Abstract
Members of the Borrelia burgdorferi paralogous gene family 54 (pgf 54) are regulated by conditions simulating mammalian infection and are thought to be instrumental in borrelial host survival and pathogenesis. To explore the activities of these genes in vivo, a comprehensive analysis of pgf 54 genes BBA64, BBA65, and BBA66 was performed to assess the genetic stability, host antibody responses, and kinetics of gene expression in the murine model of persistent infection. DNA sequencing of pgf 54 genes obtained from reisolates at 1 year postinfection demonstrated that all genes of this family are stable and do not undergo recombination to generate variant antigens during persistent infection. Antibodies against BBA64 and BBA66 appeared soon after infection and were detectable throughout the infection, suggesting that there was gene expression during infection. However, quantitative reverse transcription-PCR revealed that BBA64 gene expression was considerably decreased in Borrelia residing in the mouse ear tissue compared to the expression in cultured spirochetes by 20 days postinfection and that the levels of expression remained low throughout the infection. Conversely, transcription of the BBA65 and BBA66 genes was increased, and both of these genes were continuously expressed until 100 days postinfection; this was followed by periods of differential expression late in infection. The expression profile of the BBA64 gene suggests that this gene has an important role during tick-to-host transmission and early infection, whereas the expression profile of the BBA65 and BBA66 genes suggests that these genes have a role in persistent infection. The differential regulation of pgf 54 genes observed during infection may help confer a survival advantage during persistent infection, influencing mechanisms for B. burgdorferi dissemination, tissue tropism, or evasion of the adaptive immune response.
The Lyme disease agent, Borrelia burgdorferi, is remarkably adept at adapting to environmental changes encountered throughout its enzootic life cycle, which involves shuttling to and from mammalian reservoir hosts and vector ticks. Several studies have demonstrated this organism's ability to differentially express genes in response to the conditions confronted during transmission from ticks to warm-blooded hosts. In vitro factors that simulate these changes include shifts in the temperature, pH, cell density, oxygen concentration, and carbon dioxide concentration and addition of blood to the culture medium (6, 7, 8, 20, 21, 35, 38, 41, 43, 44). Studies addressing B. burgdorferi differential expression in vivo are best represented by the prototypical example of two outer surface proteins (OspA and OspC) demonstrating a reciprocal gene expression pattern in unfed and feeding ticks (37). A remarkable and poorly understood feature of B. burgdorferi is its ability to survive in the host despite elicitation of a strong antibody response directed against several borrelial antigens. Recent work has shown that B. burgdorferi modulates gene expression in response to selective pressure resulting from host adaptive immunity, suggesting that there is a spirochetal mechanism for immune evasion (26, 28). Therefore, B. burgdorferi host survival and pathogenicity depend on the contributions of differentially regulated gene products that function as virulence determinants essential for host dissemination, tissue tropism, and avoidance of clearance by the adaptive immune response. However, little is known about the identities of borrelial genes essential for infectivity and pathogenicity and about the mechanisms by which such vital genes are regulated in vivo.
B. burgdorferi genes belonging to paralogous gene family 54 (pgf 54) have been shown to be associated with infectious phenotypes and are differentially regulated by temperature and pH shifts during in vitro cultivation (6, 11, 32, 36). This family of genes, annotated by the B. burgdorferi genome sequencing project (15), consists of 12 members (9). Eight of the 12 pgf 54 genes are located on the 54-kb linear plasmid (lp54 or plasmid A), a plasmid containing genes that (i) display the highest ratio of differential expression induced by environmental signals, as shown by microarray studies, (ii) are expressed during mammalian infection (dbpA and dbpB), and (iii) are important for borrelial survival in the tick (ospA) (4, 10, 19, 32, 33, 36, 42). Moreover, lp54 is maintained in all naturally isolated B. burgdorferi infectious strains examined to date, emphasizing the importance of this plasmid for borrelial biological functions. The pgf 54 members localized to lp54 are designated BBA64 (encoding lipoprotein P35) (16, 21), BBA65, BBA66, BBA68 (encoding lipoprotein CRASP-1) (24), BBA69, BBA70, BBA71, and BBA73. Other plasmid-encoded gene family members include BBI36 and BBI38, which exhibit 99% DNA sequence identity, and BBI39 and BBJ41 which also are 99% identical.
Studies have shown that a subset of pgf 54 genes are induced in response to culture condition shifts to 35°C and/or pH 7.0, parameters resembling the mammalian host environment (5, 6, 35). Recent microarray studies have revealed similar regulation of these genes in in vitro-grown B. burgdorferi in response to temperature, mammalian host-specific signals, and the addition of blood to a culture (4, 32, 36, 42). Interestingly, microarray analyses have shown that selected pgf 54 genes exhibit the greatest upregulation under these conditions. In in vivo studies utilizing reverse transcription-PCR (RT-PCR) workers have qualitatively detected BBA64, BBA65, and BBA66 gene transcripts in B. burgdorferi-infected mouse tissues and fed ticks, providing evidence that there is active gene expression during infection (1, 26, 42). Serological data obtained from Lyme disease patients and tick-infected experimental mice have shown that there are antibody responses against the BBA64, BBA66, BBA68, and BBI36/38 proteins, indicating the immunogenic properties of these antigens (11, 16, 31). Finally, antibodies generated against the BBA64, BBA66, and BBA69 proteins have exhibited borreliacidal activity, leading to the hypothesis that these gene products may be vaccine candidates (5). Collectively, these data indicate that members of pgf 54 may be important factors for borrelial infectivity and pathogenesis in tick and mammalian hosts.
Because of the evidence demonstrating that pgf 54 genes are inducible, elicit humoral responses in infected hosts, and are expressed during infections, we began to test the hypothesis that pgf 54 genes encode products that are necessary for persistent infection and are transcriptionally regulated as a mechanism for antigenic variation and/or for tissue tropism following host dissemination to allow borrelial survival. The goals of this study were to more precisely determine the expression and serologic response kinetics of target pgf 54 genes and antigens during persistent infections in mice. As an initial step, we measured the in vivo transcription of the BBA64, BBA65, and BBA66 genes in B. burgdorferi-infected ear tissue while also monitoring the concomitant antibody responses to the corresponding gene products. Here we describe a dynamic pattern of differential expression of the BBA64, BBA65, and BBA66 genes throughout chronic murine infection, and this quantitative analysis should help further define borrelial gene function during infection.
(Portions of this work were presented at the 10th International Conference on Lyme Borreliosis and other Emerging Tick-Borne Infectious Diseases, Vienna, Austria, September 2005, at the 106th General Meeting of the American Society for Microbiology, Orlando, FL, 2006, and at the Gordon Conference on the Biology of Spirochetes, Il Ciocco, Italy, April 2006.)
MATERIALS AND METHODS
B. burgdorferi strains and growth conditions.
B. burgdorferi strain B31-A3 is a clonal, low-passage infectious strain (14) and was used for mouse inoculation. B. burgdorferi was grown in liquid Barbour-Stoenner-Kelly (BSK) complete medium at 35°C with 5% CO2. B. burgdorferi was isolated from infected mice by culturing ear tissue in BSK medium as previously described (39), followed by colony isolation on solid BSK medium plates.
Mouse inoculation, tissue and serum collection, and B. burgdorferi reisolation.
B. burgdorferi B31-A3 was grown to the mid-log phase (approximately 5 × 107 organisms/ml) and counted with a Petroff-Hauser counting chamber. Six-week-old female Swiss-Webster mice were inoculated subcutaneously with 1 × 104 organisms in 50 μl. Three cohorts of mice were used throughout this study; the first cohort consisted of 18 mice, and the second and third cohorts consisted of four mice each. All mice in a cohort were inoculated with B. burgdorferi concurrently at zero time (day 0 postinfection [p.i.]). Serum and tissue samples were obtained from mice in the first cohort at 9, 20, 41, 62, 90, 120, 180, 270, 365, and 513 days p.i. At each time, mice were bled, and ear tissue was removed from one or two mice, placed immediately into RNAlater (Ambion, Austin, TX), and stored at −80°C until it was used for RNA extraction. Additionally, ear tissue was inoculated into BSK medium for B. burgdorferi cultivation. Following growth in liquid culture medium, B. burgdorferi reisolated from the mouse ears was plated onto solid BSK medium, and after 7 to 10 days of incubation random colonies were selected for clonal isolation, grown in liquid culture medium, and frozen. Stocks of all B. burgdorferi reisolates were frozen in 60% glycerol at −70°C. The second and third cohorts were inoculated just like the first cohort was inoculated, and additional ear tissue samples were harvested as described above at days 100 and 151 p.i. Serum samples were obtained at times identical to the times used for the first cohort to augment the collection. The mouse experimental protocol was approved by the Division of Vector-Borne Infectious Diseases Institutional Animal Care and Use Committee.
B. burgdorferi plasmid profiles and DNA sequencing of pgf 54 genes.
Prior to mouse inoculation, the B. burgdorferi B31-A3 strain plasmid profile was analyzed. Primer pairs specific for regions of each B. burgdorferi plasmid were used for PCR amplification and have been described previously (14). The DNA template for PCR amplification was generated by centrifuging 50 to 100 μl of a log-phase B. burgdorferi culture, resuspending the pellet in 100 μl of water, and boiling the pellet for 10 min. After boiling, the tube was centrifuged to pellet the cell debris, and 2 to 5 μl of the supernatant was used for PCR. AmpliTaq Gold DNA polymerase (Roche, Branchburg, NJ) and standard reagents were used for PCR with the following parameters: one cycle 94°C for 2 min and 35 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 2 min, followed by one cycle of 72°C for 5 min.
For sequencing, the pgf 54 genes were amplified by PCR using a DNA template prepared from B. burgdorferi primary inoculant or colonies reisolated from mice as described above. pgf 54 gene-specific primers amplified the entire coding sequence and are listed in Table 1. The PCR parameters were one cycle of 94°C for 60 s, 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s, and one cycle of 72°C for 5 min. The PCR products were examined by electrophoresis on 1% Tris-acetate-EDTA agarose gels. Amplicons were sequenced using a BigDye v.3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and were electrophoresed with an Applied Biosystems 3130xl genetic analyzer. Sequences were aligned and analyzed using the Lasergene software (DNAStar, Madison, WI).
TABLE 1.
Primers used in this study
| Gene or primer | Forward primer sequence | Reverse primer sequence | Probe sequence | Sequence | Description |
|---|---|---|---|---|---|
| Primers for amplifying coding sequence | |||||
| BBA64 | TTGAAGGATAACATTTTGAAAAAT | CTGAATTGGAGCAAGAATATTGGT | |||
| BBA65 | TTGAATAAAATAAAATTATCAATA | ATTATTATTAAATTTAAATAATGT | |||
| BBA66 | GTATTTTGAAAATCAAACCATTAA | CATTATACTAATGTATGCTTCAAG | |||
| BBA68 | TTGAAAAAAGCCAAACTAAATATA | GTAAAAGGCAGGTTTTAAAGTATC | |||
| BBA69 | TTGAAAAAAGCCAAACTAAATATA | ATAAAAGGCAGATTGTAAAGAATC | |||
| BBA70 | ATGGCTCCAGAAGTAAACAGCTAC | TTTTATATTAGTCTCTTTTTCAAT | |||
| BBA71 | AATGAATAAATTAAAAGAAATTCT | GTTTTGTTTAAATTCAACACTGTA | |||
| BBA73 | TTGAAAAGAAACAAAATTTGGAAA | GTAGTGTATGTGGTCACAACAGGT | |||
| BBI36/38 | TTGAAAAACTTTAAATTAAATACT | ATCAGCTTGATCTAGTAGATAAAG | |||
| BBI39/J41 | TTGAAAAACCTTAAATTAAATATT | ATCAGCTTGGTTTAGTAGATAAAG | |||
| vlsE | AGTAGTACGACGGGGAAACCAG | ACTTTGCGAACTGCAGACT (41) | |||
| TaqMan primers | |||||
| flaB | TCTTTTCTCTGGTGAGGGAGCT | TCCTTCCTGTTGAACACCCTCT | AAACTGCTCAGGCTGCACCGGTTC | ||
| BBA64 | AGCCCGCAGCTGGAAAA | AGCTTTGCAACTTCAGGATCTAATATT | AATCCCAACGCTAATGCCAACAATGCT | ||
| BBA65 | GCATGTTTGAACTGCTTAATGTTTACA | TGCGAGGCTTGAGTTCATTTT | TCCACAAATGGCACAAGGCTTCA | ||
| BBA66 | GCAAGAGCTGCATCACTAACAAA | TGTTGGCAGCCCGCTATT | TCAAGCCGTTACAACCGTACCCG | ||
| Primers for cloning into expression vector | |||||
| bba64.3 TR | TGCTCTCTGGAAGTCAAAGACAGC | Forward BBA64 truncated at Cys28 | |||
| bba64.2 FL+H3 | AAATTTAAGCTTCTGAATTGGAGCAAGAAT | Reverse BBA64 + HindIII site | |||
| bba65.1 FL+BamHI | AAATTTGGATCCTTGAATAAAATAAAATTATCAa | Forward full-length BBA65 + BamHI site | |||
| bba65.2 FL | ATTAAATTTAAATAATGTGTC | Reverse full-length BBA65 | |||
| bba66.3 TR | TGCACGATTGATGCCAATCTAAACG | Forward BBA66 truncated at Cys19 | |||
| bba66.2 FL+Xba | AAATTTTCTAGACATTATACTAATGTATGCa | Reverse BBA66 + XbaI site | |||
| malE forward | GGTCGTCAGACTGTCGATGAAGCC | Forward PCR primer for malE |
The restriction site is underlined.
Cloning of genes and expression and purification of recombinant proteins.
Truncated (without the signal sequence and lipidation motif) or full-length members of pgf 54 were cloned from B. burgdorferi B31 genomic DNA using specific primers (Table 1) and ligated into XmnI and XbaI restriction endonuclease-digested vector pMAL-c2X (New England Biolabs, Beverly, MA). Ligation reaction products were transformed into the ER2508 strain of Escherichia coli (New England Biolabs) (lacking lon and native malE), and transformants were selected for growth on Luria-Bertani medium plates containing 100 μg/ml ampicillin. Colonies were screened by PCR using the MalE primer and a target gene-specific primer (Table 1), and plasmid DNA from a positive colony was sequenced for confirmation. For overexpression and purification of most recombinant proteins, ER2508 cultures containing pMAL recombinants were grown in Luria-Bertani broth supplemented with 100 μg/ml ampicillin and 0.2% glucose to an optical density at 600 nm (OD600) of 0.5 and then induced with isopropyl-β-d-thiogalactopyranoside (final concentration, 0.3 mM) for 2 h at 37°C; the only exception was the BBA64 truncated recombinant, which was induced for 14 h at 16°C to decrease the amount of degradation. Cultures were pelleted, suspended in column buffer (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1.0 mM dithiothreitol) with 5× Complete EDTA-free protease inhibitor (Roche Diagnostics, Indianapolis, IN), and lysed at 4°C with a French press (two or three passes at 18,000 lb/in2). The crude lysate was clarified to remove unlysed cells and other cellular material by centrifugation (8,000 × g, 15 min, 4°C). MalE-tagged recombinants were purified by passing the lysate over an amylose resin affinity column (New England Biolabs) using an AKTA prime automated liquid chromatography system (Amersham Biosciences, Piscataway, NJ). Purified proteins were eluted from the amylose resin affinity column using 10 mM maltose in column buffer, and they were concentrated using Centricon YM-30 units (Millipore, Billerica, MA), quantitated by using a modified Lowry assay, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and stained with silver (Silver Stain Plus; Bio-Rad, Hercules, CA) to monitor the purity.
Immunoblotting and ELISA.
Recombinant proteins (200 ng/blotting strip) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes, and immunoblotting was performed using mouse serum samples obtained during the infection at a 1:1,000 dilution, followed by incubation with a 1:1,000 dilution of alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Kirkegaard and Perry Laboratories, Gaithersburg, MD) using standard procedures. For an enzyme-linked immunosorbent assay (ELISA), antigen diluted in carbonate buffer (90 mM NaHCO3, 60 mM Na2CO3; pH 9.6) was bound to 96-well format plates at a concentration of 100 ng/well overnight at 4°C. The plates were washed five times with TBS-T (20 mM Tris [pH 7.4], 140 mM NaCl, 2.7 mM KCl, 0.05% Tween 20) and incubated with TBS-T containing 0.25% bovine serum albumin (blocking buffer) for 30 min at room temperature. Mouse serum samples from the infected animals were diluted 1:200 in blocking buffer, added to the plate wells, and incubated for 1 h at room temperature. Alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (1:1,000) was added to each well and incubated for 1 h at room temperature. Phosphatase substrate (p-nitrophenylphosphate) pellets (Sigma, St. Louis, MO) were dissolved in 23 mM NaHCO3-25 mM Na2CO3-10 mM MgCl2 (pH 9.8), and 100 μl was added to each well, followed by incubation at room temperature for 30 min. All preparations were subjected to mild agitation, which was followed by five washes in TBS-T. Reactions were squelched by addition of 100 μl 5N NaOH to each well. Plates were read at OD405 using an ELx808IU Ultra microplate reader and the KC4 software (version 3.2, revision 3; BioTek Instruments, Inc., Winooski, VT). Plates on which anti-BBA64 was analyzed were read at OD450 due to off-scale readings when they were read at OD405. The optical densities for triplicate sample wells were averaged, and a cutoff value was established by calculating 3 standard deviations of the mean for the preimmune control serum samples.
RNA isolation.
RNA was isolated from mouse ear tissue by Trizol (Invitrogen, Carlsbad, CA) extraction. Approximately 50 mg of tissue was homogenized with 1 ml Trizol in a Tenbroeck homogenizer until no traces of tissue were visible. The homogenate was spun through a QiaShredder spin column (QIAGEN, Valencia, CA) to complete homogenization. Following addition of 0.2 volume of chloroform, the suspension was centrifuged to separate the phases, the aqueous phase containing RNA was removed, and the RNA was precipitated with 0.5 volume of isopropanol. Following two washes in cold 75% ethanol, the RNA pellet was allowed to air dry and resuspended in RNA storage buffer (Ambion). RNA was isolated from B. burgdorferi grown in BSK culture medium to the mid-log phase using an RNAqueous 4-PCR kit (Ambion). All RNA samples were subjected to DNase treatment using Turbo DNA-free (Ambion). RNA samples were tested for contaminating DNA by PCR amplification using rig/S15 (mouse specific; a eukaryotic, highly conserved, constitutively expressed gene encoding a small ribosomal subunit protein [22, 23]) and flaB (B. burgdorferi specific; constitutively expressed flagellin gene) gene primers prior to use in quantitative RT-PCR (qRT-PCR) assays.
qRT-PCR.
Reverse transcription was performed in a 20-μl reaction mixture containing 3 μg of total RNA isolated from infected mouse ears using a Retroscript kit (Ambion) at 44°C for 60, min followed by incubation at 92°C for 10 min to inactivate the enzyme. A TaqMan real-time PCR was performed using cDNA generated from the reverse transcription as follows. TaqMan PCR primer and probe sequences were designed using the Primer Express program (Applied Biosystems) and were synthesized with a probe containing 6-carboxyfluorescein at the 5′ end and black hole quencher at the 3′ end (Table 1). Real-time PCR was performed using a 50-μl reaction mixture containing each primer at a final concentration of 1 μM, 0.15 μM probe, 1× TaqMan universal PCR master mixture (Roche), and 1 μl cDNA (from the 20-μl reverse transcription reaction mixture). The volume of cDNA used corresponded to 150 and 10 ng of RNA reverse transcribed from the infected ear tissue and cultured B. burgdorferi, respectively. All test sample PCRs were performed in triplicate in 96-well PCR iCycler plates (Bio-Rad) using one cycle of 95°C for 10 min and 50 cycles of 95°C for 30 s and 60°C for 1 min with the Bio-Rad iCycler. Crossing threshold (CT) values were determined by the iCycler software. Relative quantitation of gene expression was performed by the 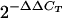 method described by Livak and Schmittgen (29), in which transcript levels were normalized using the constitutively expressed borrelial flaB gene and were analyzed relative to the levels in a mid-log-phase culture. To eliminate plate-to-plate and day-to-day variations, each plate in which qRT-PCRs were performed with infected ear tissue cDNA also contained qRT-PCR mixtures with cultured B. burgdorferi cDNA to calculate relative gene expression.
method described by Livak and Schmittgen (29), in which transcript levels were normalized using the constitutively expressed borrelial flaB gene and were analyzed relative to the levels in a mid-log-phase culture. To eliminate plate-to-plate and day-to-day variations, each plate in which qRT-PCRs were performed with infected ear tissue cDNA also contained qRT-PCR mixtures with cultured B. burgdorferi cDNA to calculate relative gene expression.
RESULTS
Stability of pgf 54 genes throughout persistent infection.
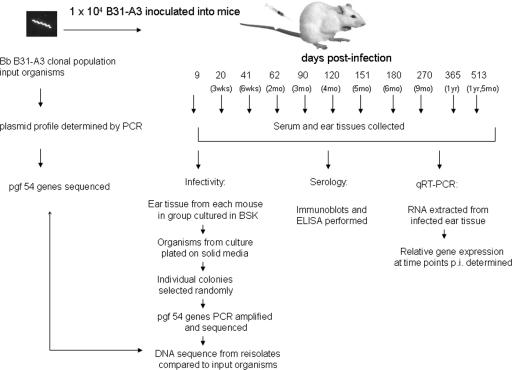
To investigate whether members of pgf 54 underwent DNA rearrangements during long-term infection to create antigenic variants, genes from B. burgdorferi reisolated at 1 year p.i. were sequenced. For this study, 26 mice in three cohorts were inoculated with 1 × 104 B. burgdorferi cells and were used as the source of serum, ear tissue, and reisolated Borrelia; one or two mice were utilized for 11 specific times p.i. The experimental design is shown in Fig. 1. Prior to inoculation of mice, the plasmid profile of the clonal B. burgdorferi B31-A3 strain used as the inoculant was determined. As expected, all plasmids except cp9 were present (data not shown).
FIG. 1.
Experimental design and procedures used in this study to assess B. burgdorferi (Bb) infectivity, the genetic stability of pgf 54 members, and gene expression during persistent infection in mice.
B. burgdorferi was successfully cultured from mouse ear tissue obtained at all times until 513 days p.i. (approximately 1 year and 5 months) except day 9, demonstrating that the mice were persistently infected. We have found that the earliest time for obtaining culture-positive ear tissues is around 14 days p.i., so a negative day 9 ear culture was not unexpected. B. burgdorferi samples cultivated from the ears were plated on solid media for clonal isolation, and three colonies were selected randomly for gene sequence analysis. pgf 54 genes BBA64, BBA65, BBA66, BBA68, BBA69, BBA70, BBA71, BBA73, and BBI36/38 were amplified by PCR from the colonies plated from the 1-year p.i. culture, and the amplicons were sequenced. The DNA sequences of the pgf 54 genes from the 1-year reisolates (three colonies/gene) were identical to the DNA sequences of the corresponding genes from the primary inoculant (data not shown). As a control, the vlsE gene, which is known to undergo genetic recombination during mouse infection (45), was also amplified and sequenced. As predicted, the vlsE gene sequence from each colony was found to differ from the input gene sequence. These data proved that recombination between pgf 54 genes did not occur and that these genes remain genetically stable during long-term infection.
Temporal analysis of the antibody response against BBA64, BBA65, and BBA66 antigens.
Immunoblotting for the BBA64, BBA65, and BBA66 recombinant proteins was performed using serum samples collected at each time to estimate when specific antibodies arose during infection. Prior to this analysis, serum samples from individual infected mice were immunoblotted with B. burgdorferi whole-cell lysate to assess seroconversion. We observed virtually identical immunoreactivity profiles for all mouse sera; therefore, samples were pooled for each time p.i. It is important to note that antibodies against OspA, the predominant protein produced by B. burgdorferi in culture, were not present in the mouse serum samples. This agrees with the findings of Barthold et al., who reported that a large inoculum (>106 organisms) is required to produce an anti-OspA response (3). Additionally, Barthold et al. also demonstrated that a heat-killed inoculum of B. burgdorferi at this concentration did not elicit detectable antibody responses to any borrelial antigens, including BBA64, BBA65, and BBA66. Collectively, the results demonstrate that the antibody responses measured in this study were due to host-adapted Borrelia and not due to antigens present on the cultured organisms used for the inoculum.
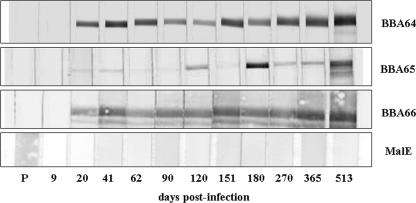
The first antibodies detected early in infection were antibodies against BBA64 and BBA66 and appeared between day 9 and day 20 p.i. The anti-BBA64 and -BBA66 antibodies remained detectable throughout the infection, maintaining strong reactivity as determined by immunoblot analysis (Fig. 2). Although not as strong, an antibody response against BBA65 was observed at day 20 p.i., and detectable reactivity was observed throughout the infection (Fig. 2). No immunoreactivity was detected when mouse serum was blotted with the recombinant MalE fusion protein alone, nor was any cross-reactivity among the BBA64, BBA65, and BBA66 recombinant proteins observed (data not shown). The elicitation of specific antibodies against these three antigens early in mammalian infection provided evidence that the BBA64, BBA65 and BBA66 genes are expressed upon transmission of B. burgdorferi from the tick to the host. Additionally, the presence of antibodies later during infection suggested that either there was continued protein synthesis from all three genes or there were sustained humoral responses in the absence of expression.
FIG. 2.
Temporal serological analysis of anti-BBA64, -BBA65, and -BBA66 during persistent infection in mice. Immunoblotting was performed for recombinant BBA64, BBA65, BBA66, and maltose-binding protein MalE. Mouse serum was obtained at specific days p.i. and was blotted with the recombinant proteins, as indicated at the bottom. P, preimmune serum.
qRT-PCR of in vivo BBA64, BBA65, and BBA66 gene expression.
Direct measurement of gene transcription by qRT-PCR was used to specifically determine whether the BBA64, BBA65, and BBA66 genes were expressed at a particular time p.i., and an ELISA was also performed to more accurately measure antibody levels. Total RNA was extracted from B. burgdorferi-infected mouse ear tissue for use in qRT-PCR assays. Ear tissue was chosen for these assays because Borrelia had been cultured from this tissue and the tissue was readily accessible. We obtained quantitative expression data for the BBA64, BBA65, and BBA66 genes at each time p.i. (normalized to the expression of the constitutively expressed gene flaB) relative to the expression in a mid-log-phase culture, which was used as the inoculum and served as the time zero reference. The relative gene expression analysis was performed by the 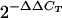 method (29). The PCR efficiency of the TaqMan primer-probe set for each gene was determined by amplification of serial dilutions of B. burgdorferi genomic DNA. PCR amplification efficiency for all TaqMan primers was calculated at 91 to 99%. Values over 90% are recommended for accuracy. Our assay could detect approximately 10 to 100 gene copies, calculated using serial dilutions of genomic DNA (data not shown). The BBA64, BBA65, and BBA66 genes are expressed at low levels during in vitro cultivation. The mid-log-phase expression levels of these genes were determined to be significantly less than the levels of expression of flaB, the gene encoding the constitutively expressed flagellin protein (Table 2). Moreover, we determined that the transcription of the BBA64 gene was greater than the transcription of the BBA66 gene, which in turn was greater than the transcription of the BBA65 gene in vitro (BBA64 > BBA66 > BBA65). The relative levels of expression of the BBA64, BBA65, and BBA66 genes in persistently infected mice through day 513 p.i. were determined and are shown in Fig. 3; the raw data are shown in Table 2. The average CT values for genes amplified from the ears at selected times approached 40. Examination of the raw data showed that the amplification signals at these CT values (in triplicate) rose exponentially and were as strong as the control amplification signals that gave lower CT values when genomic DNA was used as the template. Also, no CT values were detected when the water (no-template) control was used. Finally, preliminary experiments demonstrated that BBA64 gene transcripts could be detected in infected ear tissue by nested PCR (data not shown), providing additional evidence that borrelial mRNA was present in this tissue. Therefore, we were confident that the observed CT values for the ears were not artifacts.
method (29). The PCR efficiency of the TaqMan primer-probe set for each gene was determined by amplification of serial dilutions of B. burgdorferi genomic DNA. PCR amplification efficiency for all TaqMan primers was calculated at 91 to 99%. Values over 90% are recommended for accuracy. Our assay could detect approximately 10 to 100 gene copies, calculated using serial dilutions of genomic DNA (data not shown). The BBA64, BBA65, and BBA66 genes are expressed at low levels during in vitro cultivation. The mid-log-phase expression levels of these genes were determined to be significantly less than the levels of expression of flaB, the gene encoding the constitutively expressed flagellin protein (Table 2). Moreover, we determined that the transcription of the BBA64 gene was greater than the transcription of the BBA66 gene, which in turn was greater than the transcription of the BBA65 gene in vitro (BBA64 > BBA66 > BBA65). The relative levels of expression of the BBA64, BBA65, and BBA66 genes in persistently infected mice through day 513 p.i. were determined and are shown in Fig. 3; the raw data are shown in Table 2. The average CT values for genes amplified from the ears at selected times approached 40. Examination of the raw data showed that the amplification signals at these CT values (in triplicate) rose exponentially and were as strong as the control amplification signals that gave lower CT values when genomic DNA was used as the template. Also, no CT values were detected when the water (no-template) control was used. Finally, preliminary experiments demonstrated that BBA64 gene transcripts could be detected in infected ear tissue by nested PCR (data not shown), providing additional evidence that borrelial mRNA was present in this tissue. Therefore, we were confident that the observed CT values for the ears were not artifacts.
TABLE 2.
qRT-PCR data: BBA64, BBA65, and BBA66 gene expression in infected mice relative to the expression in culture following normalization to the flaB genea
| Gene | Days p.i. | Avg CT for flaB in cultureb | Avg CT for BBA gene in cultureb | ΔCT | Fold decrease compared to flaB | Avg CT for flaB in earb | Avg CT for BBA gene in earb | ΔCT | ΔΔCT | Fold difference compared to culture |
|---|---|---|---|---|---|---|---|---|---|---|
| BBA64 | 20 | 16.53 | 23.73 | 7.20 | 27.80 | 36.33 | 8.53 | 1.33 | −2.51 | |
| 41 | 16.93 | 24.57 | 7.64 | 32.63 | 41.00c | 8.37 | 0.73 | −1.66 | ||
| 62 | 16.40 | 23.80 | 7.40 | 31.33 | NDd | |||||
| 100 | 16.73 | 24.03 | 7.30 | 30.40 | 39.40 | 9.00 | 1.70 | −3.25 | ||
| 180 | 16.87 | 24.43 | 7.56 | 30.93 | 39.85c | 8.92 | 1.36 | −2.57 | ||
| 270 | 16.87 | 24.57 | 7.70 | 31.57 | 40.87 | 9.30 | 1.60 | −3.03 | ||
| Avg | 16.72 ± 0.21 | 24.19 ± 0.38 | 7.47 ± 0.43 | 181.00 | ||||||
| BBA65 | 20 | 16.53 | 27.03 | 10.50 | 27.80 | 36.30 | 8.50 | −2.00 | 4.00 | |
| 41 | 16.93 | 27.40 | 10.47 | 32.63 | 39.25c | 6.62 | −3.85 | 14.4 | ||
| 62 | 16.40 | 27.00 | 10.60 | 31.33 | 38.25c | 6.92 | −3.68 | 12.8 | ||
| 100 | 16.73 | 27.30 | 10.57 | 30.40 | 36.60 | 6.20 | −4.37 | 20.7 | ||
| 180 | 16.87 | 27.33 | 10.46 | 30.93 | 39.30 | 8.37 | −2.09 | 4.26 | ||
| 270 | 16.87 | 27.63 | 10.76 | 31.57 | 37.77 | 6.20 | −4.56 | 23.6 | ||
| Avg | 16.72 ± 0.21 | 27.28 ± 0.24 | 10.56 ± 0.32 | 1,509.70 | ||||||
| BBA66 | 20 | 16.53 | 26.33 | 9.80 | 27.80 | 35.83 | 8.03 | −1.77 | 3.41 | |
| 41 | 16.93 | 26.90 | 9.97 | 32.63 | 39.00c | 6.37 | −3.60 | 12.1 | ||
| 62 | 16.40 | 26.03 | 9.63 | 31.33 | 39.33 | 8.00 | −1.63 | 3.10 | ||
| 100 | 16.73 | 26.60 | 9.87 | 30.40 | 36.30 | 5.90 | −3.97 | 15.7 | ||
| 180 | 16.87 | 26.90 | 10.03 | 30.93 | 39.85c | 8.92 | −1.11 | 2.16 | ||
| 270 | 16.87 | 27.10 | 10.23 | 31.57 | 39.27 | 7.70 | −2.53 | 5.78 | ||
| Avg | 16.72 ± 0.21 | 26.64 ± 0.40 | 9.92 ± 0.45 | 968.80 |
No gene transcription was detected for RNA extracted from mice on days 120, 365, and 513.
Each value is the mean from triplicate reactions.
Average of two amplifications; the third amplification was not detectable.
ND, amplification not detectable in two of three reactions.
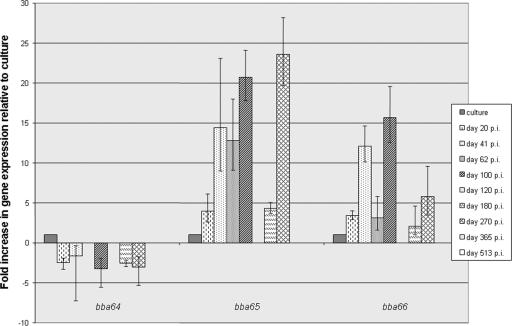
FIG. 3.
Quantitative expression of the BBA64, BBA65, and BBA66 genes throughout mouse infection. The graph shows the severalfold increases and decreases in gene expression relative to the expression in a culture (defined as 1) following normalization to the constitutively expressed flaB gene. Standard deviations of the means are indicated by the error bars. No expression of any gene, including flaB, was detected in the day 120, 365, and 513 p.i. samples, and no expression of the BBA64 gene was detected at day 62 p.i. Raw data from which the graph was derived are shown in Table 2, and calculations were done by the 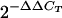 method.
method.
An unexpected finding was that BBA64 gene expression was considerably downregulated by day 20 p.i. compared with both the expression of this gene in a mid-log-phase culture and the expression of the BBA65 and BBA66 genes early in infection (Fig. 3). This result was surprising insofar as anti-BBA64 antibodies are routinely detected in mice and humans at various stages of infection, implying that the BBA64 gene is expressed during mammalian infections. Indeed, the immunoblot in Fig. 2 shows that anti-BBA64 antibodies were present long after the gene was downregulated. Therefore, this result suggests that the BBA64 gene is upregulated very early in host infection (within the first few hours or days) and/or within the tick during ingestion of the bloodmeal in preparation for transmission to the host.
In contrast to BBA64 gene expression, expression of the BBA65 and BBA66 genes was significantly upregulated compared to the expression in cultured Borrelia. On average, BBA65 gene expression was increased 4-, 14-, 13-, and 21-fold at days 20, 41, 62, and 100 p.i., respectively (Fig. 3). Similarly, on average, BBA66 gene expression was increased 3-, 12-, 3-, and 16-fold at the same times. Therefore, both BBA65 gene expression and BBA66 gene expression are highly upregulated soon after infection and remain upregulated for more than 3 months p.i. Oddly, at 4 months (120 days) p.i., expression of the BBA64, BBA65, BBA66, and flaB genes was undetectable. Analysis of the total RNA extracted from the ear tissue showed that the integrity of RNA was maintained and that the RNA was not degraded. RT-PCR was performed and amplified mRNA of the mouse gene S15, indicating that the reverse transcription reaction and PCR were not compromised (data not shown). Additionally, B. burgdorferi was cultured from this ear sample, demonstrating that organisms were present. This experiment was repeated by extracting total RNA from the ear of a second mouse that had been sacrificed at day 120 p.i., and the results were identical to the results obtained for the first mouse (i.e., no detectable transcripts of any gene). Therefore, we postulated that the numbers of Borrelia cells and the amount of corresponding mRNA at this time p.i. in ear tissue were below the level of detection (10 to 100 genomic equivalents) for this assay.
Later in the infection, BBA65 gene expression again increased on average 4- and 23-fold and BBA66 gene expression increased on average 2- and 6-fold at 180 and 270 days p.i, respectively, compared to the expression in cultured spirochetes (Fig. 3). At 365 and 513 days p.i., we were not able to detect transcripts for any genes, including flaB, similar to the results obtained with the day 120 samples and indicating that the numbers of borreliae in the ear decrease dramatically during the late stages of infection. Duplicate experiments were performed using RNA extracted at identical times from the ears of separate infected mice, and similar results were obtained, demonstrating that there was consistency between individual experiments. Again, all ear tissues at these times yielded positive cultures, demonstrating that the tissues contained viable spirochetes.
Antibody measurement by ELISA.
An ELISA was performed to determine the levels of anti-BBA64, -BBA65, and -BBA66 antibodies in order to augment the immunoblot data which provided a qualitative analysis of host antibody production. ELISA data were plotted with the qRT-PCR data shown in Fig. 3 to obtain a composite chart that allowed correlations between antibody levels and gene expression to be determined (Fig. 4). The ELISA data for anti-BBA64 showed the antibody response early in infection (by day 20), which was maintained at a fairly constant level and then increased at day 120 p.i. and in the later stages of infection (Fig. 4A). This result is consistent with the immunoblot data shown in Fig. 2. Interestingly, the strong anti-BBA64 antibody response observed corresponded to a decrease in the expression of the BBA64 gene compared to the expression in cultured spirochetes.
FIG. 4.
BBA64, BBA65, and BBA66 gene expression and anti-BBA64, -BBA65, and -BBA66 antibody levels during mouse infection. The bars show the gene expression data shown in Fig. 3, and the lines show the data for antibodies (1:200 dilution) determined by ELISA. The y axis on the left indicates the severalfold increase in gene expression relative to the expression in a culture, and the y axis on the right indicates the optical density determined by the ELISA. No qRT-PCR analysis was performed on day 151 p.i., so a space was skipped to allow for the day 151 p.i. ELISA data. No ELISA data were obtained for day 62 p.i., so a dashed line connects day 40 to day 90 for continuity. No gene expression was detected by qRT-PCR on days 120, 365, and 513 p.i. (A) Data for BBA64. The ELISA cutoff value obtained using preimmune serum was 0.31. (B) Data for BBA65. The ELISA cutoff value obtained using preimmune serum was 0.30. (C) Data for BBA66. The ELISA cutoff value obtained using preimmune serum was 0.18.
A small but detectable anti-BBA65 response was observed from approximately 3 weeks to 3 months p.i., corresponding to an increase in the expression of the BBA65 gene compared to the expression in cultured cells (Fig. 4B). The increase in BBA65 gene expression seen by day 100 p.i. was followed by an increase in the anti-BBA65 level by day 120 p.i. The anti-BBA65 antibody levels waned around day 151 p.i.; however, detectable levels of the BBA65 gene transcript were observed by day 180 p.i., and there was a continuing increase in transcription (compared to the transcription in cultured spirochetes) up to 23.6-fold by day 270 p.i. In turn, a spike in the anti-BBA65 antibody level was observed by day 180 p.i., which was followed by a decrease by day 270 p.i. and then by an increase at the late times after 1 year p.i., when BBA65 gene expression was undetectable.
The levels of anti-BBA66 antibodies steadily increased for 4 months (to day 120 p.i.), which was similar to what was observed with BBA64. Unlike BBA64 gene transcription, BBA66 gene expression increased compared to the expression in cultured spirochetes. However, BBA66 gene expression decreased after 100 days p.i. through 180 days p.i., perhaps in response to the increase in anti-BBA66 antibody during this time. As the anti-BBA66 level decreased by day 180 p.i., we observed an increase in the expression of the BBA66 gene by day 270. This in turn was followed by an increase in the anti-BBA66 levels very late in infection (days 365 and 513).
DISCUSSION
Eight of the 12 pgf 54 genes localize to lp54, a linear plasmid containing genes that display the highest ratio of differential expression induced by environmental signals, as shown by microarray studies. The relevance of the B. burgdorferi pgf 54 members has been established by several studies demonstrating that regulation of this gene family is influenced by in vitro conditions that simulate the tick and/or mammalian host environment. Furthermore, serological analysis has indicated that the antigens encoded by some pgf 54 members are immunogenic during natural and experimental infections. These findings have led to hypotheses that members of pgf 54 are involved in mechanisms that are essential for infection or pathogenesis. However, with the exception of the BBA68 gene product, CRASP-1, a factor H binding protein (24), defined functions for the pgf 54 gene products are unknown. Furthermore, there is little information regarding pgf 54 gene expression during mammalian infection. This study was designed to examine the temporal expression patterns of select pgf 54 genes in B. burgdorferi-infected tissue and the accompanying specific antibody responses in the mouse model. Our data allowed us to obtain invaluable insight into gene expression patterns, providing the groundwork for investigations into the biological functions of the gene products in the context of host and tick infections.
The annotated B. burgdorferi genome sequence includes over 100 paralogous gene families based on DNA sequence homologies within genes. The purpose of such a large number of similar genes in B. burgdorferi is not known; however, homologous sequences can serve as recombination sites to create variant genes, a mechanism that pathogens employ for antigenic variation to avoid host immunity. Therefore, to determine whether there was genetic rearrangement between these genes or mutations via other mechanisms, we amplified and sequenced the pgf 54 genes from B. burgdorferi reisolated from mice 1 year p.i. The sequencing results revealed no nucleotide base changes in the sequences of any of the nine pgf 54 genes analyzed at 1 year p.i. compared to the sequences in the input organisms. This demonstrated that these genes are stable and do not undergo coding sequence mutations due to internal recombination with other family members or gross point mutations. The stability of this gene family was not unexpected, as sequence analysis has demonstrated that there is a putative promoter region for each pgf 54 gene that indicates an active expression site, as opposed to silent pseudogene sequences. Additionally, in previous studies investigating other B. burgdorferi genes and gene families workers have also reported that there was genetic stability during long-term infection (13, 18, 30, 34, 40, 46). Therefore, if pgf 54 is involved in immune evasion, we hypothesized that the mechanism would not involve antigenic variation due to DNA rearrangements but rather would involve modulation of transcript levels (2, 12).
qRT-PCR was performed to analyze the transcript levels of specific family members during murine infection. We found that BBA64 gene expression was significantly decreased at each time assayed compared to the expression of in vitro-grown spirochetes. This was surprising considering the marked increase in the antibody response against BBA64 (P35) that was observed soon after infection and was maintained for more than 1 year p.i. The anti-BBA64 antibody response following murine and human infection with B. burgdorferi has been observed previously by other researchers, leading to the supposition that the BBA64 gene is expressed during infection and may be an important factor in this process. However, this result suggests that BBA64 gene transcription is upregulated during tick feeding and/or within hours following introduction into the host, thereby driving antigen synthesis and subsequent immune processing that leads to the production of antibodies between 9 and 20 days p.i. Supporting this idea, Anguita et al. (who used RT-PCR) and Liang et al. (who used a microarray) reported early detection of BBA64 gene transcripts at up to 14 and 33 days p.i., respectively, in mice (1, 26). Furthermore, Tokarz et al. demonstrated that BBA64 gene transcription was increased in actively feeding ticks but not in flat ticks (42). Also, we have determined that B. burgdorferi does not express the BBA64 gene in replete ticks that have dropped from the host (17), which is further evidence that the BBA64 gene may be essential during the early phases of tick-to-host transmission. Although BBA64 gene transcription was decreased in ear tissue compared to the transcription in in vitro-grown organisms, we still detected transcripts of this gene in infected mice. We assayed gene expression only in organisms localized in mouse ear tissue, and it is possible that disseminated spirochetes in other host tissues may have elevated BBA64 gene transcript levels contributing to antigen production, leading to the antibody level observed. In several studies workers have reported variation in transcription of B. burgdorferi genes among infected tissues (19, 25, 27).
The increase in BBA66 gene expression early during the infection likely resulted in the synthesis of BBA66 antigen, leading to the generation of the anti-BBA66 antibody response (Fig. 4C). However, during later stages of infection, both BBA65 and BBA66 gene expression exhibited an inverse relationship with the levels of host antibodies elicited against the corresponding antigens, similar to the relationship observed for the BBA64 gene. This observation suggests that gene expression may be modulated as a mechanism to circumvent host immunity and subsequently to select for organisms not expressing the BBA64, BBA65, or BBA66 gene during certain phases of infection. This concept has been demonstrated in general for borrelial lipoprotein-encoding genes and specifically for genes encoding OspC, DbpA, BBF01, and VlsE (27, 28). One may speculate that the decrease in borrelial numbers that occurred around days 120, 365, and 513 p.i., when we could not detect transcription, may have been due to host immune selection that caused a significant decrease in the borrelial population, leading to an adaptation process in which the organisms expressed genes differentially to change their surface structure in order to survive. Interestingly, in a recent study, Lederer et al. were also unable to detect B. burgdorferi or borrelial transcripts in tissues at day 120 p.i (25). The reason for this phenomenon is not known, but the lack of detection could reflect a decrease in the number of spirochetes in specific tissues (i.e., ear) as the population adapts to the hostile immune response.
We observed reciprocal expression patterns for the BBA64 gene and both the BBA65 and BBA66 genes in ear tissue throughout the experiment. There were pronounced increases in expression of the BBA65 and BBA66 genes, while the expression of the BBA64 gene was downregulated. These results closely corresponded to the results obtained for in vitro-cultivated B. burgdorferi when environmental conditions were shifted to simulate the change from the tick to a warm-blooded host. Clifton et al. demonstrated by using qRT-PCR that there was a dramatic increase in BBA65 and BBA66 gene transcription when the culture conditions were changed from pH 8 and 23°C to pH 7 and 35°C, while BBA64 gene expression was unchanged (11). The reciprocal gene expression patterns observed in mouse ear tissue may reflect a coordinated regulatory response to factors occurring in the mammalian host, such as the shift in environmental conditions, immune pressure, and/or interactions with host tissues.
In two independent microarray studies in which they compared B. burgdorferi transcription in spirochetes grown in vitro to transcription in host-adapted spirochetes sequestered in dialysis membrane chambers (DMC) incubated in the peritoneum of rats, researchers obtained results which contrasted with the results obtained in this study for BBA64, BBA65, and BBA66 gene transcript levels (4, 36). Brooks et al. reported a 2.7-fold increase in BBA64 gene transcription and a 1.4-fold decrease in BBA65 gene transcription as determined by microarray analysis when DMC spirochetes were compared to spirochetes grown at 23°C. Likewise, Revel et al. described 6-, 18-, and 31-fold decreases in the transcription of the BBA64, BBA65, and BBA66 genes, respectively, in DMC spirochetes compared to the transcription in spirochetes grown in vitro at 37°C and pH 6.8. Our results differ somewhat from the microarray findings obtained in these two studies, and there are several possible explanations for the discrepancies. Mechanistically, it is plausible that the BBA65 and BBA66 genes are required for borrelial dissemination and/or tissue or cellular colonization during infection. In such a case, one may speculate that the BBA65 and BBA66 genes would be upregulated either when they are required to arrive or upon arrival in the appropriate host microenvironment during the infection. Therefore, spirochetes within DMC would perhaps not encounter the regulatory signals necessary to upregulate genes essential for dissemination to and/or establishment in host tissues. Additionally, organisms were harvested from the DMC at only one time, whereas in our study we measured gene transcription at several times. We also measured gene expression directly in B. burgdorferi-infected tissue rather than in organisms obtained from an artificial infection model. Moreover, microarrays are particularly useful for providing a global analysis of gene transcription, but they are generally not as quantitative or specific as qRT-PCR for measuring gene expression. Our results partially agree with the results of the microarray analysis of Liang et al., who monitored the expression of genes encoding lipoproteins for up to 33 days following transplantation of host-adapted B. burgdorferi-infected ear tissues (26). Liang et al. detected transcription of the BBA64 and BBA65 genes in mouse ear tissue at all times tested (days 0, 11, 22, and 33) but detected BBA66 gene transcription only on days 11 and 22. Comparing the two studies is difficult as many experimental factors are dissimilar. Primarily, the method of detection was different (microarray versus qRT-PCR). While we injected a known quantity of spirochetes into mice, Liang et al. transplanted ear tissue from infected mice into naive mice. Furthermore, we did not measure gene expression at day 33 p.i., and they did not perform their analysis beyond that time. Likewise, the mice used in the two investigations were different, and it has been determined that some mouse strains are more susceptible to infection (higher spirochete loads and pronounced inflammation) than others. Ultimately, all of the studies did show that these genes encoding lipoproteins are expressed at some point during mammalian infection. The global analyses of gene expression performed by the investigators using microarrays were valuable in that they provided a basis for extended investigations focusing on specific genes of interest. Our study provided a more comprehensive approach to further defining B. burgdorferi differential gene expression during mammalian infection.
Clearly, regulation of B. burgdorferi gene expression during in vivo infection is a complex mechanism involving a variety of environmental and host-specific factors, perhaps depending on the organism's needs for survival during particular phases of infection. In this study we looked at the temporal expression of three members of pgf 54 in mouse ear tissue compared to the expression in in vitro-grown spirochetes, and we observed two genes, the BBA65 and BBA66 genes, which exhibited similar trends during infection. The early upregulation patterns for the BBA65 and BBA66 genes were distinct from the pattern for the BBA64 gene, which was inversely downregulated. The expression patterns for the BBA65 and BBA66 genes showed that there were periods of differential regulation during infection, perhaps reflecting either a mechanism for immune evasion based on modulation of transcription or a mechanism for tissue tropism following host dissemination. In ongoing work in our laboratory we are focusing on B. burgdorferi infectious phenotypic analysis of pgf 54 mutants and transcriptional expression patterns of the remaining pgf 54 genes. Characterizing the temporal expression profiles for the pgf 54 genes during infection should provide important information for defining how B. burgdorferi adapts and survives in changing environments and how pgf 54 genes influence B. burgdorferi infectivity and pathogenicity.
Acknowledgments
We thank Jill Livengood and Becky Byram for their help and constructive suggestions throughout this work. We thank Phil Stewart for supplying the B31-A3 strain.
This research was supported in part by NIH grant AI055178 (J.A.C.) and by CDC cooperative agreement CI000181 (J.A.C.). A.J.N. was supported by NIH T32 training grant HD042987.
Editor: D. L. Burns
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Anguita, J., S. Samanta, B. Revilla, K. Suk, S. Das, S. W. Barthold, and E. Fikrig. 2000. Borrelia burgdorferi gene expression in vivo and spirochete pathogenicity. Infect. Immun. 68:1222-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G., and B. I. Restrepo. 2000. Antigenic variation in vector-borne pathogens. Emerg. Infect. Dis. 6:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthold, S. W., E. Fikrig, L. K. Bockenstedt, and D. H. Persing. 1995. Circumvention of outer surface protein A immunity by host-adapted Borrelia burgdorferi. Infect. Immun. 63:2255-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks, C. S., S. R. Vuppala, A. M. Jett, and D. R. Akins. 2006. Identification of Borrelia burgdorferi outer surface proteins. Infect. Immun. 74:296-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, J. A., R. M. Cordova, and C. F. Garon. 2000. Identification of 11 pH-regulated genes in Borrelia burgdorferi localizing to linear plasmids. Infect. Immun. 68:6677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, J. A., N. El-Hage, J. C. Miller, K. Babb, and B. Stevenson. 2001. Borrelia burgdorferi RevA antigen is a surface-exposed outer membrane protein whose expression is regulated in response to environmental temperature and pH. Infect. Immun. 69:5286-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 10.Cassatt, D. R., N. K. Patel, N. D. Ulbrandt, and M. S. Hanson. 1998. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect. Immun. 66:5379-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clifton, D. R., C. L. Nolder, J. L. Hughes, A. J. Nowalk, and J. A. Carroll. 2006. Regulation and expression of bba66 encoding an immunogenic infection-associated lipoprotein in Borrelia burgdorferi. Mol. Microbiol. 61:243-258. [DOI] [PubMed] [Google Scholar]
- 12.Deitsch, K. W., E. R. Moxon, and T. E. Wellems. 1997. Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol. Mol. Biol. Rev. 61:281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Hage, N., L. D. Lieto, and B. Stevenson. 1999. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect. Immun. 67:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore, R. D., Jr., K. J. Kappel, and B. J. Johnson. 1997. Molecular characterization of a 35-kilodalton protein of Borrelia burgdorferi, an antigen of diagnostic importance in early Lyme disease. J. Clin. Microbiol. 35:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 18.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, and D. R. Akins. 2002. Changes in temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 70:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodzic, E., S. Feng, K. J. Freet, and S. W. Barthold. 2003. Borrelia burgdorferi population dynamics and prototype gene expression during infection of immunocompetent and immunodeficient mice. Infect. Immun. 71:5042-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyde, J. A., J. P. Trzeciakowski, and J. T. Skare. 2007. Borrelia burgdorferi alters its gene expression and antigenic profile in response to CO2 levels. J. Bacteriol. 189:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Indest, K. J., R. Ramamoorthy, M. Sole, R. D. Gilmore, B. J. Johnson, and M. T. Philipp. 1997. Cell-density-dependent expression of Borrelia burgdorferi lipoproteins in vitro. Infect. Immun. 65:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue, C., K. Shiga, S. Takasawa, M. Kitagawa, H. Yamamoto, and H. Okamoto. 1987. Evolutionary conservation of the insulinoma gene rig and its possible function. Proc. Natl. Acad. Sci. USA 84:6659-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitagawa, M., S. Takasawa, N. Kikuchi, T. Itoh, H. Teraoka, H. Yamamoto, and H. Okamoto. 1991. rig encodes ribosomal protein S15. The primary structure of mammalian ribosomal protein S15. FEBS Lett. 283:210-214. [DOI] [PubMed] [Google Scholar]
- 24.Kraiczy, P., J. Hellwage, C. Skerka, H. Becker, M. Kirschfink, M. M. Simon, V. Brade, P. F. Zipfel, and R. Wallich. 2004. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 279:2421-2429. [DOI] [PubMed] [Google Scholar]
- 25.Lederer, S., C. Brenner, T. Stehle, L. Gern, R. Wallich, and M. M. Simon. 2005. Quantitative analysis of Borrelia burgdorferi gene expression in naturally (tick) infected mouse strains. Med. Microbiol. Immunol. 194:81-90. [DOI] [PubMed] [Google Scholar]
- 26.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang, F. T., J. Yan, M. L. Mbow, S. L. Sviat, R. D. Gilmore, M. Mamula, and E. Fikrig. 2004. Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect. Immun. 72:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
29.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the
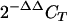 method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar] - 30.McDowell, J. V., S. Y. Sung, G. Price, and R. T. Marconi. 2001. Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 69:4831-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowalk, A. J., R. D. Gilmore, Jr., and J. A. Carroll. 2006. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect. Immun. 74:3864-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal, U., A. M. de Silva, R. R. Montgomery, D. Fish, J. Anguita, J. F. Anderson, Y. Lobet, and E. Fikrig. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Investig. 106:561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Persing, D. H., D. Mathiesen, D. Podzorski, and S. W. Barthold. 1994. Genetic stability of Borrelia burgdorferi recovered from chronically infected immunocompetent mice. Infect. Immun. 62:3521-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramamoorthy, R., and D. Scholl-Meeker. 2001. Borrelia burgdorferi proteins whose expression is similarly affected by culture temperature and pH. Infect. Immun. 69:2739-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-29013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seshu, J., J. A. Boylan, F. C. Gherardini, and J. T. Skare. 2004. Dissolved oxygen levels alter gene expression and antigen profiles in Borrelia burgdorferi. Infect. Immun. 72:1580-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinsky, R. J., and J. Piesman. 1989. Ear punch biopsy method for detection and isolation of Borrelia burgdorferi from rodents. J. Clin. Microbiol. 27:1723-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevenson, B., L. K. Bockenstedt, and S. W. Barthold. 1994. Expression and gene sequence of outer surface protein C of Borrelia burgdorferi reisolated from chronically infected mice. Infect. Immun. 62:3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole-genome DNA array. Infect. Immun. 72:5419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 44.Yang, X. F., A. Hubner, T. G. Popova, K. E. Hagman, and M. V. Norgard. 2003. Regulation of expression of the paralogous Mlp family in Borrelia burgdorferi. Infect. Immun. 71:5012-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 46.Zuckert, W. R., and A. G. Barbour. 2000. Stability of Borrelia burgdorferi bdr loci in vitro and in vivo. Infect. Immun. 68:1727-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]