 0.37 | 9 | Different mathematical approaches; KCN |
VTT, viral turnover time.
0.2-μm FSW, seawater filtered with a 0.2-μm-pore-size filter; USW, unfiltered seawater; KCN, natural seawater with added potassium cyanide.
FIG. 6.
Distributions of picoplankton abundance (PA; gray squares), PPP (full circles), and viral abundance (VA; open circles) throughout the water columns of all the stations occupied in the subtropical North Atlantic.
The abundance of potential prokaryotic hosts in the lower mesobathypelagic layer and the entire bathypelagic realm is close to the reported threshold of 104 cells ml−1 required to sustain a prokaryotic phage community (28). However, infection and replication of bacteriophages have been reported at host abundances and phage concentrations as low as 102 cells and viruses ml−1 (22). From the viral abundance and the decay rate, viral production and turnover time can be estimated assuming that viral abundance remains constant (44) (Table 5). In the deep-water masses of the North Atlantic, viral production ranges from 1.7 × 103 to 4.5 × 103 viruses ml−1 h−1. These viral production estimates result in a low burst size ranging from 2 to 4 (Table 5, 100% PA VC). In contrast, Weinbauer et al. (51) found increasing burst size with depth between 100 m and 2,000 m in the Mediterranean Sea by enumerating the number of virus-like particles in visibly infected bacteria by use of transmission electron microscopy. These authors report burst sizes ranging from 31 to 37 for the meso- and bathypelagic waters of the Mediterranean.
Not all of the detected picoplankton cells are available for viral infection, since a certain fraction is comprised of inactive or dead cells. Recently, it has been shown that about 70% of all DAPI (4′,6′diamidino-2-phenylindole)-stained cells in the deep ocean can be detected by the highly sensitive catalyzed reporter deposition fluorescence in situ hybridization method (CARD-FISH), indicating that these CARD-FISH-stainable cells contain RNA and can therefore be considered active (48). A more conservative estimate of active prokaryotic cells is obtained by microautoradiography using radiolabeled substrates such as leucine (18). By microautoradiography in combination with CARD-FISH (MICRO-CARD-FISH), about 16 to 20% of the heterotrophic prokaryotic community of the meso- and bathypelagic North Atlantic was taking up leucine and can be considered as metabolically active (46, 47). If we use this MICRO-CARD-FISH-based conservative estimate of active deep-water prokaryotes potentially available for viral infection and replication, the burst size estimates would increase to 9 to 27 (Table 5, 16% PA VC), values in the lower range commonly reported for marine environments in general (34) and for the deep waters of the Mediterranean by Weinbauer et al. (51).
However, the low prokaryotic production and estimated viral production based on the viral decay rates and the calculated burst size cannot explain the high viral abundance found in the deep North Atlantic. Assuming that every contact of a virus results in successful infection and that 100% of all the prokaryotic cells are infected, the maximum potential viral production can be calculated. Taking the mean prokaryotic production and the burst size calculated from the abundance of leucine-active cells based on microautoradiography in the AAIW/MSOW as an example (given in Table 5, 16% PA VC) and assuming that all newly produced prokaryotes are infected, viral production amounts to 740 viruses ml−1 day−1 and a viral turnover time (viral abundance/viral production) of 1,800 days. This estimated viral turnover time is 2 orders of magnitude higher than the viral turnover time based on the viral decay time of 12 days (Table 5, 16% PA VC). This comparison indicates an apparent mismatch between the viral production deduced from decay rate measurements and viral production estimates based on prokaryotic activity.
Several factors might be responsible for this discrepancy. The obtained viral decay rates might be too low. This is unlikely, however, since the filtration steps involved in preparing the incubations and the possible decompression effects would more likely result in elevated decay rather than in lower decay rates. The calculated burst size might be grossly underestimated due to overestimating the infection rate or because of a high viral production or contact rate calculated. The viral production calculation involves the viral decay rate and the viral abundance. It is unlikely that viral abundance determined by flow cytometry is overestimated, because enumeration of virus-like particles by epifluorescence microscopy was in good agreement with flow cytometry-based counts (R2 = 0.77). The contact rate calculations are based on transport theory, which is fairly well understood (28).
One possibility to resolve this discrepancy would be substantial allochthonous input of viruses from the overlying water column via sedimenting marine snow-type particles. The lack of a direct relation between viral abundance and prokaryotic abundance as observed in our study might also indicate allochthonous input of viruses (Table 3 and Fig. 6). Generally, marine snow harbors viruses, prokaryotes, and eukaryotes in abundances exceeding the abundances of their free-living counterparts by orders of magnitude (21, 37, 56). Hara et al. (16) discussed the possibility of allochthonous input of viruses from surface waters into the deep layers of the Pacific to explain the high bathypelagic viral abundance they found. The aggregation-sedimentation mechanism of colloidal particles has been hypothesized as a way for viral transport into the deep sea but remains untested (38). Aggregation of nonsinking small colloidal particles (<0.2 μm) found in concentrations of 105 aggregates ml−1 is common, scavenging picoplankton and viruses from the free-living phase (56). These microscopic colloidal particles collide and finally form macroscopic marine snow-type particles sufficiently large to sediment. Phages embedded in these macroscopic particles might exhibit decay rates lower than those seen for freely suspended forms (20). There is also evidence that infected cells are attached to sinking particles (38). Besides this vertical flux of macroaggregates to the bathypelagic zone, lateral transport of matter from the more productive continental shelves into the bathypelagic realm along isopycnals might represent an important source of deep-water viruses as well. For the region of our study, considerable lateral input of organic matter from the continental slope underlying the productive Mauritanian upwelling into the mesopelagic subtropical North Atlantic was reported by Aristegui et al. (1), explaining the high mesopelagic respiration rates found there.
The bathypelagic viral diversity reported here also might provide some information to test this “sedimenting virus hypothesis.” If viruses attached to particles derived from near-surface waters are an important source for deep-water viruses, viral diversity in the deep ocean should be similar to that of near-surface waters. The number of PFGE bands obtained from surface-water viral communities can be as high as 35 (63), and still each band can include different types of viruses with the same genome size. In the deep North Atlantic, we detected only four bands, indicating that the richness of the bathypelagic viral community, or at least that detectable by PFGE, is rather limited (Fig. 4). However, it might well be that the richness of prokaryotic taxa on sedimenting macroaggregates is substantially lower than that in the free-living prokaryotic community given the stability of the particle system, thereby also reducing the richness of the associated viral community.
There is evidence that a considerable fraction of the phages in aquatic environments are temperate rather than lytic (5, 36), especially in oligotrophic environments. If the bathypelagic viral community is lysogenic, the production of viruses depends on the rate of induction. Reported inducing agents of prokaryotic communities inhabiting the euphotic layers of the ocean, such as UV radiation, are not present in the deep ocean. Nutrient limitation found to induce phage production in the surface-water prokaryotic communities might be an inducing agent in the dark ocean as well, since deep-water heterotrophic prokaryotic communities are generally carbon limited. Using a high spontaneous induction frequency for the meso- and bathypelagic region of the Mediterranean Sea, where lysogenic cells dominate the prokaryotic community, prophage induction rates did not contribute significantly to total virus abundance (51). Thus, as long as no major inducing agents in deep waters are identified, prophage induction cannot solve the problem of the high viral abundance. Pseudolysogeny is a viral lifestyle that can potentially add to the explanation of high viral abundance (59). Pseudolysogeny is defined as “sustained phage production along with a thriving population of host cells” (59). If low nutrient concentrations result in a pseudolysogenic interaction or prophages with their hosts, the high number of lysogens in deep marine waters might result in a constant virus production. In this case, viral production would be decoupled from lytic infection.
In summary, the apparent discrepancy between high viral abundance and low prokaryotic and viral production in the meso- and bathypelagic waters of the subtropical eastern basin of the North Atlantic cannot be explained by lytic infection. A substantial allochthonous input of viruses attached to particles from adjacent water masses or pseudolysogenic interactions between viruses and hosts are potential explanations to solve that discrepancy. We showed that the decay rates of viruses in the deep ocean are about 1 order of magnitude lower than in surface waters and that the VPR increases with depth in the water column. High viral abundance and VPR are commonly interpreted as indications of intense control of prokaryotic abundance and taxon richness by lytic phages. We propose, however, two deviating but not mutually exclusive explanations. First, the prokaryotic community in deep waters is controlled mainly by the availability of suitable substrate, and lysis by lytic viruses plays only a marginal role in controlling prokaryotic diversity, as a substantial fraction of the viral community might be allochthonously derived. Due to the low decay rates, a high viral abundance is maintained in the dark ocean. Second, the high viral abundance in the deep ocean might originate from pseudolysogenic rather than lytic interactions. In this case, viruses might exert a substantial control over PPP.
Acknowledgments
We thank the captain and the crew of the R/V Pelagia for their excellent support and the atmosphere on board. The Dutch Marine Research Facilities provided excellent logistic support.
Seagoing research was supported by two grants from the Dutch Science Foundation, Earth and Life Sciences branch (NWO-ALW project no. 835.20.023 and 812.03.001, both to G.J.H.). The work was carried out within the framework of the “Networks of Excellence” MarBef and EurOceans supported by the 6th Framework Program of the European Union.
This paper is in partial fulfillment of the requirements for a Ph.D. degree from the University of Groningen by V.P.
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Aristegui, J., C. M. Duarte, J. M. Gasol, and L. Alonso-Sáez. 2005. Active mesopelagic prokaryotes support high respiration in the subtropical northeast Atlantic Ocean. Geophys. Res. Lett. 32:L03608. doi: 10.1029/2004GL021863. [DOI] [Google Scholar]
- 2.Bongiorni, L., M. Magagnini, M. Armeni, R. Noble, and R. Danovaro. 2005. Viral production, decay rates, and life strategies along a trophic gradient in the North Adriatic Sea. Appl. Environ. Microbiol. 71:6644-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Børsheim, K. Y. 1993. Native marine bacteriophages. FEMS Microb. Ecol. 102:141-159. [Google Scholar]
- 4.Børsheim, K. Y., G. Bratbak, and M. Heldal. 1990. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl. Environ. Microbiol. 56:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratbak, G., M. Heldal, S. Norland, and T. F. Thingstad. 1990. Viruses as partners in spring bloom microbial trophodynamics. Appl. Environ. Microbiol. 56:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bratbak, G., M. Heldal, T. F. Thingstad, B. Riemann, and O. H. Haslund. 1992. Incorporation of viruses into the budget of microbial C-transfer. A first approach. Mar. Ecol. Prog. Ser. 83:273-280. [Google Scholar]
- 7.Brussaard, C. P. D. 2004. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70:1506-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and C. A. Suttle. 1995. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol. Oceanogr. 40:730-739. [Google Scholar]
- 9.Fischer, U. R., and B. Velimirov. 2002. High control of bacterial production by viruses in a eutrophic oxbow lake. Aquat. Microb. Ecol. 27:1-12. [Google Scholar]
- 10.Fuhrman, J. 2000. Impact of viruses on bacterial processes, p. 327-350. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, NY.
- 11.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., and R. T. Noble. 1995. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 40:1236-1242. [Google Scholar]
- 13.Gobler, C. J., D. A. Hutchins, N. S. Fisher, E. M. Cosper, and S. A. Sañudo-Wilhelmy. 1997. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol. Oceanogr. 42:1492-1504. [Google Scholar]
- 14.Guixa-Boixereu, N., D. Vaque, J. M. Gasol, and C. Pedros-Alio. 1999. Distribution of viruses and their potential effect on bacterioplankton in an oligotrophic marine system. Aquat. Microb. Ecol. 19:205-213. [Google Scholar]
- 15.Guixa-Boixereu, N., D. Vaque, J. M. Gasol, J. Sanchez-Camara, and C. Pedros-Alio. 2002. Viral distribution and activity in Antarctic waters. Deep-Sea Res. Part II 49:827-845. [Google Scholar]
- 16.Hara, S., I. Koike, K. Terauchi, H. Kamiya, and E. Tanoue. 1996. Abundance of viruses in deep oceanic waters. Mar. Ecol. Prog. Ser. 145:269-277. [Google Scholar]
- 17.Heldal, M., and G. Bratbak. 1991. Production and decay of viruses in aquatic environments. Mar. Ecol. Prog. Ser. 72:205-212. [Google Scholar]
- 18.Herndl, G. J., T. Reinthaler, E. Teira, H. van Aken, C. Veth, A. Pernthaler, and J. Pernthaler. 2005. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl. Environ. Microbiol. 71:2303-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hewson, I., J. A. Steele, D. G. Capone, and J. A. Fuhrman. 2006. Remarkable heterogeneity in meso- and bathypelagic bacterioplankton assemblage composition. Limnol. Oceanogr. 51:1274-1283. [Google Scholar]
- 20.Kapuscinski, R. B., and R. Mitchell. 1980. Processes controlling virus inactivation in coastal waters. Water Res. 14:363-371. [Google Scholar]
- 21.Kepkay, P. E. 1994. Particle aggregation and the biological reactivity of colloids. Mar. Ecol. Prog. Ser. 109:293-304. [Google Scholar]
- 22.Kokjohn, T. A., G. S. Sayler, and R. V. Miller. 1991. Attachment and replication of Pseudomonas aeruginosa bacteriophages under conditions simulating aquatic environments. J. Gen. Microbiol. 137:661-666. [Google Scholar]
- 23.Lu, J., F. Chen, and R. E. Hodson. 2001. Distribution, isolation, host specificity, and diversity of cyanophages infecting marine Synechococcus spp. in river estuaries. Appl. Environ. Microbiol. 67:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marie, D., C. P. D. Brussard, R. Thyrhaug, G. Bratbak, and D. Vaulot. 1999. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 65:45-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathias, C. B., A. K. T. Kirschner, and B. Velimirov. 1995. Seasonal variations of virus abundance and viral control of the bacterial production in a backwater system of the Danube River. Appl. Environ. Microbiol. 61:3734-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Middelboe, M. 2000. Bacterial growth rate and marine virus-host dynamics. Microb. Ecol. 40:114-124. [DOI] [PubMed] [Google Scholar]
- 27.Moeseneder, M. M., C. Winter, and G. J. Herndl. 2001. Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol. Oceanogr. 46:95-107. [Google Scholar]
- 28.Murray, A. G., and G. A. Jackson. 1992. Viral dynamics: a model of the effects of size, shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Prog. Ser. 89:103-116. [Google Scholar]
- 29.Nagata, T., H. Fukuda, R. Fukuda, and I. Koike. 2000. Bacterioplankton distribution and production in the deep Pacific waters: large-scale geographic variations and possible coupling with sinking particle fluxes. Limnol. Oceanogr. 45:426-435. [Google Scholar]
- 30.Noble, R. T., and J. A. Fuhrman. 1998. Use of Sybr Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 31.Noble, R. T., and J. A. Fuhrman. 1997. Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 63:77-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortmann, A. C., and C. A. Suttle. 2005. High abundances of viruses in a deep-sea hydrothermal vent system indicate viral mediated microbial mortality. Deep-Sea Res. Part I 52:1515-1527. [Google Scholar]
- 33.Paepe, M. D., and F. Taddei. 2006. Viruses’ life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parada, V., G. J. Herndl, and M. G. Weinbauer. 2006. Viral burst size of heterotrophic prokaryotes in aquatic systems. J. Mar. Biol. Assoc. U.K. 86:613-621. [Google Scholar]
- 35.Paul, J. H. 2001. Methods in microbiology, vol. 30. Marine microbiology. Academic Press, San Diego, CA.
- 36.Paul, J. H., and S. C. Jiang. 2001. Lysogeny and transduction, p. 106-125. In J. H. Paul (ed.), Marine microbiology. Methods in microbiology, vol. 30. Academic Press, San Diego, CA. [Google Scholar]
- 37.Peduzzi, P., and M. G. Weinbauer. 1993. Effect of concentrating the virus-rich 2-200-nm size fraction of seawater on the formation of algal flocs (marine snow). Limnol. Oceanogr. 38:1562-1565. [Google Scholar]
- 38.Proctor, L. M., and J. A. Fuhrman. 1991. Roles of viral infection in organic particle flux. Mar. Ecol. Prog. Ser. 69:133-142. [Google Scholar]
- 39.Reinthaler, T., H. van Aken, C. Veth, P. J. L. B. Williams, J. Aristegui, C. Robinson, P. Lebaron, and G. J. Herndl. 2006. Prokaryotic respiration and production in the meso- and bathypelagic realm of the eastern and western North Atlantic basin. Limnol. Oceanogr. 51:1262-1273. [Google Scholar]
- 40.Riemann, B., H. M. Sørensen, P. K. Bjørnsen, S. J. Horsted, L. M. Jensen, T. G. Nielsen, and M. Søndergaard. 1990. Carbon budgets of the microbial food web in estuarine enclosures. Mar. Ecol. Prog. Ser. 65:159-170. [Google Scholar]
- 41.Sano, E., S. Carlson, L. Wegley, and F. Rohwer. 2004. Movement of viruses between biomes. Appl. Environ. Microbiol. 70:5842-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, M., and F. Azam. 1989. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201-213. [Google Scholar]
- 43.Suttle, C. A., and A. M. Chan. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suttle, C. A., and F. Chen. 1992. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, G. T., M. Iabichella, T.-Y. Ho, M. I. Scranton, R. C. Thunell, F. Muller-Karger, and R. Varela. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant mid-water source of organic carbon production. Limnol. Oceanogr. 46:148-163. [Google Scholar]
- 46.Teira, E., H. van Aken, C. Veth, and G. J. Herndl. 2006. Archaeal uptake of enantiomeric amino acids in the meso- and bathypelagic waters of the North Atlantic. Limnol. Oceanogr. 51:60-69. [Google Scholar]
- 47.Teira, E., P. Lebaron, H. van Aken, and G. J. Herndl. 2006. Distribution and activity of Bacteria and Archaea in the deep water masses of the North Atlantic. Limnol. Oceanogr. 51:2131-2144. [Google Scholar]
- 48.Teira, E., T. Reinthaler, A. Pernthaler, J. Pernthaler, and G. J. Herndl. 2004. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl. Environ. Microbiol. 70:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 50.Tomczak, M., and J. S. Godfrey. 1994. Regional oceanography. Elsevier Science Ltd., Oxford, United Kingdom.
- 51.Weinbauer, M., I. Brettar, and M. G. Höfle. 2003. Lysogeny and virus-induced mortality of bacterioplankton in surface, deep, and anoxic marine waters. Limnol. Oceanogr. 48:1457-1465. [Google Scholar]
- 52.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 53.Weinbauer, M. G., D. Fuks, S. Puskaric, and P. Peduzzi. 1995. Diel, seasonal, and depth-related variability of viruses and dissolved DNA in the Northern Adriatic Sea. Microb. Ecol. 30:25-41. [DOI] [PubMed] [Google Scholar]
- 54.Weinbauer, M. G., and C. A. Suttle. 1996. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the Gulf of Mexico. Appl. Environ. Microbiol. 62:4374-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinbauer, M. G., S. W. Wilhelm, C. A. Suttle, and D. R. Garza. 1997. Photoreactivation compensates for UV damage and restores infectivity to natural marine virus communities. Appl. Environ. Microbiol. 63:2200-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells, M. L., and E. D. Goldberg. 1994. The distribution of colloids in the North Atlantic and Southern Oceans. Limnol. Oceanogr. 39:286-302. [Google Scholar]
- 57.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea. Viruses play critical roles in the structure and function of aquatic food webs. BioScience 49:781-788. [Google Scholar]
- 58.Wilhelm, S. W., M. G. Weinbauer, C. A. Suttle, and W. H. Jeffrey. 1998. The role of sunlight in the removal and repair of viruses in the sea. Limnol. Oceanogr. 43:586-592. [Google Scholar]
- 59.Williamson, S. J., M. R. McLaughlin, and J. H. Paul. 2001. Interaction of the φHSIC virus with its host: lysogeny or pseudolysogeny? Appl. Environ. Microbiol. 67:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winter, C., G. J. Herndl, and M. G. Weinbauer. 2004. Diel cycles in viral infection of bacterioplankton in the North Sea. Aquat. Microb. Ecol. 35:207-216. [Google Scholar]
- 61.Winter, C., A. Smit, G. J. Herndl, and M. G. Weinbauer. 2004. Impact of virioplankton on archaeal and bacterial community richness as assessed in seawater batch cultures. Appl. Environ. Microbiol. 70:804-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winter, C., A. Smit, T. Szoeke-Denes, G. J. Herndl, and M. G. Weinbauer. 2005. Modelling viral impact on bacterioplankton in the North Sea using artificial neural networks. Environ. Microbiol. 7:881-893. [DOI] [PubMed] [Google Scholar]
- 63.Wommack, K. E., J. Ravel, R. T. Hill, J. Chun, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wommack, K. E., S. J. Williamson, A. Sundbergh, R. R. Helton, B. T. Glazer, K. Portune, and S. C. Cary. 2004. An instrument for collecting discrete large-volume water samples suitable for ecological studies of microorganisms. Deep-Sea Res. Part I 511:1781-1792. [Google Scholar]
- 65.Zachary, A. 1976. Physiology and ecology of bacteriophages of marine bacterium Beneckea natriegens: salinity. Appl. Environ. Microbiol. 31:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]