Abstract
Erythromycin and tylosin are commonly used in animal production, and such use is perceived to contribute to the overall antimicrobial resistance (AR) reservoirs. Quantitative measurements of this type of AR reservoir in microbial communities are required to understand AR ecology (e.g., emergence, persistence, and dissemination). We report here the development, validation, and use of six real-time PCR assays for quantifying six classes of erm genes (classes A through C, F, T, and X) that encode the major mechanism of resistance to macrolides-lincosamides-streptogramin B (MLSB). These real-time PCR assays were validated and used in quantifying the six erm classes in five types of samples, including those from bovine manure, swine manure, compost of swine manure, swine waste lagoons, and an Ekokan upflow biofilter system treating hog house effluents. The bovine manure samples were found to contain much smaller reservoirs of each of the six erm classes than the swine manure samples. Compared to the swine manure samples, the composted swine manure samples had substantially reduced erm gene abundances (by up to 7.3 logs), whereas the lagoon or the biofilter samples had similar erm gene abundances. These preliminary results suggest that the methods of manure storage and treatment probably have a substantial impact on the persistence and decline of MLSB resistance originating from food animals, thus likely affecting the dissemination of such resistance genes into the environment. The abundances of these erm genes appeared to be positively correlated with those of the tet genes determined previously among these samples. These real-time PCR assays provide a rapid, quantitative, and cultivation-independent measurement of six major classes of erm genes, which should be useful for ecological studies of AR.
There is a growing interest in ecological studies of antimicrobial resistance (AR) owing to the increasing concern over the potential risk associated with AR originating from animals intended for food (2, 4, 12, 20, 47). Although mostly commensals, the microbes in the intestines and manures of food animals (estimated at >1010/g of manure) can serve as larger reservoirs of AR genes than pathogens (16, 32). These large AR gene reservoirs likely increase both dissemination of AR genes to the environment and resistance gene transfer, not only among commensals (22) but also to pathogens (45). To assess the potential risk associated with AR originating from agricultural use of antibiotics, these resistance gene reservoirs need to be measured. Whole-community analysis was proposed as providing the greatest ability to assess the resistance gene reservoir in a microbial community (16).
Both tylosin and erythromycin belong to the structurally distinct, yet functionally related, macrolide-lincosamide-streptogramin B (MLSB) superfamily of antibiotics. Erythromycin is used on both human and food animals, whereas tylosin is exclusively used on food animals (23). In fact, tylosin is one of the most commonly used antimicrobials in poultry, swine, and beef cattle (42). The use of tylosin on animals significantly increased the resistance by gut commensal bacteria to MLSB (9, 17). Resistance to tylosin in a food animal production environment was found to be encoded by erm genes (1, 18, 19, 43). The erm genes encode 23S rRNA adenine-specific N6-methyltransferases, which methylate the 23S rRNA of bacteria (28). Such methylation results in decreased binding of all MLSB drugs to their target (bacterial ribosomes) and thus resistance to all MLSB antibiotics. The erm genes are among the most common AR genes of MLSB, and 32 classes of erm genes (≥80% amino acid sequence identity within each class) have been identified and sequenced to date among many different genera of bacteria (http://faculty.washington.edu/marilynr/ermwebA.pdf) (29). Additionally, erm genes are among the most common acquired resistance genes in bacteria and the only genes conferring resistance to MLSB currently found in anaerobes (28, 31).
Given the difficulties in cultivating most of the bacteria in intestines and manures of mammalians (44), DNA-based techniques, especially PCR, are often used to examine resistance genes in these microbial communities. Both PCR and real-time PCR have been used in detecting and quantifying, respectively, tet genes in various environments (3, 8, 34, 47). These studies yielded interesting new knowledge on the distribution and reservoirs of many tet gene classes in several types of microbial communities. Because of both the widespread use of erythromycin and tylosin and cross selection among different MLSB, erm genes are among the most widely distributed AR genes (28, 29). However, their distribution and abundance in entire microbial communities, including animal manure (the major reservoir of erm genes derived from food animals), remain to be determined. Although a few publications reported the detection of erm genes in pathogenic isolates (5, 6, 11, 25, 35, 36), no PCR-based assay has been reported to quantify the erm gene reservoirs in entire microbial communities.
To complement the emerging efforts to understand AR ecology and dynamics, we are undertaking an effort to develop capabilities for quantitative measurements of AR gene reservoirs in entire microbiomes. In a previous study (47), we developed three real-time PCR assays that permit quantification of 10 major tet gene classes present in entire microbiomes. In this report, we described the development of six real-time PCR assays specific for erm(A), erm(B), erm(C), erm(F), erm(T), and erm(X) and their utility in quantifying the reservoirs of these erm genes present in bovine manures, swine manures, composted swine manures, swine waste lagoons, and an Ekokan upflow biofilter (EUB) system.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Staphylococcus aureus::Tn554 carrying erm(A) and Bacillus subtilis carrying erm(C) on plasmid pE194 (15) were grown in trypticase soy broth containing 15 μg/ml erythromycin. Three Escherichia coli DH5α strains carrying erm(B), erm(F), and erm(X) on plasmids pJIR229 (kindly provided by M. C. Roberts, University of Washington), pFD292, and pFK12 (kindly provided by A. Tauch, Universität Bielefeld, Germany), respectively, were grown in Luria-Bertani (LB) broth containing 30 μg/ml erythromycin. Overnight cultures were centrifuged, and the biomass was resuspended in Tris-EDTA buffer. These cell suspensions and an aliquot of plasmid p121BS (43) carrying erm(T) were used as positive controls in optimizing respective PCR assays.
Microbial community samples and DNA extraction.
In addition to the same sets of community DNA samples (being stored at −80°C in separate aliquots) previously used in the development of real-time PCR assays specific for tet genes (47), another eight fresh bovine manure samples collected from a beef herd in Ohio were added to the bovine manure set. As described for the previous sets of DNA samples (47), the community DNA from these eight bovine manure samples was extracted using the RBB+C method, which was shown to substantially increase DNA yields (48). The quality and quantity of these DNA samples were also determined by agarose gel electrophoresis and fluorospectrometry (47). In total, 55 samples belonging to five types were analyzed: samples from bovine manure (n = 16), swine manure (n = 10), compost of swine manure (n = 13), lagoons with swine manure (n = 6), and throughout an EUB system treating swine manure (n = 10).
Phylogenetic analysis of erm gene sequences, primer design, and specificity tests.
Thirty-two classes of erm genes have been identified so far. All the erm gene sequences belonging to these 32 classes currently available in GenBank were retrieved and then aligned using ClustalX (40). We attempted to design a single primer pair that permits detection of all known erm genes by PCR, but such a universal primer pair is not possible, because of the high degrees of sequence divergence among erm classes (data not shown). Thus, we chose to design specific primers for individual erm gene classes. The classes chosen were A, B, C, F, T, and X, because they, based on previous studies of resistant bacterial isolates, are common and/or have been detected in bacteria of animal origin. The sequences of these six classes were dereplicated after alignment using ClustalX (40). Using the erm(Y) gene from Staphylococcus aureus as an outgroup, a neighbor-joining tree was inferred using the program TreeCon as described previously (46). Each class of sequences was separated, and one class-specific primer pair was designed using the approach described previously (47). The erm(C)-specific primer pair reported by Chung et al. (10) matches all the known erm(C) sequences and allows for suitable amplicon length. Thus, it was used in real-time PCR to quantify erm(C). All the primers used in this study are described in Table 1.
TABLE 1.
PCR primer sequences, targets, annealing temperatures, and amplicon lengths
| Primer | Class targeted | Primer sequence (5′→3′) | Amplicon size (bp) | Primer annealing temp (°C)a | TFA (°C) | Source or reference(s) |
|---|---|---|---|---|---|---|
| erm(A)-106fb | A | GAA ATY GGR TCA GGA AAA GG | 332 | 55 | 80 | This study |
| erm(A)-437rb | AAY AGY AAA CCY AAA GCT C | |||||
| erm(B)-91fc | B | GAT ACC GTT TAC GAA ATT GG | 364 | 58 | 78 | This study |
| erm(B)-454rc | GAA TCG AGA CTT GAG TGT GC | |||||
| erm(C)-43fd | C | TCA AAA CAT AAT ATA GAT AAA | 642 | 50 | 77 | 10, 36 |
| erm(C)-684rd | GCT AAT ATT GTT TAA ATC GTC AAT | |||||
| erm(F)-189fe | F | CGA CAC AGC TTT GGT TGA AC | 309 | 56 | 80 | This study |
| erm(F)-497re | GGA CCT ACC TCA TAG ACA AG | |||||
| erm(T)-52ff | T | CAT ATA AAT GAA ATT TTG AG | 369 | 51 | 77 | This study |
| erm(T)-420rf | ACG ATT TGT ATT TAG CAA CC | |||||
| erm(X)-112fg | X | GAG ATC GGR CCA GGA AGC | 488 | 58 | 86 | This study |
| erm(X)-599rg | GTG TGC ACC ATC GCC TGA |
For touchdown PCR. See the text for details.
Numbered according to the erm(A) gene in Staphylococcus aureus transposon Tn554 (GenBank accession no. X03216).
Numbered according to the erm(B) gene in Streptococcus pneumoniae transposon Tn1545 (GenBank accession no. X52632).
Numbered according to the erm(C) gene on plasmid pT48 (from S. aureus strain T48) (GenBank accession no. M19652). This primer set was first designed by Sutcliffe et al. (36) and then named CF and CR by Chung et al. (10).
Numbered according to the erm(F) gene on plasmid pBF4 (from Bacteroides fragilis) (GenBank accession no. M14730).
Numbered according to the erm(T) gene on plasmid p121BS (from Lactobacillus spp.) (GenBank accession no. AF310974).
Numbered according to the erm(X) gene on plasmid pNG2 (from Corynebacterium diphtheriae) (GenBank accession no. X51472).
“Regular” PCR was done using a PTC-100 thermocycler (MJ Research, Waltham, MA) with 50-μl volumes containing 1× PCR buffer (20 mM Tris-HCl [pH 8.4] and 50 mM KCl), 200 μM deoxynucleoside triphosphates, 500 nM of each primer, 1.75 mM MgCl2, 670 ng/μl bovine serum albumin, 1.0 μl community DNA (about 50 ng) or cell suspensions (the positive controls), and 1.25 U Platinum Taq DNA polymerase (Invitrogen Corporation, Carlsbad, CA), which allows hot-start PCR. After an initial denaturation at 94°C for 4 min, five cycles of touchdown PCR (denaturation at 94°C for 30 s, annealing for 30 s with a 1°C-per-cycle decrement from 5°C above to the final annealing temperature indicated in Table 1, and extension at 72°C for 1 min) was performed, followed by 30 regular cycles of PCR (94°C for 30 s, 30 s at the respective annealing temperature, and 72°C for 45 s) and a final extension for 7 min at 72°C. The above-mentioned optimal annealing temperatures were predetermined by gradient PCR using a RoboCycler (Stratagene, La Jolla, CA) and the plasmid DNA or the DNA from the pure cultures. No-template controls were included in parallel.
To confirm primer specificity, the PCR products from one community DNA sample were cloned into the TOPO-TA cloning vector (Invitrogen). Randomly selected clones were sequenced (one strand) by the Plant and Microbe Genome Facility at The Ohio State University. Base-calling examination and comparisons with GenBank sequences were performed as described previously (47). The BLASTn search output alignments were also examined for the presence of breakage, which can indicate chimeric sequences.
Preparation of sample-derived real-time PCR standards.
One sample-derived standard was prepared for each of the six erm classes from each of the two sets of community DNA: (i) the DNA extracted from the bovine manure samples and (ii) the DNA derived from the swine manure, swine waste lagoon, swine manure compost, and EUB system samples, as done previously (47) with minor modification. Instead of amplifying the target erm genes from individual community DNA samples and then pooling the PCR products together, we amplified the erm genes by using respective specific primers and a DNA mixture containing approximately 100 ng of the individual DNA samples within each sample set. Then, the PCR product was purified using a QIAquick PCR purification kit (QIAGEN, Inc., Valencia, CA) and quantified using a Quant-iT kit (Invitrogen) as done previously (47). One sample-derived real-time PCR standard was also prepared from each set of the samples by using the pooled DNA and the universal bacterial primer pair 27f/1525r (21) for quantification of total bacteria by real-time PCR. The conditions of this PCR are the same as those described elsewhere (49). For each sample-derived standard, copy number concentration was calculated based on the length of the PCR product and the mass concentration. Tenfold serial dilutions were made in Tris-EDTA prior to real-time PCR (47). In total, 14 real-time PCR standards were prepared from the two sets of community DNA samples for the seven (six for erm gene classes and one for total bacteria) real-time PCR assays. Each of these standards was used in respective real-time PCR assays.
Real-time PCR.
The conditions of the real-time PCR assays of erm genes were the same as those of the regular PCR described above, with the following exceptions: decreased primer concentrations (250 nM each) and inclusion of 0.133× of SYBR green I (Molecular Probes, Eugene, OR) and 30 nM of the reference dye ROX (Stratagene). As done previously (47), the thermal profiles consisted of four segments: (i) initial denaturation at 95°C for 4 min; (ii) five touch-down cycles of 94°C for 30 s, 5°C above the respective annealing temperature (Table 1) for 30 s with a 1°C decrement per cycle, and 72°C for 40 s; (iii) 45 cycles of 94°C for 30 s, the respective annealing temperature for 30 s, 72°C for 30 s, and 18 s at the temperature for fluorescence acquisition (TFA) (Table 1); and (iv) 95°C for 2 min, 55°C for 30 s, and 95°C for 30 s. Fluorescence data were collected at the 72°C and TFA steps (end point) of the third segment and during the ramping from 55°C to 95°C (all point) of the last segment. Quantification of total bacteria was performed as described by Nadkarni et al. (24), except for the use of the sample-derived standards instead of genomic DNA from a single bacterial strain. All the real-time PCR assays were performed using an Mx3000p real-time PCR system (Stratagene). Baseline and threshold calculations were performed with Mx3000p software using the fluorescence signals acquired at TFA, at which primer dimmers completely denatured and did not adversely affect the quantification accuracy. Following real-time PCR, the products were confirmed by agarose gel electrophoresis and exclusion at TFA of fluorescence resulting from possible primer dimmers was verified by melting curve analysis (except for the real-time PCR assays for total bacteria, which employed the universal TaqMan probe) (24). All the real-time PCRs were done in triplicate for both the standards and the microbial community DNA samples.
Exactly as described previously (47), each of the real-time PCR assays for erm genes were validated by quantifying a series of known copies of erm gene standards spiked into a swine manure community DNA sample against respective sample-derived real-time PCR standards. The detection limit of each real-time PCR assay was also determined from the serial dilutions of the sample-derived standard templates (47). Following these validation experiments, the abundance of each erm gene class present in each community DNA sample was quantified against that of its respective sample-derived standard by using the real-time PCR conditions described above. The identity of each sample was concealed during real-time PCR for a blind test and was revealed only after quantification was completed to avoid influencing results. For ease of description, erm gene abundance expressed as the number of copies g−1 (or copies ml−1 in the case of liquid samples) is referred to as absolute abundance, whereas abundance expressed as the number of erm copies per million copies (cpmc) of total bacterial rrs genes is referred to as relative abundance. The absolute abundance was calculated by multiplying the number of copies per real-time PCR and the number of reactions that can be done with the DNA derived from 1 gram or ml of each sample (47), while the relative abundance was calculated by dividing the absolute abundance of each erm gene class by the corresponding total bacterial abundance (the number of rrs gene copies per g or ml of sample) in each sample and then multiplying by 1 million.
The real-time PCR assays were also used to determine the prevalence of each erm gene class among different types of samples. A sample was considered positive for an erm gene when at least two of the three replicate real-time PCRs yielded a threshold cycle value and a PCR product of the expected size (based on the agarose electrophoresis) in the respective real-time PCR assay of that sample. The prevalence of the erm gene among each type of sample was calculated as the percentage of samples that yielded the expected PCR product.
Statistical analysis.
The data were log10 transformed and analyzed using the Mixed Procedure of SAS 9.1 (SAS Institute, Cary, NC). Least-squares means (LSM) were calculated for all the data sets. Mean separation was conducted by using Fisher's protected least-significant-difference test, with significance declared at P values of ≤0.05. The absolute abundance and relative abundance of erm genes were graphed as boxes and whiskers by using GraphPad Prism 4 (GraphPad Software, San Diego, CA).
Nucleotide sequence accession numbers.
The erm gene sequences determined in this study have been deposited in GenBank under the accession numbers listed in Table 2.
TABLE 2.
Affiliations of the sequenced erm genes as determined by comparison to GenBank sequences
| erm gene | erm clone | GenBank accession no. | Prevalence | Most similar match (GenBank accession no.) | Identity (%) |
|---|---|---|---|---|---|
| erm(A) | ermA-SM1 | DQ887617 | 5/5 | Oceanobacillus iheyensis HTE831 erm(A) gene (BA000028) | 95.5 |
| erm(B) | ermB-BM3 | DQ887618 | 6/7 | Streptococcus pyogenes erm(B) gene (AJ972606) and 20 other erm(B) sequences in GenBank | 100 |
| ermB-BM5 | DQ887619 | 1/7 | Enterococcus faecium erm(B) gene (AY827541) and three other erm(B) sequences in GenBank | 99.2 | |
| erm(C) | ermC-SM1 | DQ887620 | 3/3 | Staphylococcus hyicus plasmid pSES21 erm(C) (Y09003) | 99.5 |
| erm(F) | ermF-SM2 | DQ887621 | 5/5 | Bacteroides fragilis R plasmid pBF4 erm(F) (M14730) and three other erm(F) sequences in GenBank | 98.7 |
| erm(T) | ermT-BM6 | DQ887622 | 7/7 | Lactobacillus fermentum plasmid pLME300 erm(T) (AJ188494) and two other erm(T) sequences in GenBank | 99.7 |
| erm(X) | ermX-BM1 | DQ887623 | 6/6 | Corynebacterium striatum strain M82B R plasmid pTP10 erm(X) (AF024666) | 99.4 |
RESULTS
Primer specificity and erm gene diversity.
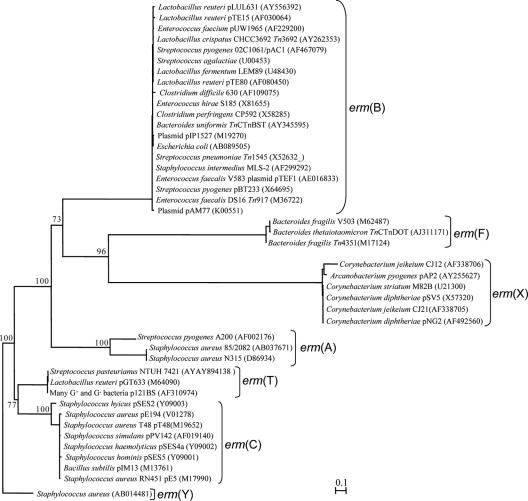
Our in silico analysis suggested that no universal erm primer is possible, due to sequence divergency (Fig. 1), but at least one primer pair can be designed for each erm gene class. In this study, one specific primer pair was designed for each of the six classes of erm genes: erm(A), erm(B), erm(C), erm(F), erm(T), and erm(X). Because of the high degree of sequence similarity within these classes, degenerate bases were needed only for the erm(A)- and erm(X)-specific primers (Table 1). Amplification of the intended erm gene by PCR using each of the primer pairs from the pure culture carrying the target erm gene and from one of the community DNA samples all produced a band of the expected size (data not shown). The sequencing of the randomly selected clones from each clone library of the community DNA produced sequences that match known erm sequences of the intended class in GenBank with high sequence identity (Table 2). The sequence identities with known erm gene sequences are within the homology ranges proposed for each class (31). Additionally, none of our sequences was broken into two segments in the BLASTn search alignments, suggesting a very low probability of chimeric sequences among our erm sequences. Collectively, these sequencing results confirmed the specificity of the erm class-specific primers and their utility with complex microbiome DNA samples. As indicated by clone ermA-SM1, these primer pairs may also prime amplification of heretofore unidentified members of the respective erm gene classes. These preliminary results suggest a greater diversity of erm genes than what has been identified in bacterial isolates.
FIG. 1.
A neighbor-joining tree of six classes of erm genes. The tree was inferred from DNA sequences, and it was arbitrarily rooted with the erm(Y) gene of Staphylococcus aureus. Bootstrap values were calculated from 100 replicates, and the number at each node indicates the number of times that the node was supported in the bootstrap analysis. The bar represents a 0.1 estimated change per nucleotide. Each primer pair listed in Table 1 targets a corresponding cluster.
Validation of real-time PCR assays and quantification of erm genes.
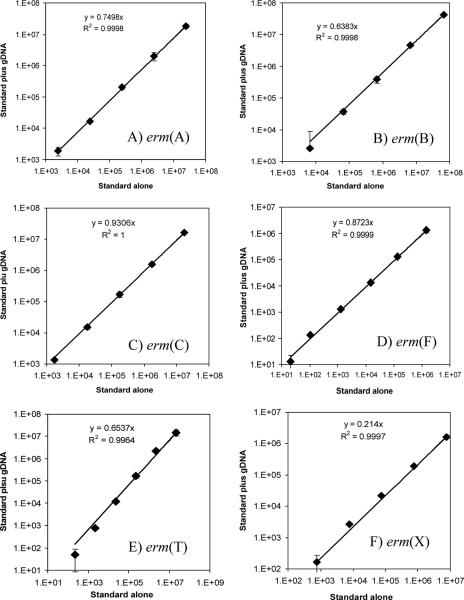
The accuracy of each real-time PCR assay was validated by quantifying known numbers of respective erm gene templates mixed into microbiome DNA samples, essentially as described previously (47). When the copy numbers of the erm genes spiked into the samples were plotted against the corresponding copy numbers of the erm genes quantified in the validation experiments, after correction for the background numbers of the erm genes present in the microbiome DNA itself, high r2 values over at least 5 orders of magnitude were obtained for all six assays (Fig. 2). However, the slopes of all six regression plots are less than 1, suggesting suppression of PCR amplification of the targets in the presence of community DNA, especially in the case of erm(X). The value of each slope, referred to as the “suppression coefficient” hereafter, was factored into the calculation of abundance of the respective erm class in all the samples by dividing the quantification values by the respective suppression coefficient. These real-time PCR assays detected fewer than 10 erm gene copies per real-time PCR (data not shown).
FIG. 2.
Validation curves plotting the copy numbers of the spiked erm gene standard (x axis) against the corresponding quantification values for that erm gene, after correction for background copies present in the community DNA sample, which contained (in numbers of copies/reaction) the following: erm(A), 4.18 × 103; erm(B), 6.32 × 104; erm(C), 88.6; erm(F), 11.4, erm(T), 208; and erm(X), 75.4. Error bars indicate standard deviations (n = 3). gDNA, genomic community DNA.
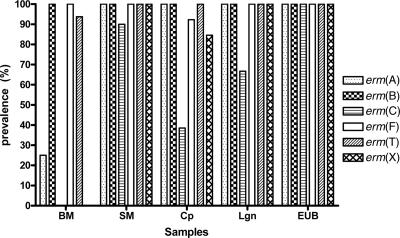
Different classes of the erm genes differed in prevalence among the five types of samples analyzed (Fig. 3). The erm(B) genes are the most prevalent and were detected in all 55 samples, while erm(C) genes were the least prevalent. The prevalence of erm(F) was only slightly lower than that of erm(B). The five different types of samples also differed in prevalence of different erm gene classes (Fig. 3). The swine manure samples have much higher prevalences of the erm genes than the bovine manure samples. Compared to the swine manure samples, the composted manure samples, but not the lagoon samples and the samples collected from the Ekokan biofilter system, had lower prevalences of erm(C), erm(F), and erm(X).
FIG. 3.
Prevalence (percent positive samples) of erm genes in the five types of samples analyzed. Prevalence is indicated as a percentage of positive samples among all the samples within each sample type. BM, fresh bovine manure samples (n = 16); SM, fresh swine manure samples (n = 10); Cp, composted swine manure samples (n = 13); Lgn, samples from lagoons receiving hog house effluent (n = 6); EUB, samples from an EUB system treating hog house effluent and a lagoon receiving its effluent (n = 10).
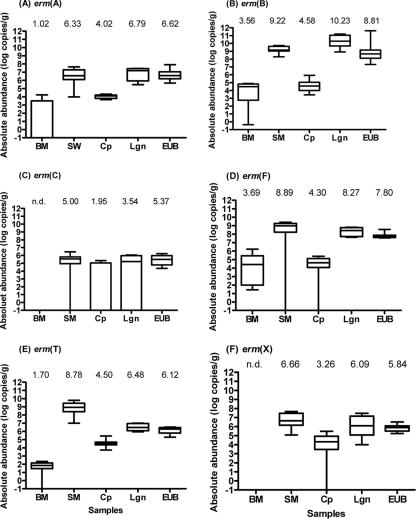
The absolute abundances of the erm genes varied considerably among the five types of samples as well as among different erm gene classes within individual sample types (Fig. 4 and Table 3). LSM for the bovine manure samples were substantially (>4.0 logs, P < 0.001) lower for erm genes of all six classes than LSM for the swine manure samples. Compared to the swine manure samples, the composted swine manure samples had substantially reduced (P < 0.001) erm gene abundances: erm(A), by 2.3 logs; erm(B), by 5.7 logs; erm(C), by 5.0 logs; erm(F), by 4.3 logs; erm(T), by 7.1 logs; and erm(X), by 7.3 logs. In contrast to the composted swine manure samples, the lagoon samples and the EUB samples did not exhibit significantly reduced erm gene abundances, except for the lagoon samples, which had 1.5 logs fewer erm(A) (P = 0.03) and 2.3 logs fewer erm(T) (P < 0.001), and the Ekokan biofilter samples, which had 2.7 logs fewer erm(T) (P < 0.0001) than the swine manure samples.
FIG. 4.
Box-and-whisker plots of absolute abundance of erm genes. All erm data are expressed as log10 numbers of copies per gram (wet weight) or ml of samples. See the legend to Fig. 3 for the acronyms of the sample types. Error bars indicate maximum and minimum values, horizontal lines indicate median values, and boxes indicate values between the 25th and 75th percentiles. The value above each box-and-whisker plot is the LSM for each type of sample. n.d., not detected.
TABLE 3.
Proportion of each erm gene class (percentage of total erm genes) in the erm gene reservoirs consisting of the six erm gene classes among the five types of samples (based on LSM for absolute abundance)
| Sample typea | Total erm genes (no. of copies/g) | Proportion (%)b of indicated gene class
|
|||||
|---|---|---|---|---|---|---|---|
| erm(A) | erm(B) | erm(C) | erm(F) | erm(T) | erm(X) | ||
| BM | 9.99 × 103 | 0.11 ± 23.75 | 56.48 ± 37.21 | 0.01 ± <0.01 | 42.65 ± 37.85 | 0.74 ± 3.89 | 0.01 ± <0.01 |
| SM | 3.63 × 109c | 0.08 ± 0.16 | 71.53 ± 14.81 | 0.00 ± 0.00 | 2.45 ± 9.56 | 25.36 ± 15.40 | 0.58 ± 0.56 |
| Cp | 1.60 × 105 | 8.80 ± 5.48 | 37.71 ± 22.16 | 0.06 ± 10.29 | 10.83 ± 6.14 | 30.42 ± 15.67 | 12.17 ± 22.35 |
| Lgn | 2.67 × 1010c | 0.03 ± 0.08 | 99.33 ± 1.55 | 0.00 ± 0.00 | 0.60 ± 1.32 | 0.02 ± 0.06 | 0.02 ± 0.16 |
| EUB | 1.07 × 109c | 0.52 ± 1.92 | 93.74 ± 15.86 | 0.02 ± 0.13 | 5.23 ± 14.35 | 0.19 ± 0.51 | 0.30 ± 0.57 |
BM, bovine manure; SM, swine manure; Cp, compost; Lgn, lagoon.
Data shown are means ± standard deviations.
Significantly (P < 0.05) higher than values for BM and Cp.
The total erm gene reservoir consisting of the six erm gene classes is summarized in Table 3. Collectively, the swine manure samples had almost 6 logs more of the erm genes than the bovine manure samples. In the swine manure samples, erm(B) was the most abundant, accounting for approximately 72% of the erm gene reservoir, whereas erm(B) and erm(F) accounted for approximately 56% and 43%, respectively, of the erm gene reservoir in the bovine manure samples. Among the three types of treated swine manure samples, composted swine manure samples had at least 4 logs fewer (P < 0.05) erm genes than the swine manure samples collected from the lagoons and the Ekokan biofilter system. The proportions of each of the six erm gene classes also differed among the four types of swine manure samples. Relative to the swine manure samples, the composted swine manure samples had increased proportions of erm(A), erm(F), erm(T), and erm(X) but decreased proportions of erm(B). Interestingly, in both the lagoon and the Ekokan biofilter samples, erm(B) accounts for most (approximately 99% and 94%, respectively) of the erm gene reservoir.
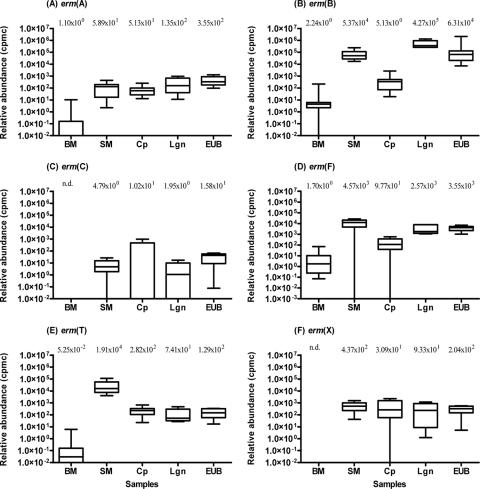
In order to determine the relative abundances of individual erm gene classes, the total bacteria in each sample were quantified and expressed as the number of rrs copies per gram or ml of the sample. The total bacterial abundances in the bovine manure, swine manure, compost, lagoon, and Ekokan biofilter system samples were 2.53 × 109, 4.86 × 1010, 2.74 × 108, 6.20 × 1010, and 1.58 × 1010 rrs copies g−1 or ml−1, respectively. Different erm gene classes exhibited different relative abundances (Fig. 5). Apparently, erm(B) had the greatest (P < 0.01) relative abundance among the six classes measured, reaching as high at 4.27 × 105 cpmc (equivalent to 42.7% of the total bacterial rrs copies) in the lagoon samples (Fig. 5B). The other five erm gene classes were much less abundant, amounting to fewer than 2.0 × 104 cpmc (or 2.0% of the total bacterial rrs copies). The relative abundances of all six erm gene classes also varied considerably among the five different types of manure samples. The bovine manure samples had the lowest relative abundances of all six erm gene classes, several orders of magnitude lower than those found in the four types of swine manure samples. Except for the erm(C) genes, the swine manure samples had considerably greater (P ≤ 0.0001) relative abundances for all the erm genes analyzed than the bovine manure samples (Fig. 5).
FIG. 5.
Box-and-whisker plots of relative abundance of erm genes. All erm data are expressed as log10 numbers of transformed erm cpmc of total 16S rrs genes. See the legend to Fig. 3 for the acronyms of the sample types. See the legend to Fig. 4 for a detailed explanation of the plots.
The three types of treated swine manure samples had variable relative abundances of erm genes (Fig. 5). Except for the erm(A) and erm(C) genes, the composted swine manure samples had lower relative abundances of erm(B) (by 2.38 logs, P < 0.001), erm(F) (by 2.26 logs, P < 0.0001), erm(T) (by 2.03 logs, P < 0.005), and erm(X) (by 1.1 logs, P < 0.01) than the swine manure samples. On the contrary, the lagoon and the Ekokan biofilter samples were found to have relative abundances for all the quantified erm genes similar (P > 0.05) to those for the swine manure samples, except those for erm(B), which were more than 2 logs lower (P < 0.0001) in these two types of treated manure samples than in the swine manure samples.
DISCUSSION
Using a similar approach reported previously (47), we developed six real-time PCR assays that permit quantification of six major classes of erm genes in animal manures, which constitute the major reservoirs of AR originating from food animal production. As done previously (47), we used respective sample-derived standards in validating each of the real-time PCR assays and in quantifying the erm gene classes. The use of such standards is important because differences in target sequence diversity (and thus differences in amplification efficiencies) between the samples and the real-time PCR standards can lead to inaccuracies (24). By using sample-derived standards, the real-time PCR assays were validated, and the target genes quantified, against potentially all target rrs or erm genes present in the samples analyzed rather than a few selected MLSB-resistant laboratory strains, which may or may not be present in the samples. Additionally, the validation against sample-derived standards eliminates the effects of cell lysis and DNA recovery, which probably vary with different DNA extraction methods and stains used. All the assays were shown to be robust with five types of samples and specific for the intended erm gene classes. As validated against known copies of respective erm genes spiked into microbiome DNA, these real-time PCR assays were found to be precise and accurate (Fig. 2). Given the physiochemical and microbial complexity of the samples tested in this study, these real-time PCR assays may be applicable to other types of samples, such as soil, aquatic, and sewage samples.
The validation experiments identified different suppression coefficients, as indicated by the different slopes (Fig. 2), for different real-time PCR assays. All the suppression coefficients are smaller than 1, suggesting that the targets in the standards are amplified more efficiently than those present in the samples. This is probably attributable to a higher complexity of the DNA templates in the community DNA than in the real-time PCR standards. The differences in suppression coefficients among the real-time PCR assays may be explained, at least partially, by differences in efficiencies in primer annealing and in secondary structures of the amplicons and primers. Since suppression coefficients vary for different real-time time PCR assays, even with the same set of samples, new real-time PCR assays should be validated and the suppression coefficients factored into quantification by real-time PCR for improved accuracy.
Different erm gene classes had considerably variable abundances within each of the five types of samples. However, several general trends appeared to be evident. Except for erm(T) being slightly more abundant than erm(F) in the compost samples, erm(B) and erm(F) exhibited the greatest abundances in nearly all the sample types (Fig. 4 and 5). This is in concordance with cultivation-based studies that revealed the distribution of erm(B) and erm(F) genes in the highest numbers of bacterial genera (21 and 20 genera, respectively) (30). Moreover, erm(B) was also found in 95% of the erythromycin-resistant enterococci isolated from three swine farms (17). Hence, the high abundance of these two erm genes is associated, or at least concurrent, with their wide occurrence in a large number of bacterial populations. The finding that these two erm genes often reside on mobile genetic elements (31) may explain, at least partially, their wide distribution and high abundance in microbiomes. Of interest is the high prevalence (Fig. 3) and abundance (Fig. 4E) of erm(T) found in the swine manure samples. This gene was first identified in a Lactobacillus strain from swine manure in 2001 (43) and has been identified only in Streptococcus pasteurianus (41) and Streptococcus bovis (38) as well as on plasmids p121BS in a Lactobacillus sp. (43), pLME300 in Lactobacillus fermentum (14), and pGT633 in Lactobacillus reuteri. Since none of these species is typically predominant in manure, other species may be the host for the erm(T) gene. Further studies are needed to confirm whether the erm(T) gene is universally abundant in swine manures. It also remains to be determined why only swine manures had large erm(T) gene reservoirs. On the other end of the spectrum, erm(C) exhibited the lowest abundance in all of the five types of samples. Its wide occurrence (so far found in 16 bacterial genera) (http://faculty.washington.edu/marilynr/ermweb4.pdf) and frequent residence on mobile genetic elements (31) seem to contradict its low abundance in these samples of animal origin.
All of the swine manure samples had significantly greater abundances of all six classes of erm genes than the bovine manure samples, either in absolute terms or in relative terms (Fig. 4 and 5). This is consistent with the previous finding with respect to tet genes present in these samples. The use of antibiotics (including tetracycline and erythromycin) in these swine farms was suggested to be the main contributing factor (47), but further studies are required, perhaps through the examination of conventional and organic swine farms, to determine if these differences can be attributed solely to differences in the use of erythromycin (or tylosin) or if differences in community composition in fecal microflora also play a role. The preliminary results also suggest that treatment of hog house effluents by either the EUB system or the lagoons tends not to appreciably reduce the erm gene reservoirs. These findings are consistent with the previous study of tet gene abundance (47) and corroborate a recent report (13) describing the low efficacy of eliminating erythromycin-resistant enterococci in an urban wastewater treatment plant. More in-depth studies are required to assess reduction and dissemination of AR in various types of wastewater treatment facilities and to identify potential factors that can affect the reduction and dissemination of AR during the treatment processes.
AR derived from food animals disseminates into the environment primarily via application and discharge of animal manures. Therefore, AR dissemination to the environment can be prevented or minimized through adequate and proper treatment and management of animal manures. In our previous study (47), we found that composted swine manure samples had substantially reduced tet gene reservoirs, whereas the samples from the lagoons and the EUB system did not. Interestingly, the erm genes quantified in this study also showed such a trend (Fig. 4). Thus, the results of these two independent studies tend to support our previous hypothesis that composting can effectively reduce AR to a variety of antimicrobials (47). However, it remains to be determined why composting, but not using lagoons or the Ekokan system, can effectively reduce AR reservoirs in animal manures. Inactivation of anaerobic erm-carrying bacteria during the composting process, which is largely aerobic and often has a thermophilic phase, and the low efficiency of lateral gene transfer in a solid-compost matrix to bacteria that can survive the composting process may collectively contribute to effective AR reduction during composting.
The prevalence of AR to a particular antimicrobial has been exclusively reported as a percentage of resistant isolates. The prevalences of individual resistant bacterial species obtained from swine farms are rather high and vary considerably. For instance, approximately 55% of the Campylobacter coli isolates (26, 39) and up to 98% of the airborne enterococcus isolates (7) from conventional swine farms were found to be resistant to erythromycin. Jackson et al. (17) also found that 95% of the erythromycin-resistant enterococci isolated from three swine farms carry erm(B). The relative abundances of all six classes of erm genes as determined in this study were much lower (less than 43%). These results further indicate that resistance prevalence likely varies among different bacterial species within a microbiome and imply that prevalence data obtained from specific cultivated species probably are not reflective of the overall prevalence in entire microbiomes.
Among the five different types of samples analyzed, patterns and/or magnitudes of differences differed between the relative and the absolute abundance measurements (Fig. 4 and 5). There are at least two possible explanations for such a disparity. First, because total bacterial abundance in the samples also affects the relative erm gene abundance data, any difference in total bacterial abundance will change the relative erm gene abundance. Second, different types of samples may harbor different bacterial populations carrying different erm genes, conceivably possessing different ecologies, and such differences could contribute to the incongruity observed between absolute and relative erm gene abundance. Further studies are needed to test the latter hypothesis.
Interestingly, we noticed rather similar patterns of abundance between the tet genes determined previously (47) and the erm genes determined in this study among these sets of samples. Multidrug resistance is common among resistant isolates due to the occurrence of multiple resistance genes on the same mobile elements (27, 33, 37). It has been shown that the erm(F) gene is often associated with conjugative transposons and linked to tet(M), tet(Q), and tet(X) (29), whereas erm(B) is often linked with tet(M) on Tn917-like conjugative transposons (30, 33) and with tet(Q) (29). Some staphylococci strains were also found to have both tet and erm genes (5). Although staphylococci are not likely predominant microbes in the samples analyzed, this type of linkage between tet and erm genes may also exist in other microbes. We postulate that the similar patterns of abundance observed between tet and erm genes in these five types of samples are probably attributable, at least partially, to the physical linkage between these two types of resistant genes and/or the carriage of these genes by the same bacteria. The results of this study provided some preliminary community-level clues that erm and tet genes, and maybe other AR genes, may be linked together and/or carried by the same bacteria, so they can exhibit similar dynamics in microbial communities. Consequently, reduction of one type of AR by a specific manure treatment may indicate the reduction of other types of AR present in the manure. Further studies are needed to test this hypothesis.
In conclusion, we developed and validated real-time PCR assays that can be used to accurately quantify the reservoirs of six classes of erm genes present in manure, compost, lagoon, and bioreactor samples. Our preliminary data suggest that different erm genes may have different reservoir sizes in microbial communities and different methods of manure treatments may have different efficiencies in reducing erm gene abundance. These should be evaluated in greater detail so that effective mitigation strategies can be developed to reduce dissemination of AR originating from food animals. Additionally, AR prevalence determined from bacterial isolates probably does not reflect the overall prevalence or abundance in the microbial community where the isolates are isolated.
Acknowledgments
We gratefully acknowledge the receipt of Staphylococcus aureus::Tn554, Bacillus subtilis, and p121BS provided by T. R. Whitehead (USDA/ARS, Peoria, IL). We thank S. Karnati and C. Reveneau for their assistance with statistical analysis, S. Sreevatsan (UMN) for providing the DNAs extracted from the swine manure samples, and M. Williams and B. Sheldon (NCSU) for providing samples collected from the EUB system and the lagoons. We also thank J. Firkins for helpful discussions during the preparation of the manuscript.
The work was supported by a Board of Regents award from The Ohio State University and a USDA-CSREES award (M.M. and Z.Y.; 2003-45050-01616) as well as by a USDA-IFAFS award (F.C.M.).
Footnotes
Published ahead of print on 11 May 2007.
REFERENCES
- 1.Aarestrup, F., H. Kruse, E. Tast, A. Hammerum, and L. Jensen. 2000. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb. Drug Resist. 6:63-70. [DOI] [PubMed] [Google Scholar]
- 2.American Society for Microbiology. 2002. Antimicrobial resistance: an ecological perspective. Report from the American Academy of Microbiology colloquium. http://www.asm.org/ASM/files/CCPAGECONTENT/docfilename/0000003761/Antimicrobialrpt[1].pdf. [PubMed]
- 3.Aminov, R. I., J. C. Chee-Sanford, N. Garrigues, B. Teferedegne, I. J. Krapac, B. A. White, and R. I. Mackie. 2002. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microbiol. 68:1786-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardic, N., M. Ozyurt, B. Sareyyupoglu, and T. Haznedaroglu. 2005. Investigation of erythromycin and tetracycline resistance genes in methicillin-resistant staphylococci. Int. J. Antimicrob. Agents 26:213-218. [DOI] [PubMed] [Google Scholar]
- 6.Cerda Zolezzi, P., M. C. Rubio Calvo, L. Millan, P. Goni, M. Canales, S. Capilla, E. Duran, and R. Gomez-Lus. 2004. Macrolide resistance phenotypes of commensal viridans group streptococci and Gemella spp. and PCR detection of resistance genes. Int. J. Antimicrob. Agents 23:582-589. [DOI] [PubMed] [Google Scholar]
- 7.Chapin, A., A. Rule, K. Gibson, T. Buckley, and K. Schwab. 2005. Airborne multidrug-resistant bacteria isolated from a concentrated swine feeding operation. Environ. Health Perspect. 113:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee-Sanford, J. C., R. I. Aminov, I. J. Krapac, N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie, P. J., J. N. Davidson, R. P. Novick, and G. M. Dunny. 1983. Effects of tylosin feeding on the antibiotic resistance of selected Gram-positive bacteria in pigs. Am. J. Vet. Res. 44:126-128. [PubMed] [Google Scholar]
- 10.Chung, W. O., C. Werckenthin, S. Schwarz, and M. C. Roberts. 1999. Host range of the ermF rRNA methylase gene in bacteria of human and animal origin. J. Antimicrob. Chemother. 43:5-14. [DOI] [PubMed] [Google Scholar]
- 11.Duarte, R. S., O. P. Miranda, B. C. Bellei, M. A. Brito, and L. M. Teixeira. 2004. Phenotypic and molecular characteristics of Streptococcus agalactiae isolates recovered from milk of dairy cows in Brazil. J. Clin. Microbiol. 42:4214-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.FAAIR Scientific Advisory Panel. 2002. Policy recommendations. Clin. Infect. Dis. 34:S76-S77. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira da Silva, M. F., I. Tiago, A. Verissimo, R. A. Boaventura, O. C. Nunes, and C. M. Manaia. 2006. Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 55:322-329. [DOI] [PubMed] [Google Scholar]
- 14.Gfeller, K. Y., M. Roth, L. Meile, and M. Teuber. 2003. Sequence and genetic organization of the 19.3-kb erythromycin- and dalfopristin-resistance plasmid pLME300 from Lactobacillus fermentum ROT1. Plasmid 50:190-201. [DOI] [PubMed] [Google Scholar]
- 15.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 150:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacson, R. E., and M. E. Torrence. 2002. The role of antibiotics in agriculture. American Academy of Microbiology, Washington, DC. [PubMed]
- 17.Jackson, C. R., P. J. Fedorka-Cray, J. B. Barrett, and S. R. Ladely. 2004. Effects of tylosin use on erythromycin resistance in enterococci isolated from swine. Appl. Environ. Microbiol. 70:4205-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jost, B. H., A. C. Field, H. T. Trinh, J. G. Songer, and S. J. Billington. 2003. Tylosin resistance in Arcanobacterium pyogenes is encoded by an Erm X determinant. Antimicrob. Agents Chemother. 47:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jost, B. H., H. T. Trinh, J. G. Songer, and S. J. Billington. 2004. A second tylosin resistance determinant, Erm B, in Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 48:721-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kummerer, K. 2004. Resistance in the environment. J. Antimicrob. Chemother. 54:311-320. [DOI] [PubMed] [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. D. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 22.Luo, H., K. Wan, and H. H. Wang. 2005. High-frequency conjugation system facilitates biofilm formation and pAMβ1 transmission by Lactococcus lactis. Appl. Environ. Microbiol. 71:2970-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellon, M., C. M. Benbrook, and K. L. Benbrook. 2001. Hogging it: estimates of antimicrobial abuse in livestock. UCS Publications, Cambridge, MA.
- 24.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 25.Novais, C., T. M. Coque, M. J. Costa, J. C. Sousa, F. Baquero, and L. V. Peixe. 2005. High occurrence and persistence of antibiotic-resistant enterococci in poultry food samples in Portugal. J. Antimicrob. Chemother. 56:1139-1143. [DOI] [PubMed] [Google Scholar]
- 26.Payot, S., S. Dridi, M. Laroche, M. Federighi, and C. Magras. 2004. Prevalence and antimicrobial resistance of Campylobacter coli isolated from fattening pigs in France. Vet. Microbiol. 101:91-99. [DOI] [PubMed] [Google Scholar]
- 27.Poutanen, S. M., J. de Azavedo, B. M. Willey, D. E. Low, and K. S. MacDonald. 1999. Molecular characterization of multidrug resistance in Streptococcus mitis. Antimicrob. Agents Chemother. 43:1505-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, M. C. 2003. Acquired tetracycline and/or macrolide-lincosamides-streptogramin resistance in anaerobes. Anaerobe 9:63-69. [DOI] [PubMed] [Google Scholar]
- 29.Roberts, M. C. 2004. Distribution of macrolide, lincosamide, streptogramin, ketolide and oxazolidinone (MLSKO) resistance genes in Gram-negative bacteria. Curr. Drug Targets Infect. Disord. 4:207-215. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, M. C. 2004. Resistance to macrolide, lincosamide, streptogramin, ketolide, and oxazolidinone antibiotics. Mol. Biotechnol. 28:47-62. [DOI] [PubMed] [Google Scholar]
- 31.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salyers, A. A. 1995. Antibiotic resistance transfer in the mammalian intestinal tract: implications for human health, food safety, and biotechnology. Springer-Verlag, New York, NY.
- 33.Seral, C., F. Castillo, C. Garcia, M. Rubio-Calvo, and R. Gomez-Lus. 2000. Presence of conjugative transposon Tn1545 in strains of Streptococcus pneumoniae with mef(A), erm(B), tet(M), catpC194 and aph3′-III genes. Enferm. Infecc. Microbiol. Clin. 18:506-511. (In Spanish.) [PubMed] [Google Scholar]
- 34.Smith, M. S., R. K. Yang, C. W. Knapp, Y. Niu, N. Peak, M. M. Hanfelt, J. C. Galland, and D. W. Graham. 2004. Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl. Environ. Microbiol. 70:7372-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strommenger, B., C. Kettlitz, G. Werner, and W. Witte. 2003. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 41:4089-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szczepanowski, R., I. Krahn, B. Linke, A. Goesmann, A. Puhler, and A. Schluter. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613-3630. [DOI] [PubMed] [Google Scholar]
- 38.Teng, L.-J., P.-R. Hsueh, S.-W. Ho, and K.-T. Luh. 2001. High prevalence of inducible erythromycin resistance among Streptococcus bovis isolates in Taiwan. Antimicrob. Agents Chemother. 45:3362-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakur, S., and W. A. Gebreyes. 2005. Prevalence and antimicrobial resistance of Campylobacter in antimicrobial-free and conventional pig production systems. J. Food Prot. 68:2402-2410. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai, J. C., P. R. Hsueh, H. J. Chen, S. P. Tseng, P. Y. Chen, and L. J. Teng. 2005. The erm(T) gene is flanked by IS1216V in inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus. Antimicrob. Agents Chemother. 49:4347-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.U.S. Department of Agriculture. 2000. Part III: health management and biosecurity in U.S. feedlots, 1999. USDA:APHIS:VS, CEAH, report no. N336.1200. National Animal Health Monitoring System, Fort Collins, CO.
- 43.Whitehead, T. R., and M. A. Cotta. 2001. Sequence analyses of a broad host-range plasmid containing erm(T) from a tylosin-resistant Lactobacillus sp. isolated from swine feces. Curr. Microbiol. 43:0017-0020. [DOI] [PubMed] [Google Scholar]
- 44.Wilson, K., and R. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witte, W. 2000. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents 16:S19-S24. [DOI] [PubMed] [Google Scholar]
- 46.Yu, Z., V. J. J. Martin, and W. W. Mohn. 1999. Occurrence of two resin acid-degrading bacteria and a gene encoding resin acid degradation in pulp and paper mill effluent biotreatment systems assayed by PCR. Microbiol. Ecol. 38:114-125. [DOI] [PubMed] [Google Scholar]
- 47.Yu, Z., F. C. Michel, Jr., G. Hansen, T. Wittum, and M. Morrison. 2005. Development and application of real-time PCR assays for quantification of genes encoding tetracycline resistance. Appl. Environ. Microbiol. 71:6926-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, Z., and M. Morrison. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 36:808-812. [DOI] [PubMed] [Google Scholar]
- 49.Yu, Z., G. R. Stewart, and W. W. Mohn. 2000. Apparent contradiction: psychrotolerant bacteria from hydrocarbon-contaminated Arctic tundra soils that degrade diterpenoids synthesized by trees. Appl. Environ. Microbiol. 66:5148-5154. [DOI] [PMC free article] [PubMed] [Google Scholar]