Abstract
The respiratory pathogen Chlamydia (Chlamydophila) pneumoniae is associated with chronic diseases, including atherosclerosis and giant-cell arteritis, which are accompanied by the occurrence of these obligate intracellular bacteria in blood vessels. There, C. pneumoniae seems to be present in a persistent state. Persistence is characterized by modified bacterial metabolism and morphology, as well as a reversible arrest of chlamydial development. In cell culture, this persistent state can be induced by gamma interferon (IFN-γ). To elucidate this long-term interaction between chlamydiae and their host cells, microarray screening on epithelial HeLa cells was performed. Transcription of persistently (and productively) infected cells was compared with that of mock-infected cells. Sixty-six host cell genes were regulated at 24 h and/or 96 h of IFN-γ-induced persistence. Subsequently, a set of 17 human host cell genes related to apoptosis, cell cycle, or metabolism was identified as permanently up- or down-regulated by real-time PCR. Some of these chlamydia-dependent host cell responses were diminished or even absent in the presence of rifampin. However, other expression patterns were not altered by the inhibition of bacterial RNA polymerase, suggesting two different modes of host cell activation. Thus, in the IFN-γ model, the persisting bacteria cause long-lasting changes in the expression of genes coding for functionally important proteins. They might be potential drug targets for the treatment of persistent C. pneumoniae infections.
The spectrum of illness caused by Chlamydia (also Chlamydophila) pneumoniae ranges from severe community-acquired pneumonia to bronchitis, pharyngitis, laryngitis, or sinusitis (31). Chlamydial infections show high rates of recurrence (3, 11, 12). The increasingly strong association between C. pneumoniae and several chronic conditions, including asthma, follicular conjunctivitis, chronic bronchitis, giant-cell arteritis, and atherosclerosis, has been studied by many research groups (9, 18, 19, 32, 47, 53, 58) and supports the assumption that the organism can persist for extended periods in its human host. Several studies have shown that C. pneumoniae can cause long-lasting and chronic infections that do not respond to chlamydicidal antibiotic treatment (1, 16, 17, 20, 21, 48, 55). Consequently, there is an urgent need to elucidate the mechanisms of persistence, to reveal markers of the persistent infection, and to identify new potential drug targets in host cells and bactericidal drugs that are effective against persisting chlamydia.
The obligate intracellular bacterium C. pneumoniae has a unique productive developmental cycle. An infectious, metabolically inert form called an elementary body (EB) initiates the infection by attaching to and stimulating uptake by the host cell. The internalized EB remains within a host-derived vacuole (inclusion) and differentiates into a larger, metabolically active, and replicative form termed the reticulate body (RB). The RB multiplies by 8 to 12 rounds of binary division. Then, it differentiates asynchronously back into an infectious EB (35), which is released from the host cell 2 to 3 days postinfection (34, 35, 54). During recent years, this classical paradigm of a biphasic developmental cycle of chlamydia had to be revised and supplemented. Under suboptimal growth conditions and in certain cell types (such as monocytes), the chlamydial development from RBs back into infectious EBs is arrested (or at least slowed to below the detection limit of infectious EBs). In this persistent state, chlamydiae exhibit an aberrant morphology and a modified metabolic activity. When conditions become more favorable, persisting chlamydiae can be reactivated to complete their growth cycle.
Such a long-term interaction with the infected host cell can be optimally investigated in cell culture (2, 26). Usually, it can be achieved by a deviation from conventional growth conditions, i.e., by addition of gamma interferon (IFN-γ) or antibiotics (penicillin G) or deprivation of essential nutrients, including iron. In murine pneumonia models, IFN-γ plays a central role in the Th1-dominated immune response to C. pneumoniae infection (42-44). IFN-γ exposure of chlamydial infections in cell culture provides a well-defined model of persistence (39). It reflects in vivo situations where this cytokine is present in concentrations that already modify chlamydial growth but that are not sufficient to achieve bacterial elimination. The most important mechanism responsible for the intracellular effects of IFN-γ is tryptophan depletion by activation of indoleamine 2,3-dioxygenase (38). Additionally, other mechanisms, such as the inducible nitric oxide synthase effector pathway and iron deprivation, are associated with IFN-γ (28).
In productive C. pneumoniae infection, host cell gene expression in epithelial HeLa cells is drastically altered, as was recently shown by our group. These induced host cell responses were strictly dependent on the viability of the chlamydiae: UV-inactivated or heat-treated chlamydiae do not activate HeLa cells (24). Most likely, this biological effect is due to an active bacterial mechanism, such as the translocation of chlamydial effector proteins into the host cell cytosol by the bacterial type III secretion system. Moreover, an apoptosis-inhibitory effect of chlamydia was shown in productive infection. Chlamydia efficiently blocks the release of cytochrome c from mitochondria upon the induction of apoptosis by external stimuli (12). In this context, a new mechanism has been described targeting the proapoptotic BH3-only proteins Bim, Puma, and Bad for proteasomal destruction (13). In leukocytes, additional signaling cascades are triggered via Toll-like receptors. Thus, in contrast to HeLa cells and several other cell types, monocytes respond to UV-inactivated or heat-inactivated chlamydia (41).
However, it is still not clear how host cells are modified in persistence. Only a few responses of HeLa cells have recently been investigated by us in three models of long-term (24 h, 96 h, and 7 days postinfection) C. pneumoniae persistence: interleukin 6 (IL-6), IL-8, IL-11, LIF, connective tissue growth factor, and the transcription factors EGR-1 and ETV-4. Intriguingly, this direct comparison of persistence induced by IFN-γ, penicillin G, or depletion of iron revealed two different modes of host cell reaction 96 h postinfection (i.e., at a later time point, when the persistence-causing conditions are achieved) (41). In the IFN-γ model (as well as in the penicillin G model), regulation of the above-mentioned host cell genes returns to basal expression levels, i.e., their mRNA levels are indistinguishable from those of mock-infected cells. Additionally, the host cell signal transduction triggered by other stimuli is altered in persistently infected HeLa cells in the IFN-γ model (41). This suggested a general silencing of the infected host cells in the presence of IFN-γ, which may help to suppress inflammation and to prevent recognition of persistently infected cells by the immune system. However, the number of analyzed genes was very small.
In the present study, we wanted to clarify in a broader approach whether infected host cells are exclusively silenced at a time point late in IFN-γ-induced persistence or if they show chlamydia-dependent up- or down-regulation of host cell genes (compared to mock-infected cells). To evaluate this gene expression pattern and to identify such responses during IFN-γ-induced persistence, we analyzed HeLa cells with Affymetrix microchips (HG-U133A) 24 h and, more importantly, 96 h postinfection, comparing persistent and productive C. pneumoniae infections (versus mock-infected cells). Several human genes were selected for detailed expression profiling by semiquantitative real-time PCR. Our study demonstrates that C. pneumoniae causes long-lasting changes in the expression of genes involved in antiapoptosis, cell cycle control, and host cell metabolism in IFN-γ-induced persistence.
MATERIALS AND METHODS
Cell and chlamydial culture.
Human cervical epithelial HeLa-cells (kindly provided by R. Heilbronn, Berlin, Germany) were cultured in Earle's minimal essential medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 0.1 M nonessential amino acids, and 1 mM sodium pyruvate (Biochrom, Berlin, Germany). Native cells were grown at 37°C and 5% CO2. C. pneumoniae CWL-029 (ATCC) was propagated in HeLa cells as recently published (41). To obtain buffer samples for an optimal mock control, the complete purification procedure for EBs was performed in parallel in the absence of C. pneumoniae.
IFN-γ model of persistence in HeLa cells.
Infection with C. pneumoniae (multiplicity of infection [MOI] = 30 or MOI = 3) was performed in RPMI 1640 medium (10% fetal calf serum, 0.1 M nonessential amino acids, and 10 mM HEPES) by centrifugation (55 min; 35°C; 2,000 × g). To obtain a defined starting point for kinetic studies (time zero), the infected cells were washed with 1× phosphate-buffered saline and incubated with fresh medium. After 30 min postinfection, the medium was replaced with medium containing 100 U ml−1 of IFN-γ (R&D Systems, Wiesbaden, Germany) in order to induce persistence. The minimal concentration of the persistence-inducing IFN-γ was selected, as evaluated by Peters et al. (41), as the concentration at which less than 1% infectious EBs were recoverable after 4 days compared to productive infection. Daily exchange of the medium with IFN-γ assured constant concentrations of growth factors and the persistence inducer. If not otherwise indicated, an MOI of 30 was used in all HeLa cell experiments. Under these conditions, an infection efficiency of more than 95% was achieved, as determined by immunofluorescence. With an MOI of 3 used in one experiment, an infection efficiency of more than 90% was achieved. Infections were performed in 12-well cell culture dishes with 8.6E5 cells and in 1.5 ml medium per well. For productive infection compared to persistence, RPMI 1640 including 10% fetal calf serum, 0.1 M nonessential amino acids, and 10 mM HEPES was used. In experiments comparing the effects of metabolically active and inactive C. pneumoniae, rifampin (1 μg/ml) was added to the cell culture medium directly after infection, as well as together with the daily exchanged medium. Control experiments showed no propagation of C. pneumoniae under these conditions. Infected cells were always grown at 35°C and 5% CO2.
Electron microscopy.
For transmission electron microscopy, monolayers of HeLa cells were infected with C. pneumoniae (MOI = 30) and treated with IFN-γ or left untreated (productive infection). The infected cells were trypsinized at 48 h postinfection, pelleted by centrifugation, and fixed with 2.5% glutaraldehyde buffered in 0.1 M Na-cacodylate-HCl (pH 7.3) for 12 h at 4°C. After postfixation with 2% osmium tetroxide dissolved in 0.1 M Na-cacodylate-HCl (pH 7.3) for 1 h, the specimens were dehydrated in graded ethanols and embedded in Epon. Thin sections stained with uranyl acetate and lead citrate were examined in a Philips EM 301 electron microscope. The electron micrographs were selected, digitized, and processed using Adobe Photoshop 6.0. The relative amounts of EBs, intermediate bodies, and RBs as described previously (22, 39), as well as of aberrant large bodies, were calculated.
Nucleic acid extraction.
DNA used for bacterial-load analysis was isolated using the RNeasy Mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Cells were harvested by removal of the supernatant and addition of 350 μl RLT buffer per well. After the first round of extraction, 10 μl eluate was stored to measure the bacterial load. The rest of the eluate was reextracted by additional on-column DNase digestion. RNA was eluted in 50 μl RNase-free water.
RNA quality assurance and quantitation.
RNA quality and quantity were analyzed on the Agilent Bioanalyzer using the RNA 6000 Nano chip (Agilent Technologies Deutschland GmbH, Böblingen, Germany). In addition, RNA was quantitated by optical density measurement.
cDNA synthesis.
RNA used for transcript analysis by real-time PCR was reverse transcribed with a RevertAid H− First Strand cDNA Synthesis kit (MBI Fermentas, St. Leon-Rot, Germany). As a reference, FirstChoice Human Cell Line Total Cervical Adenocarcinoma RNA (Ambion Europe Ltd., Huntingdon, Cambridgeshire, United Kingdom) was used. For microarray analysis, cDNA was synthesized using the ExpressArt mRNA Amplification Micro Kit (Artus GmbH, Hamburg, Germany) according to the manufacturer's instructions. cDNA was quantitated by optical density measurement. For all experiments, samples without the addition of reverse transcriptase were run in parallel as an additional control.
Microarray analysis.
RNA transcription labeling was performed using the BioArray HighYield RNA Transcript Labeling kit (T7) (Enzo, Farmingdale, NY). cRNA from two independent preparations was hybridized onto Affymetrix human genome U133A chips (duplicates). Scanning and image analysis were done with Affymetrix Microarray Suite software (version 5.0). The Affymetrix standard normalization technique was used (global scaling; normalization factor, 1,000). Microarray data analysis was performed using Spotfire Software (Spotfire, Somerville, MA). The results were filtered using the following values: change call = increase (I) or decrease (D), I and signal/log ratio (SLR) > 1.32 (i.e., a factor of 2.5), or D and SLR < −1.32; α1 = 0.05; α2 = 0.065; τ = 0.015; TGT = 1,000. The detection algorithm used probe pair intensities to assign a present, marginal, or absent call. A score was calculated for each probe pair and compared to a predefined threshold, tau (τ = 0.015). Probe pairs with scores higher than tau voted for the presence of the transcript. Probe pairs with scores lower than tau voted for the absence of the transcript. In a comparison analysis, two samples, hybridized to two microarrays of the same type, were compared against each other in order to detect and quantify changes in gene expression. One array was designated as the baseline and the other as an experiment (treatment versus mock treatment at the corresponding time point). As an additional filter mechanism, only altered gene expressions with present calls in both samples were selected. Expression changes between two arrays were designated “change” (n-fold) and were defined as the ratio between the normalized intensities of the two arrays. Comparison analysis was done with Microsoft Excel X for Mac (Microsoft GmbH, Unterschleißheim, Germany).
Statistical analysis.
Statistical analysis was performed using SPSS 11 for Mac OS X (SPSS Inc., Chicago, IL) and Microsoft Excel X for Mac. A probit analysis to determine the limit of detection for the RealArt C. pneumoniae TM PCR Kit (Artus GmbH) was performed using PriProbit version 1.63 designed by Masayuki Sakuma (Kyoto University, Kyoto, Japan) (http://bru.gmprc.ksu.edu/proj/priprobit/index.asp).
Real-time PCR.
The bacterial load was measured in each sample for C. pneumoniae DNA, as well as for cDNA. DNAs and cDNAs from three independent preparations were analyzed quantitatively according to the manufacturer's instructions using the RealArt C. pneumoniae TM PCR Kit (Artus GmbH), with each sample run in triplicate. All real-time PCRs were run on the ABI PRISM 7000 SDS (Applied Biosystems, Weiterstadt, Germany).
The real-time PCR primer sequences can be found in Table S1 in the supplemental material. Primers and probes were designed using Primer Express software (version 1.0; Applied Biosystems). TaqMan probes were labeled at the 5′ end with the reporter dye molecule 6-carboxyfluorescein (emission wavelength, 518 nm) and at the 3′ end with the black-hole quencher dye BHQ1. A list of the TaqMan predeveloped assay reagents (Applied Biosystems) used can be seen in Table S2 in the supplemental material. Each real-time PCR sample was run in quadruplicate (and in triplicate in the rifampin experiment [see Fig. 5]) on each of three independent experiments at an MOI of 30 and two independent experiments at an MOI of 3. Statistical evaluation for gene regulation by using a twofold regulation cutoff was performed by one-way analysis of variance (P ≤ 0.05).
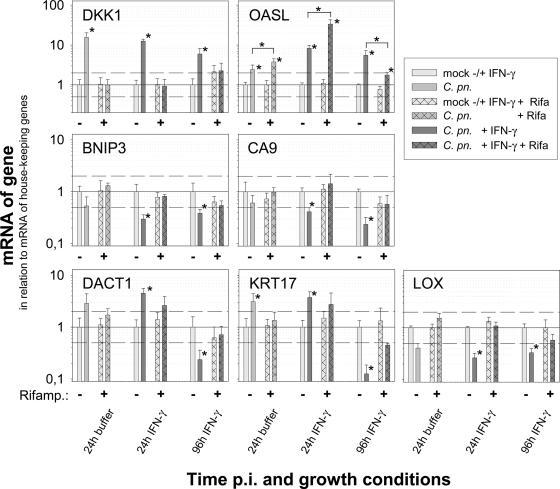
FIG. 5.
The effect of rifampin on host cell gene regulation induced by C. pneumoniae in productive infection (24 h) and IFN-γ-induced persistence (24 and 96 h postinfection [p.i.]). Depicted are means plus standard deviations (SD) of the normalized log10 of the relative amounts of mRNA of genes determined by two-step real-time PCR in mock-infected and infected HeLa cells. The figures summarize the results of three independent infection experiments (A, B, and C). In each experiment, the real-time PCR was performed in triplicate for seven selected candidate genes and in duplicate for the two genes that were used for normalization (TBP and 18S rRNA). For better comparison of the rifampin effect, the amount of mRNA determined for each experimental condition was set to 1. The results for mock-infected cells and the corresponding C. pneumoniae-infected cells are depicted as pairs for each condition. The changes (n-fold) of the mock control in the absence of rifampin caused by 24 h or 96 h of IFN-γ treatment were as follows: 0.9/1.1 for DKK1, 2.6/2.0 for OASL, 1.2/2.5 for BNIP3, 0.8/6.2 for CA9, 0.8/5.3 for DACT1, 3.5/8.4 for KRT17, and 1.4/3.7 for LOX. To provide some orientation, dotted lines are drawn at relative mRNA amounts of 2.0 and 0.5. An asterisk indicates a significant change (P ≤ 0.05). The two genes with the highest factor of regulation are in the top row; the others are in alphabetical order.
Real-time PCR data were usually normalized to the levels of three simultaneously used endogenous controls (beta-glucuronidase [GUS], TATA-box binding protein [TBP], and 18S rRNA), which were run in triplicate in each experiment. In the rifampin experiment, only GUS and TBP were used as housekeeping genes. Relative quantitation was performed using the standard-curve method on FirstChoice Human Cell Line Total Cervical Adenocarcinoma RNA. For each experimental sample, the amounts of target and endogenous control were determined from the appropriate standard curve. Then, the target amount was divided by the endogenous control amount to obtain a normalized target value. As three or two endogenous controls were used for the target gene, three or two different normalized target values were obtained. Moreover, each of the normalized target values was divided by the normalized corresponding mock-infected control sample to generate the relative expression sample. For each analyzed gene, negative and no-reverse transcriptase (RT) controls were performed to rule out genomic-DNA contamination. The PCR efficiencies of all assays were greater than 90%, and the calculated R2 value for each standard curve was at least 0.99.
From a total of 66 human genes found to be regulated in the microarray (see above), 19 genes were selected for real-time PCR analysis based on the following criteria. (i) The selected genes had to belong to different important functional groups. (ii) For a persistence model, 96 h in the presence of IFN-γ is considered the most important time point and experimental condition, respectively. Nevertheless, genes were also selected when they were regulated under other conditions in the screening, because by applying a stringent selection procedure, regulation observed in the microarray can almost always be confirmed by real-time PCR. However, negative results in the array cannot be equally trusted, as the more sensitive real-time PCR will often be able to show regulation for samples with seemingly negative results in the microarray (24). (iii) Within these groups, genes with a relatively high factor of regulation were selected, assuming that a drastic change in the regulation of a gene should have a larger influence on the host cell. (iv) To focus on novel information, genes which had already been described as being regulated by chlamydia were excluded.
RESULTS
IFN-γ persistence model in epithelial HeLa cells.
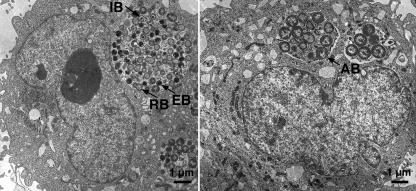
A cell culture model of chlamydial persistence in HeLa cells (in the absence of cycloheximide) was optimized to permit observation periods of up to 1 week, thereby best characterizing the time point 4 days (i.e., 96 h) postinfection. Details of this IFN-γ model have recently been published by us, clearly demonstrating key features of chlamydial persistence (41). In summary, these were (i) almost complete lack of infectious EBs on day 4 or day 7, (ii) the persisting chlamydia could be reactivated after removal of IFN-γ and addition of l-tryptophan on day 4 postinfection by 40% compared to productive infection, and (iii) typical changes in morphology of the altered RB-like forms became apparent within the first 2 to 3 days and were sustained during the entire observation period (as depicted here in Fig. 1 and Table 1). Moreover, the expression of four analyzed host cell genes did not alter between days 4 and 7, thus indicating that a new steady state in transcription was achieved during this period.
FIG. 1.
Typical changes in morphology of the RB-like aberrant forms of C. pneumoniae in IFN-γ-induced persistence (right) compared to productive infection (left) determined by transmission electron microscopy in HeLa cells at 48 h postinfection. A statistical evaluation can be found in Table 1.
TABLE 1.
Relative amounts of developing C. pneumoniae forms in inclusions
| HeLa cells infected with C. pneumoniae for 48 h | Amt (%)a
|
|||
|---|---|---|---|---|
| EB | Intermediate bodies | RB | Aberrant bodies | |
| Untreated | 19 ± 2 | 28 ± 2 | 53 ± 4 | 0 |
| Treated with IFN-γ | 0 | 5 | 37 ± 2 | 58 ± 2 |
Relative amounts of developing C. pneumoniae forms in inclusions of HeLa cells analyzed by transmission electron microscopy 48 h postinfection in productive or persistent infection, i.e., in the presence of IFN-γ. The results are highly significant, as shown by variance analysis.
To enable direct comparison with the data obtained in our previous investigation (41) and to achieve high sensitivity in the screening, an MOI of 30 was used again in the present study for microarrays and most real-time PCRs. Additionally, one real-time PCR experiment was performed with an MOI of 3, as indicated. Based on our knowledge of this persistence model, the analyses were performed on three pairs of samples within this study: (i) after 24 h of productive infection, as well as (ii) after 24 h and (iii) after 96 h of IFN-γ-induced persistent infection. The analyses always compared infected HeLa cells and the corresponding mock-infected HeLa cells.
Determination of bacterial load by real-time PCR of chlamydial DNA.
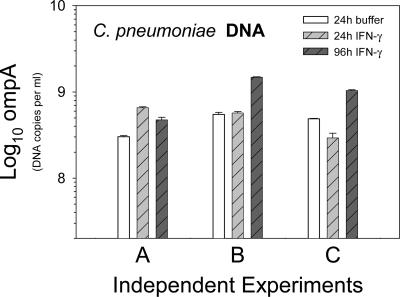
Total RNA, obtained in three sets (A, B, and C) of biologically independent experiments, was later analyzed by real-time PCR. To confirm similar rates of infection in the corresponding samples, ompA DNA levels were determined by the Artus C. pneumoniae TM PCR Kit. The bacterial loads differed only slightly between the samples of one biological replicate, and they were also very similar when the three independent sets were compared (Fig. 2). In the mock-infected control samples, no C. pneumoniae DNA was detected. Thus, contamination during DNA preparation could be ruled out.
FIG. 2.
Determination of DNA for ompA of C. pneumoniae demonstrating similar bacterial loads in the three biologically independent infection experiments (A, B, and C) that were analyzed by real-time PCR. The amount of ompA DNA was determined in triplicate for each of the three infection experiments. Depicted are means plus standard deviations.
Screening of host cell gene regulation in persistence using microarray technology.
To test the hypothesis that as-yet-unknown pathways might be differentially regulated by C. pneumoniae, the transcriptional response of HeLa cells was examined in two independent experiments using Affymetrix U133A human genome chips consisting of 22,284 human gene probe sets. This analysis was performed on the three pairs of samples obtained 24 h or 96 h postinfection.
The two independent microarray experiments correlated with a mean Pearson coefficient of 0.99 (standard deviation, 0.0028). Sixty-six human genes were differentially regulated during C. pneumoniae infection (see Table S3 in the supplemental material) under at least one of the three experimental conditions (treatment versus mock treatment at the corresponding time point) by ≥2.5-fold (and present calls in both samples). The calculated mean changes for the 19 host cell genes that were selected for real-time PCR analysis are given in Table 2.
TABLE 2.
Gene expression data in microarray for the 19 selected probe sets
| Homo sapiens gene or gene product | Probe set | Accession no. | Mean change (n-fold) vs. mock (n = 2)a
|
||
|---|---|---|---|---|---|
| 24 h Cpn | 24 h Cpn + IFN-γ | 96 h Cpn + IFN-γ | |||
| 2′-5′oligoadenylate synthetase-like (OASL) | 205660_at | NM_198213 | NC | I (5.2/5.5) | I (1.9/3.7) |
| 2′-5′oligoadenylate synthetase-like (OASL) | 210797_s_at | NM_198213 | NC | I (5.6/6.2) | I (2.2/3.0) |
| Adlican | 209596_at | AF245505.1 | D (1.7/1.8) | D (NC/1.5) | D (3.5/3.2) |
| BCL2 adenovirus E1B 19-kDa interacting protein 3 (BNIP3) | 221478_at | NM_004052 | D (2.6/2.1) | D (2.2/1.8) | D (2.8/2.6) |
| BCL2 adenovirus E1B 19-kDa interacting protein 3 (BNIP3) | 201848_s_at | NM_004052 | D (3.4/1.5) | NC | D (3.0/3.3) |
| Carbonic anhydrase IX (CA9) | 205199_at | NM_001216 | D (2.3/2.2) | D (NC/1.8) | D (15.6/7.9) |
| Cystathionase (cystathionine gamma-lyase) (CTH) | 206085_s_at | NM_001902 | D (1.2/1.1) | D (4.5/2.9) | D (NC/1.8) |
| Cystathionase (cystathionine gamma-lyase) (CTH) | 217127_at | NM_001902 | D (2.1/1.3) | D (5.7/3.9) | D (NC/1.3) |
| Cysteine-rich, angiogenic inducer, 61 (CYR61) | 210764_s_at | AF003114.1 | I (5.7/4.1) | I (2.5/1.8) | I (1.6/NC) |
| Dapper homolog 1 (DACT1) | 219179_at | NM_016651 | NC | D (1.9/2.7) | D (2.9/2.5) |
| Dickkopf (Xenopus laevis) homolog 1 (DKK1) | 204602_at | NM_012242 | I (4.0/4.2) | I (2.5/2.4) | I (3.5/4.1) |
| Hairy/enhancer-of-split related with YRPW motif 1 (HEY1) | 44783_s_at | NM_012258 | D (4.2/3.7) | D (4.9/5.7) | D (1.4/NC) |
| Heat shock 70-kDa protein 1A (HSPA1A) | 200799_at | NM_005345 | NC | I (10.3/7.3) | D (2.0/1.7) |
| Heat shock 70-kDa protein 1A (HSPA1A) | 200800_s_at | NM_005345 | NC | I (14.4/11.8) | D (1.6/1.7) |
| Heat shock 70-kDa protein 1B (HSPA1B) | 202581_at | NM_005346 | D (NC/1.5) | I (13.6/9.0) | D (NC/1.7) |
| Insulin-induced gene 1 (INSIG1) | 201626_at | NM_005542 | D (3.9/3.2) | D (2.3/1.9) | D (1.8/3.5) |
| Insulin-induced gene 1 (INSIG1) | 201627_s_at | NM_005542 | D (3.0/3.8) | D (2.6/2.3) | NC |
| Interferon-induced, hepatitis C-associated microtubular aggregate protein (44 kDa) (MTAP44 or IFI44) | 214453_s_at | NM_006417 | I (1.7/NC) | I (5.4/5.0) | I (2.4/3.8) |
| Interferon-induced, hepatitis C-associated microtubular aggregate protein (44 kDa) (MTAP44 or IFI44) | 214059_at | NM_006417 | NC | I (3.3/4.4) | NC |
| Keratin 17 (KRT17) | 212236_x_at | NM_000422 | I (2.1/2.0) | I (1.9/2.0) | D (3.5/3.9) |
| Keratin 17 (KRT17) | 205157_s_at | NM_000422 | I (2.4/2.0) | I (1.6/.03) | D (3.0/5.4) |
| Lysyl oxidase (LOX) gene | 215446_s_at | L16895 | D (2.1/2.3) | D (2.5/2.1) | D (4.2/3.6) |
| N-myc downstream regulated (NDRG1) | 200632_s_at | NM_006096 | D (4.1/2.2) | D (3.0/2.1) | D (7.1/4.1) |
| Prostaglandin E receptor 4 (subtype EP4) (PTGER4) | 204897_at | NM_000958 | I (2.2/2.0) | I (2.4/2.3) | I (2.8/3.9) |
| Retinoic acid induced 3 (RAI3) | 203108_at | NM_003979 | I (3.8/2.7) | I (2.8/3.4) | I (2.0/1.5) |
| Serum-inducible kinase (SNK) or polo-like kinase 2 (PLK2) | 201939_at | NM_006622 | I (2.4/4.2) | NC | I (3.7/4.8) |
Altered gene expression data for selected Affymetrix HG-U133A probe sets. All 19 genes were selected for real-time two-step RT-PCR analysis. Expression changes between two arrays are designated “change (n-fold)” (treatment versus mock treatment at the corresponding time point). Depicted are the changes (D, decrease; I, increase; NC, no change) in two independent microarray experiments of C. pneumoniae-infected HeLa-cells (24 h Cpn = productive infection) in the IFN-γ persistence model (24 h and 96 h Cpn + IFN-γ). All selected genes had signal log ratios greater than 1.32 or lower than −1.32 (α1 = 0.05; α2 = 0.065; τ = 0.015) under at least one experimental condition and present calls in both samples.
Host cell gene expression profiling of persistent C. pneumoniae infection by real-time PCR.
To further characterize our DNA microarray findings, we performed semiquantitative real-time PCRs for 19 HeLa cell genes representing different functional groups (Table 2 lists the complete gene name, abbreviation, probe set, and accession number). Additional criteria for the selection of these genes can be found in Materials and Methods. This analysis was performed on RNAs obtained in three independent experiments. Three eukaryotic housekeeping genes were simultaneously used in each experiment as endogenous controls (see Materials and Methods) in order to obtain accurate normalization and to compensate for small variations. Real-time PCRs (two-step RT-PCRs) were set up for these endogenous controls and for eight genes from microarray screening (for primer and probe sequences, see Table S1 in the supplemental material). For the remaining 11 genes, TaqMan predeveloped assay reagents were used (Applied Biosystems) (see Table S2 in the supplemental material).
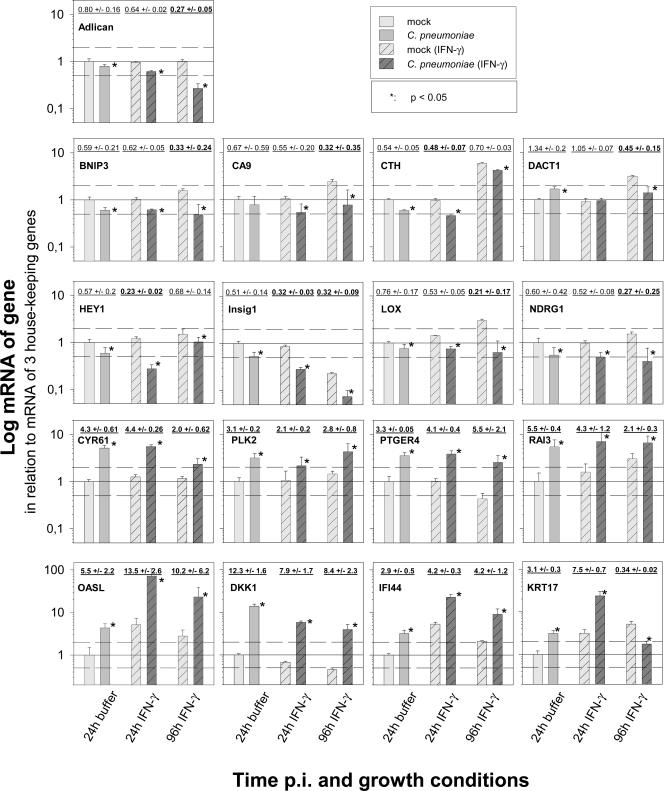
As expected, the basal expression level of several host cell genes after mock infection changed slightly due to the treatment with IFN-γ. Therefore, mRNA levels for mock-infected controls and for C. pneumoniae-infected cells are depicted in parallel for direct comparison in Fig. 3. Statistically significant changes between infected cells and the corresponding mock-infected controls are indicated. The calculated factors of regulation are also depicted above the pair of corresponding columns. The real-time PCR results revealed two main groups of C. pneumoniae-induced gene regulation. Seven host cell genes were permanently up-regulated in IFN-γ-induced persistence (CYR61, PLK2, PTGER4, RAI3, OASL, DKK1, and IFI44) (Fig. 3, bottom). Eight genes were permanently down-regulated compared to mock-infected HeLa cells (Adlican, BNIP3, CA9, CTH, HEY1, Insig1, LOX, and NDRG1) (Fig. 3, top). Chlamydia-induced down-regulation was less pronounced, with a minimum regulation of approximately 20% compared to mock-infected cells. Only two genes (KRT17 and DACT1) (Fig. 3; also see Fig. 5) were up-regulated 24 h postinfection but down-regulated 96 h postinfection. The regulation of two genes (HASPA1-A and HSPA1-B) observed in the microarray could not be confirmed, either by real-time PCR, even using three different sets of primers, or by Northern blotting (data not shown). Thus, they were most likely microarray artifacts.
FIG. 3.
Host cell gene regulation induced by C. pneumoniae in productive infection (24 h) and IFN-γ-induced persistence (24 and 96 h postinfection). Depicted are means plus standard deviations (SD) of the normalized log10 of the relative amount of mRNA of genes determined by two-step real-time PCR in mock-infected and C. pneumoniae-infected HeLa cells. The figures summarize the results of three independent infection experiments (A, B, and C). In each experiment, the real-time PCR was performed in quadruplicate for the 17 candidate genes and in triplicate for the 3 genes that were used for normalization (GUS, TBP, and 18S rRNA). For better comparison, the amount of mRNA determined after 24 h of productive infection was set to 1. To provide some orientation, dotted lines are drawn at relative mRNA amounts of 2.0 and 0.5. Additionally, the calculated factor of regulation (mean ± SD), in comparing chlamydia-infected cells and mock-infected cells, can be found above the corresponding pair of columns. Calculated factors of differential regulation above 2.0 and below 0.5 are indicated in boldface. An asterisk indicates a significant change (P ≤ 0.05). There was no regulation of HASPA1-A and HSPA1-B as confirmed by Northern blotting (data not shown). Therefore, the regulation observed in the microarray was most likely an artifact.
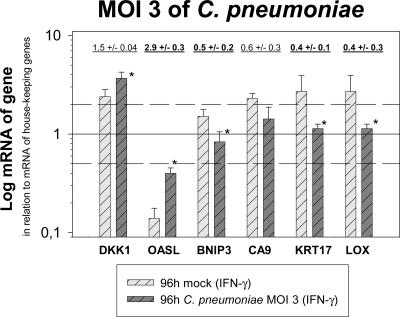
The experiments performed with an MOI of 30 demonstrate the principal ability of chlamydia to alter host cell gene expression under conditions where these regulations can be optimally observed. Such a large number of EBs might also be found locally when productively infected cells lyse. To ensure that the results obtained in these experiments also provide information for other situations (i.e., at a greater distance to lysed productively infected cells, leading to a decreased number of infectious EBs), real-time PCR analysis was repeated for some of the genes, using an MOI of 3 for C. pneumoniae in the IFN-γ persistence model. Three genes that were strongly regulated at an MOI of 30 (OASL, DKK1, and KRT17), as well as three genes with a lower factor of regulation (BNIP3, CA9, and LOX), were selected. Four of these genes were down-regulated in the high-MOI real-time PCR experiment 96 h postinfection in the persistence model, and two were up-regulated. This comparison focused on the best-characterized time point of persistence. The modified experiment revealed concordant results and the same pattern of functional responses to persisting C. pneumoniae 96 h postinfection as described above for a higher MOI. However, as expected, the factor of regulation was smaller: OASL and DKK1 were again up-regulated, while KRT17, BNIP3, CA9, and LOX were again down-regulated compared to the corresponding mock controls (Fig. 4). The effect of regulation in infected HeLa cells is most likely stronger than calculated, because the mRNA of noninfected and therefore still dividing cells (∼30 to 40% after 96 h in the absence of cycloheximide at the lower MOI) was included in the analysis of the host cell gene expression. Taken together, the results obtained with an MOI of 3 confirm the pattern of gene expression observed with an MOI of 30.
FIG. 4.
Host cell gene regulation induced by C. pneumoniae in IFN-γ-induced persistence at the most relevant time point, 96 h postinfection with an MOI of 3, confirms the data obtained with an MOI of 30. Depicted are means plus standard deviations (SD) of the normalized log10 of the relative amounts of mRNA of a selection of genes determined by two-step real-time PCR in mock-infected and infected (MOI = 3) HeLa cells. The figures summarize the results of two independent infection experiments. In each experiment, the real-time PCR was performed in quadruplicate for the candidate genes and in triplicate for the three genes that were used for normalization (GUS, TBP, and 18S rRNA). Calculated factors of regulation (mean ± SD), comparing chlamydia-infected and mock-infected cells above 2.0 and below 0.5, are indicated in boldface. An asterisk indicates significant changes (P ≤ 0.05). As examples, three genes with a high factor of regulation at an MOI of 30 (OASL, DKK1, and KRT17), as well as three genes with relatively low factors of regulation (BNIP3, CA9, and LOX), were selected for the experiment. Four of these genes were down-regulated in the high-MOI real-time PCR 96 h postinfection in the persistence model, and two were up-regulated. The genes are in the same order as in Fig. 5.
Moreover, to clarify whether host cell responses were dependent on chlamydial metabolism, the infection experiment at an MOI of 30 was repeated for the same six host cell genes with a new preparation of C. pneumoniae in the absence or presence of rifampin (Fig. 5). DACT1 was also included because the gene (and KRT17) had shown opposite regulation in productive infection and the late time point of persistent infection (Fig. 3), an observation that could be confirmed in this new set of experiments. Intriguingly, different patterns were observed in evaluating three independent rounds of infection. The up-regulation of DKK1 mRNA at 24 h of productive infection, as well as at 24 h and 96 h of persistent infection, was completely inhibited by the antibiotic blocking bacterial RNA polymerase. Similarly, the C. pneumoniae-induced decreases in BNIP3 and CA9 mRNAs were completely dependent on bacterial metabolism. In contrast, the strong up-regulation of OASL by C. pneumoniae 24 h postinfection in the absence or the presence of IFN-γ were not influenced (or sometimes were even higher) when the bacterial RNA polymerase was blocked. For DACT1, KRT17, and LOX, the response seemed to be to some extent dependent on rifampin and thus on chlamydial metabolism.
DISCUSSION
In recent years, it has become apparent that chlamydiae can switch in cell culture and in the living organism to a metabolically and morphologically altered and difficult to eliminate persistent form (26). Thus, persistence has recently become a central issue of chlamydial research. To elucidate the biology of persistence and its implications, cell culture models can provide valuable information. Experimental conditions are precisely defined, high rates of relatively synchronized infection can be achieved, and signaling pathways can be easily dissected. Therefore, the molecular interaction between chlamydiae and their host cells can be investigated in a very sensitive system. Additionally, due to relatively strong cell responses and relatively low variations, screening methods, such as microarrays, can be applied with less difficulty. Of course, the cell culture models cannot replace animal models or the analysis of human tissue; however, they can guide us in the design of such studies. In particular, they can help to select good candidates for specific investigations in situ. For example, up-regulation of Egr-1 by chlamydiae was first identified by microarray and RT-PCR in HeLa cells in our laboratory (25). Meanwhile, this regulation has been confirmed for C. pneumoniae in vascular tissue (46).
IFN-γ induction, used in this study to identify such candidates in cell culture, is often considered a key model for chlamydial persistence and is certainly the best-studied one. It has several advantages compared to others models. In mice, IFN-γ plays a central role in the defense against C. pneumoniae (15, 36, 43-45, 52). The penicillin G model leads to similar host cell responses, suggesting that both models represent expression patterns that can be induced by various conditions. In the IFN-γ model, persistence is clearly reversible, as aberrant bodies can be efficiently reactivated into the productive cycle. However, it should be noted that variations in cell biology between different persistence models can and will occur, perhaps representing different in situ situations (41).
The present study was conducted to determine whether C. pneumoniae silences host cell responses in general, as formerly observed for individual genes, or if other changes in the expression of host cell genes can be identified in persistence which might ameliorate the survival of the bacteria and might influence the pathogenesis. The latter hypothesis of a modified expression pattern of persistently infected cells (compared to mock-infected cells) was confirmed. The observed changes on the mRNA level concern genes that participate in key responses and pathways of the infected cells: apoptosis and antiapoptosis, respectively; the cell cycle; and host cell metabolism and other cellular functions.
Regulated host cell genes that modify apoptosis and the cell cycle.
The proapoptotic regulator BNIP3 (59) was found to be down-regulated. The permanently up-regulated cysteine-rich 61 (CYR61/CCN1) is an immediate-early gene of the CCN family. CYR61 is not only able to control the growth of fibroblasts and epithelial cells, but also to induce or suppress apoptosis in a cell-type-dependent manner by activating Wnt, NF-κB, or the tyrosine kinase signaling pathway (6). Intriguingly, several factors that interfere with the Wnt/β-catenin pathway were found to be regulated in the C. pneumoniae infection model. The binding of Wnt to the membrane Frizzled-LRP5/6 receptor complex causes the stabilization of β-catenin, the major effector of the canonical Wnt signaling pathway, which forms heterodimeric complexes with transcription factors of the TCF family (4) and leads to the expression of a variety of TCF target genes. Wnt signaling controls a wide variety of cell processes, including cell fate specification, differentiation, migration, and polarity (40). Dickkopf-1 (DKK1), up-regulated by C. pneumoniae, encodes a secreted Wnt antagonist that binds to LRP-5/6, resulting in the inhibition of the canonical pathway and the absence of expression of TCF target genes (37). At 96 h of persistent C. pneumoniae infection, Dapper homolog 1 (DACT1; also called LOC51339), an inhibitor of Wnt/beta-catenin signaling, was down-regulated. Thus, up-regulation of DKK1 and down-regulation of DACT1 seem to counteract each other in respect to Wnt inhibition. In the rifampin experiment, only the up-regulation of DKK1 was completely dependent on chlamydial metabolism. This suggests that the two seemingly counteracting responses of DKK1 and DACT-1 might be induced differently—on one hand directly by metabolically active chlamydiae, and on the other hand by a compensating reaction of the host cell. In any case, our data indicate that the Wnt/β-catenin pathway is modified during chlamydial infection. However, the biological net effect at a certain time point cannot be predicted. The observed down-regulation of DACT1 might result in β-catenin stabilization and up-regulation of c-myc and might therefore be responsible for the observed down-regulation of n-myc downstream regulated (NDRG1), which is necessary for p53-dependent apoptosis (49).
Activation of the tumor suppressor protein p53 can lead either to cell cycle arrest and DNA repair or to apoptosis. The regulation of p53 is controlled by various mechanisms (51). A common feature in the regulation of p53 activity is its stability control. One key molecule in this process is HDM2 (the human homolog of MDM2), which directs the nuclear export and degradation of p53 in a ubiquitin-dependent way (23). Hairy/E(spl)-related with YRPW motif 1 (HEY1) activates p53 through repression of HDM2 transcription (27). During C. pneumoniae infection, HEY1 was down-regulated. This decreased expression of HEY1 might support the activation of HDM2 and thus inhibit p53-induced apoptosis. Recently, Wu and colleagues (56) revealed the retinoic acid-induced protein 3 gene (RAI3) to be a novel p53 transcriptional target functioning as an antiapoptotic gene. Thus, up-regulation of antiapoptotic RAI3 seems to be part of the C. pneumoniae strategy for long-term intracellular survival. The permanently up-regulated polo-like kinase 2 gene (PLK2/SNK) is a transcriptional target of p53 induced by genotoxic stress and seems to be a mitotic checkpoint (5). Summarizing the observed regulations involving p53, this proto-oncogene seems to participate on several levels in the C. pneumoniae-induced changes. However, it is difficult to predict how these signal cascades interact with each other and how they functionally influence the cell fate.
Dorai and colleagues (8) discovered that the carbonic anhydrase 9 protein (CA9) seems to be involved in the activation of the Akt pathway, which has a distinct antiapoptotic function and promotes cell survival. The observed down-regulation of CA9 might lead to suppression of this antiapoptotic effect.
Clearly, various host genes that modify apoptosis are differentially regulated during C. pneumoniae infection. The delicate balance of pro- and antiapoptotic effects might decide the fate of the infected cell. Most changes suggest the dominance of an antiapoptotic response in chlamydial persistence. However, a different type of study must address this issue.
Changes involving host cell metabolism and other functions.
C. pneumoniae seems to interact with the cysteine biosynthesis of the host cell by the down-regulation of cystathionase (CTH). Insig1 is a protein of the endoplasmic reticulum. It blocks proteolytic activation of sterol regulatory element-binding proteins (SREBPs), membrane-bound transcription factors that activate the synthesis of cholesterol and fatty acids (57). Thus, the observed decrease in Insig1 might lead to activation of cholesterol and lipid synthesis. Adlican, which was also down-regulated, is predicted to be a cell adhesion proteoglycan (50).
Prostaglandin receptors are involved in inflammation and defense. The up-regulation of prostaglandin E receptor 4 gene (PTGER4) expression is consistent with the observations of Fukuda et al. (14), who described a C. trachomatis LGV2-induced increase in PTGER4 expression in HeLa cells. Lysyl oxidase (LOX), which was down-regulated in our C. pneumoniae infection model, is not only the initiator of cross-linking of elastin and collagen, but also acts as a potent chemoattractant for human peripheral blood mononuclear cells (33).
The intermediate filament protein keratin 17 gene (KRT17) showed a changing gene expression from increase (24 h) to decrease (96 h) during C. pneumoniae infection in this study. KRT17 seems to participate in C. pneumoniae-induced modifications of the cytoskeleton (29). Notably, it has been shown recently that related keratin 8 is cleaved by chlamydial secreted protease (7). As suggested by the down-regulation at a late time point in the persistence model, cytoskeleton modifications might no longer be necessary when persistence is accomplished.
In our experiments, the interferon-induced 2′-5′-oligoadenylate synthetase-like gene (OASL) was permanently (i.e., still 96 h postinfection in persistence) up-regulated by C. pneumoniae in HeLa cells by one of the highest factors observed (compared to IFN-γ-treated HeLa cells that were mock infected). Members of the OAS family are critical components of innate immunity in mammals and are important for the antiviral activity of interferons (10). The interferon-induced hepatitis C-associated microtubular aggregate 44-kDa protein gene (IFI44) is also a member of the family of interferon-α/β-inducible genes (30). Its up-regulation might participate in the inflammatory process observed during persistence.
Our results clearly demonstrate that C. pneumoniae-induced host cell responses are not simply shut down at later time points of persistent infection. On the contrary, genes related to apoptosis, the cell cycle, or host cell metabolism are permanently differentially regulated by this intracellular bacterium. Functional studies are needed to show how the corresponding proteins influence the complex biology of an infected cell. Our results obtained under inhibition of bacterial protein synthesis indicate two different mechanisms influencing host cell responses in persistent and productive infection. Part of the response depends on chlamydial RNA polymerase, suggesting the involvement of bacterial effector proteins, which are synthesized and secreted during chlamydial development. Responses, such as the increase in OASL mRNA, which were not influenced or were even greater in the presence of chlamydial metabolism, suggest stimulation by bacterial cell wall components or heat shock proteins or by chlamydial effector proteins, which are already preformed in EBs and which are injected at the beginning of the infection.
Our results permit a rational design for future functional experiments to further characterize and elucidate C. pneumoniae infection. As an intracellular pathogen, C. pneumoniae relies on host cells in all aspects of its survival. The molecules interacting with the host could be attractive targets for therapeutic intervention, particularly in persistence, where antibiotic drugs are ineffective. The results of this gene expression study cast light on host-pathogen relations that are essential for chlamydial survival. Using this knowledge, strategies interfering with essential interactions between C. pneumoniae and the host cell, like apoptosis inhibition, can be exploited in the future to develop an innovative arsenal of therapeutic compounds.
Supplementary Material
Acknowledgments
We thank L. Klein-Hitpass (Institute of Cell Biology [Tumor Research], BioChip Laboratory, University Hospital of Essen, Essen, Germany) for his professional help with the microarray analysis. We also thank P. Scheinert and A. Hanne (Artus GmbH, Germany) for their excellent technical support with the Northern blot analysis. We also thank H. Geerlings (Medical School Hannover, Hannover, Germany) for his professional help in the statistical evaluation of the real-time PCR results. We thank the head of the Department of Medical Microbiology, S. Suerbaum (Medical School Hannover, Hannover, Germany), for his support.
Part of this study was financed by a grant from the German Research Council (DFG) for Special Research Area 566, Project A04.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 12 March 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Baetz, D., K. M. Regula, K. Ens, J. Shaw, S. Kothari, N. Yurkova, and L. A. Kirshenbaum. 2005. Nuclear factor-κB-mediated cell survival involves transcriptional silencing of the mitochondrial death gene BNIP3 in ventricular myocytes. Circulation 112:3777-3785. [DOI] [PubMed] [Google Scholar]
- 2.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58:686-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blythe, M. J., B. P. Katz, B. E. Batteiger, J. A. Ganser, and R. B. Jones. 1992. Recurrent genitourinary chlamydial infections in sexually active female adolescents. J. Pediatr. 121:487-493. [DOI] [PubMed] [Google Scholar]
- 4.Brantjes, H., N. Barker, J. van Es, and H. Clevers. 2002. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol. Chem. 383:255-261. [DOI] [PubMed] [Google Scholar]
- 5.Burns, T. F., P. Fei, K. A. Scata, D. T. Dicker, and W. S. El Deiry. 2003. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol. Cell. Biol. 23:5556-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y., and X. Y. Du. 2006. Functional properties and intracellular signaling of CCN1/Cyr61. J. Cell Biochem. [DOI] [PubMed]
- 7.Dong, F., H. Su, Y. Huang, Y. Zhong, and G. Zhong. 2004. Cleavage of host keratin 8 by a Chlamydia-secreted protease. Infect. Immun. 72:3863-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorai, T., I. S. Sawczuk, J. Pastorek, P. H. Wiernik, and J. P. Dutcher. 2005. The role of carbonic anhydrase IX overexpression in kidney cancer. Eur. J. Cancer 41:2935-2947. [DOI] [PubMed] [Google Scholar]
- 9.Duhaut, P., S. Bosshard, and J. P. Ducroix. 2004. Is giant cell arteritis an infectious disease? Biological and epidemiological evidence. Presse Med. 33:1403-1408. [DOI] [PubMed] [Google Scholar]
- 10.Eskildsen, S., J. Justesen, M. H. Schierup, and R. Hartmann. 2003. Characterization of the 2′-5′-oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res. 31:3166-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 12.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, S. F., J. Vier, S. Kirschnek, A. Klos, S. Hess, S. Ying, and G. Hacker. 2004. Chlamydia inhibit host cell apoptosis by degradation of proapoptotic BH3-only proteins. J. Exp. Med. 200:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda, E. Y., S. P. Lad, D. P. Mikolon, M. Iacobelli-Martinez, and E. Li. 2005. Activation of lipid metabolism contributes to interleukin-8 production during Chlamydia trachomatis infection of cervical epithelial cells. Infect. Immun. 73:4017-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng, Y., K. Berencsi, Z. Gyulai, T. Valyi-Nagy, E. Gonczol, and G. Trinchieri. 2000. Roles of interleukin-12 and gamma interferon in murine Chlamydia pneumoniae infection. Infect. Immun. 68:2245-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gieffers, J., H. Fullgraf, J. Jahn, M. Klinger, K. Dalhoff, H. A. Katus, W. Solbach, and M. Maass. 2001. Chlamydia pneumoniae infection in circulating human monocytes is refractory to antibiotic treatment. Circulation 103:351-356. [DOI] [PubMed] [Google Scholar]
- 17.Gieffers, J., J. Rupp, A. Gebert, W. Solbach, and M. Klinger. 2004. First-choice antibiotics at subinhibitory concentrations induce persistence of Chlamydia pneumoniae. Antimicrob. Agents Chemother. 48:1402-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grayston, J. T., C. C. Kuo, S. P. Wang, and J. Altman. 1986. A new Chlamydia psittaci strain, TWAR, isolated in acute respiratory tract infections. N. Engl. J. Med. 315:161-168. [DOI] [PubMed] [Google Scholar]
- 19.Hahn, D. L., R. W. Dodge, and R. Golubjatnikov. 1991. Association of Chlamydia pneumoniae (strain TWAR) infection with wheezing, asthmatic bronchitis, and adult-onset asthma. JAMA 266:225-230. [PubMed] [Google Scholar]
- 20.Hammerschlag, M. R. 2003. Advances in the management of Chlamydia pneumoniae infections. Exp. Rev. Anti. Infect. Ther. 1:493-503. [DOI] [PubMed] [Google Scholar]
- 21.Hammerschlag, M. R., K. Chirgwin, P. M. Roblin, M. Gelling, W. Dumornay, L. Mandel, P. Smith, and J. Schachter. 1992. Persistent infection with Chlamydia pneumoniae following acute respiratory illness. Clin. Infect. Dis. 14:178-182. [DOI] [PubMed] [Google Scholar]
- 22.Hatch, T. P. 1999. Developmental biology 12321, p. 29-68. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, D.C., USA
- 23.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 24.Hess, S., J. Peters, G. Bartling, C. Rheinheimer, P. Hegde, M. Magid-Slav, R. Tal-Singer, and A. Klos. 2003. More than just innate immunity: comparative analysis of Chlamydophila pneumoniae and Chlamydia trachomatis effects on host-cell gene regulation. Cell Microbiol. 5:785-795. [DOI] [PubMed] [Google Scholar]
- 25.Hess, S., C. Rheinheimer, F. Tidow, G. Bartling, C. Kaps, J. Lauber, J. Buer, and A. Klos. 2001. The reprogrammed host: Chlamydia trachomatis-induced up-regulation of glycoprotein 130 cytokines, transcription factors, and antiapoptotic genes. Arthritis Rheum. 44:2392-2401. [DOI] [PubMed] [Google Scholar]
- 26.Hogan, R. J., S. A. Mathews, S. Mukhopadhyay, J. T. Summersgill, and P. Timms. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect. Immun. 72:1843-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, Q., A. Raya, P. DeJesus, S. H. Chao, K. C. Quon, J. S. Caldwell, S. K. Chanda, J. C. Izpisua-Belmonte, and P. G. Schultz. 2004. Identification of p53 regulators by genome-wide functional analysis. Proc. Natl. Acad. Sci. USA 101:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igietseme, J. U., I. M. Uriri, S. N. Kumar, G. A. Ananaba, O. O. Ojior, I. A. Momodu, D. H. Candal, and C. M. Black. 1998. Route of infection that induces a high intensity of gamma interferon-secreting T cells in the genital tract produces optimal protection against Chlamydia trachomatis infection in mice. Infect. Immun. 66:4030-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, S., P. Wong, and P. A. Coulombe. 2006. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 441:362-365. [DOI] [PubMed] [Google Scholar]
- 30.Kitamura, A., K. Takahashi, A. Okajima, and N. Kitamura. 1994. Induction of the human gene for p44, a hepatitis-C-associated microtubular aggregate protein, by interferon-alpha/beta. Eur. J. Biochem. 224:877-883. [DOI] [PubMed] [Google Scholar]
- 31.Kuo, C. C., L. A. Jackson, L. A. Campbell, and J. T. Grayston. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo, C. C., A. Shor, L. A. Campbell, H. Fukushi, D. L. Patton, and J. T. Grayston. 1993. Demonstration of Chlamydia pneumoniae in atherosclerotic lesions of coronary arteries. J. Infect. Dis. 167:841-849. [DOI] [PubMed] [Google Scholar]
- 33.Lucero, H. A., and H. M. Kagan. 2006. Lysyl oxidase: an oxidative enzyme and effector of cell function. Cell Mol. Life Sci. 63:2304-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moulder, J. W. 1966. The relation of the psittacosis group (Chlamydiae) to bacteria and viruses. Annu. Rev. Microbiol. 20:107-130. [DOI] [PubMed] [Google Scholar]
- 35.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Netea, M. G., B. J. Kullberg, L. E. Jacobs, T. J. Verver-Jansen, D. Van, V., J. M. Galama, A. F. Stalenhoef, C. A. Dinarello, and J. W. Van Der Meer. 2004. Chlamydia pneumoniae stimulates IFN-gamma synthesis through MyD88-dependent, TLR2- and TLR4-independent induction of IL-18 release. J. Immunol. 173:1477-1482. [DOI] [PubMed] [Google Scholar]
- 37.Niehrs, C. 2006. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 25:7469-7481. [DOI] [PubMed] [Google Scholar]
- 38.Pantoja, L. G., R. D. Miller, J. A. Ramirez, R. E. Molestina, and J. T. Summersgill. 2000. Inhibition of Chlamydia pneumoniae replication in human aortic smooth muscle cells by gamma interferon-induced indoleamine 2,3-dioxygenase activity. Infect. Immun. 68:6478-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pantoja, L. G., R. D. Miller, J. A. Ramirez, R. E. Molestina, and J. T. Summersgill. 2001. Characterization of Chlamydia pneumoniae persistence in HEp-2 cells treated with gamma interferon. Infect. Immun. 69:7927-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 41.Peters, J., S. Hess, K. Endlich, J. Thalmann, D. Holzberg, M. Kracht, M. Schaefer, G. Bartling, and A. Klos. 2005. Silencing or permanent activation: host-cell responses in models of persistent Chlamydia pneumoniae infection. Cell Microbiol. 7:1099-1108. [DOI] [PubMed] [Google Scholar]
- 42.Rothfuchs, A. G., D. Gigliotti, K. Palmblad, U. Andersson, H. Wigzell, and M. E. Rottenberg. 2001. IFN-αβ-dependent, IFN-γ secretion by bone marrow-derived macrophages controls an intracellular bacterial infection. J. Immunol. 167:6453-6461. [DOI] [PubMed] [Google Scholar]
- 43.Rothfuchs, A. G., M. R. Kreuger, H. Wigzell, and M. E. Rottenberg. 2004. Macrophages, CD4+ or CD8+ cells are each sufficient for protection against Chlamydia pneumoniae infection through their ability to secrete IFN-gamma. J. Immunol. 172:2407-2415. [DOI] [PubMed] [Google Scholar]
- 44.Rothfuchs, A. G., C. Trumstedt, H. Wigzell, and M. E. Rottenberg. 2004. Intracellular bacterial infection-induced IFN-γ is critically but not solely dependent on toll-like receptor 4-myeloid differentiation factor 88-IFN-αβ-STAT1 signaling. J. Immunol. 172:6345-6353. [DOI] [PubMed] [Google Scholar]
- 45.Rottenberg, M. E., R. A. Gigliotti, D. Gigliotti, M. Ceausu, C. Une, V. Levitsky, and H. Wigzell. 2000. Regulation and role of IFN-gamma in the innate resistance to infection with Chlamydia pneumoniae. J. Immunol. 164:4812-4818. [DOI] [PubMed] [Google Scholar]
- 46.Rupp, J., T. Hellwig-Burgel, V. Wobbe, U. Seitzer, E. Brandt, and M. Maass. 2005. Chlamydia pneumoniae infection promotes a proliferative phenotype in the vasculature through Egr-1 activation in vitro and in vivo. Proc. Natl. Acad. Sci. USA. [DOI] [PMC free article] [PubMed]
- 47.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Makela, J. K. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet 2:983-986. [DOI] [PubMed] [Google Scholar]
- 48.Stamm, W. E. 2000. Potential for antimicrobial resistance in Chlamydia pneumoniae. J. Infect. Dis. 181(Suppl. 3):S456-S459. [DOI] [PubMed] [Google Scholar]
- 49.Stein, S., E. K. Thomas, B. Herzog, M. D. Westfall, J. V. Rocheleau, R. S. Jackson, M. Wang, and P. Liang. 2004. NDRG1 is necessary for p53-dependent apoptosis. J. Biol. Chem. 279:48930-48940. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, P. D., M. J. Campbell, A. Kejariwal, H. Mi, B. Karlak, R. Daverman, K. Diemer, A. Muruganujan, and A. Narechania. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13:2129-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 52.Vuola, J. M., V. Puurula, M. Anttila, P. H. Makela, and N. Rautonen. 2000. Acquired immunity to Chlamydia pneumoniae is dependent on gamma interferon in two mouse strains that initially differ in this respect after primary challenge. Infect. Immun. 68:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner, A. D., H. C. Gerard, T. Fresemann, W. A. Schmidt, E. Gromnica-Ihle, A. P. Hudson, and H. Zeidler. 2000. Detection of Chlamydia pneumoniae in giant cell vasculitis and correlation with the topographic arrangement of tissue-infiltrating dendritic cells. Arthritis Rheum. 43:1543-1551. [DOI] [PubMed] [Google Scholar]
- 54.Wolf, K., E. Fischer, and T. Hackstadt. 2000. Ultrastructural analysis of developmental events in Chlamydia pneumoniae-infected cells. Infect. Immun. 68:2379-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worm, H. C., G. H. Wirnsberger, A. Mauric, and H. Holzer. 2004. High prevalence of Chlamydia pneumoniae infection in cyclosporin A-induced post-transplant gingival overgrowth tissue and evidence for the possibility of persistent infection despite short-term treatment with azithromycin. Nephrol. Dial. Transplant. 19:1890-1894. [DOI] [PubMed] [Google Scholar]
- 56.Wu, Q., W. Ding, A. Mirza, T. Van Arsdale, I. Wei, W. R. Bishop, A. Basso, T. McClanahan, L. Luo, P. Kirschmeier, E. Gustafson, M. Hernandez, and S. Liu. 2005. Integrative genomics revealed RAI3 is a cell growth-promoting gene and a novel P53 transcriptional target. J. Biol. Chem. 280:12935-12943. [DOI] [PubMed] [Google Scholar]
- 57.Yang, T., P. J. Espenshade, M. E. Wright, D. Yabe, Y. Gong, R. Aebersold, J. L. Goldstein, and M. S. Brown. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110:489-500. [DOI] [PubMed] [Google Scholar]
- 58.Yucesan, C., and S. Sriram. 2001. Chlamydia pneumoniae infection of the central nervous system. Curr. Opin. Neurol. 14:355-359. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, H. M., P. Cheung, B. Yanagawa, B. M. McManus, and D. C. Yang. 2003. BNips: a group of pro-apoptotic proteins in the Bcl-2 family. Apoptosis 8:229-236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.