Abstract
Escherichia coli containing the K1 capsule is the leading cause of gram-negative meningitis, but the pathogenesis of this disease is not completely understood. Recent microarray experiments in which we compared the gene expression profile of E. coli K1 associated with human brain microvascular endothelial cells (HBMEC) to the gene expression profile of E. coli K1 not associated with HBMEC revealed that there was a threefold increase in the expression of the fliI gene, encoding an ATP synthase involved in flagellar synthesis and motility, in HBMEC-associated E. coli. In this study, we examined the role of flagella in E. coli K1 association with and invasion of HBMEC by constructing isogenic ΔflhDC, ΔfliI, ΔfliC, and ΔcheW mutants that represented each class of flagellar genes. Mutations that affected the flagellum structure and flagellum formation (ΔflhDC, ΔfliI, and ΔfliC) resulted in significant defects in motility, as well as in HBMEC association and invasion, compared to the characteristics of the wild-type strain when preparations were examined with or without centrifugation. Transcomplementation with the corresponding genes restored the levels of these mutants to the levels of the parent strain. These findings suggest that the HBMEC association and invasion defects of the mutants are most likely related to flagella and less likely due to their motility defects. This conclusion was supported by our demonstration that the cheW mutant was not motile but was able to associate with and invade HBMEC. In addition, purified recombinant flagellin reduced the association of the wild-type strain with HBMEC by ∼40%, while it had no effect on the fliC mutant's association with HBMEC. Together, these findings indicate that flagella promote E. coli K1 binding to HBMEC.
Neonatal meningitis caused by Escherichia coli, particularly E. coli containing the K1 capsule (E. coli K1) and serotype O18:K1:H7, is associated with high mortality and morbidity rates (24, 32). A factor that contributes to this is our incomplete understanding of the pathogenesis of this disease. The microbe-host interactions that are relevant for the pathogenesis of E. coli meningitis include (i) a high level of bacteremia (>104 CFU/ml of blood), (ii) association with and invasion of the blood-brain barrier (BBB), and (iii) traversal of the BBB as live bacteria (21). In vivo, the BBB comprises brain microvascular endothelial cells, astrocytes, and pericytes. However, as the contribution of the latter cells to microbial translocation was found to be minimal (20), an in vitro model of BBB was established in our laboratory, which is composed of human brain microvascular endothelial cells (HBMEC) alone (35). This model has been extensively examined to determine brain endothelial characteristics, morphology, and purity (20) and is currently used by many laboratories around the world to study the pathogenesis of meningitis caused by various microbes (20, 21).
For E. coli K1, the microbial determinants that have been shown to contribute to meningitis include type 1 fimbriae, the K1 capsule, OmpA, Ibe proteins, and CNF1 (39). Type 1 fimbriae and OmpA have been shown to be involved in E. coli K1 association with HBMEC (34, 36). For instance, bacteria that did not express type 1 fimbriae had a markedly reduced ability to adhere to HBMEC in vitro (36). The presence of the K1 capsule inhibits phagocytosis and opsonization in the blood, contributing to a high level of bacteremia (5, 22), while survival during HBMEC trafficking is dependent on the ability of E. coli K1-containing vacuoles to avoid phagosome-lysososme fusion (19). CNF1, an AB type toxin, and Ibe proteins have been shown to play an important role in invasion of HBMEC (15-18, 28).
Flagella, the external whip-like structures involved in bacterial motility, have been shown to contribute to the virulence of many enteric bacteria, including E. coli O157:H7, Campylobacter jejuni, Yersinia, and Salmonella (8, 23, 33). These external structures have been shown to be required for colonization of the gastrointestinal tract, including adhesion and subsequent invasion. In some instances, flagellar motility has also been shown to be essential for dispersal to different tissues of the host (31). However, the role of flagellar structure and/or motility in the pathogenesis of E. coli K1 meningitis is not known. Recent studies in which we compared the gene expression profile of E. coli K1 associated with HBMEC to the gene expression profile of E. coli K1 not associated with HBMEC using our E. coli DNA microarrays revealed that there was a threefold increase in the expression of the fliI gene, encoding an ATPase involved in flagellar synthesis and motility, in HBMEC-associated E. coli K1 (Y. Xie, G. Parthasarathy, F. DiCello, C. Teng, M. Paul-Satyaseela, and K. S. Kim, unpublished data). We hypothesized that flagella, either through motility or their structure, play a role in the pathogenesis of E. coli K1 meningitis. To examine this possibility, defined deletion mutants with mutations in flagellar genes were constructed to determine the role of flagella in E. coli K1 association with and invasion of HBMEC in vitro. The flagellum is synthesized from three classes of genes. The class I region consists of the master operon flhDC, the class II region comprises the genes involved in the synthesis of the flagellar basal body and hook (flgMNBCDEFGHIJKL, flhBAE, and fliAZYDSTEFGHIJKLMNOPQR), and the class III region consists of genes involved in flagellin synthesis (fliC) and motility (motAB, cheAW, and cheRBYZ). The class I genes control the synthesis of the class II genes, while the class II fliA gene encoding the alternative sigma factor (σ28) controls the synthesis of the class III genes (7). Isogenic mutants were constructed for each class of genes; a ΔflhDC mutant was constructed to obtain a mutant in which the overall control of flagellar synthesis was affected, ΔfliI and ΔfliC mutants were constructed to obtain mutants in which flagella and their motility were affected, and a ΔcheW mutant was constructed to obtain a mutant in which only motility was affected. We assessed the roles of the genes in E. coli K1 association with and invasion of HBMEC using the in vitro BBB model.
We found that the deletion mutants with mutations that affected the flagellar structure exhibited significant defects in the association with and invasion of HBMEC, while deletion of gene that affected only motility (ΔcheW) had no effect on the HBMEC association or invasion frequencies compared to those of the wild type. In addition, preincubation with purified flagellin decreased the association of the wild type with HBMEC by ∼40%, while it had no effect on the association of the fliC mutant with HBMEC. This is the first demonstration that bacterial flagella play an important role in E. coli K1 association with and invasion of HBMEC.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli K1 strain RS218 (O18:K1:H7), isolated from the cerebrospinal fluid of a neonate with meningitis (2, 29), was used for the experiments. The nonpathogenic E. coli K-12 strain HB101 was used as a negative control in HBMEC association and invasion assays. E. coli K-12 strain DH5α, DH10B, or 10G E.cloni Elite cells (Lucigen, Middleton, WI) were used as hosts for plasmid vectors. E. coli BL21(DE3) (Stratagene, La Jolla, CA) was used for expression of recombinant flagellin. Bacteria were routinely grown on Trypticase soy blood agar plates, and isolated colonies were grown statically in LB broth overnight at 37°C. When growth rates were determined, bacteria were grown at 37°C with agitation (200 rpm) in LB broth, and the optical density at 620 nm (OD620) was determined over a 6-h period.
HBMEC culture.
The primary HBMEC were cultured as previously described (35). Briefly, HBMEC were grown in RPMI 1640 medium containing 2 mM glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), streptomycin (100 μg/ml), 10% heat-inactivated fetal bovine serum, 10% Nu serum, essential amino acids (1%), and vitamins (1%) in 75-cm2 tissue culture flasks. All the reagents were obtained from Mediatech, Inc. (Herndon, VA).
Construction of isogenic flagellar mutants.
Mutants with deletions of the flhDC, fliI, fliC, and cheW genes were constructed by the method described by Datsenko and Wanner (9). Briefly, wild-type strain RS218 was transformed with the pKD46 vector, which contains genes coding for the arabinose-induced λ Red recombinase system that promotes recombination between linear pieces of DNA (PCR product) and the host chromosome. This recombination is based on short stretches of homology (30 to 50 nucleotides) on the linear DNA to the site of recombination. The 5′ ends of the primers used for amplification of the PCR product (the FH and RH primers [Table 1]) contain 50-nucleotide stretches homologous to the target gene, while the 3′ ends of the primers include regions homologous to the chloramphenicol cassette on the pKD3 plasmid. Each PCR product (∼1 kb) was amplified, gel extracted, and electroporated into competent strain RS218 containing pKD46, allowing recombination in the presence of arabinose. The temperature-sensitive pKD46 plasmid was cured by incubation at 37°C with agitation. The deletion mutants were verified by PCR using primers adjacent to the gene region (the FV and RV primers [Table 1]) and sequencing of the PCR products.
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| FliI-FH | ATGACCACGCGCCTGACTCGCTGGCTAACCACGCTGGATAACTTTGAAGCGTGTAGGCTGGAGCTGCTTC |
| FliI-RH | GACACTGTCGGGAAAATACGCTCCAGCCCCTGGAGAGACGCTTCCCAGTCCATATGAATATCCTCCTTAG |
| FliI-FV | CTCGGCACTGATCAAACAG |
| FliI-RV | TGTTGTCTCGCCTGCTAATG |
| FliI-FC | GCCGAATTCTTATCAGGCGGGGATTGC |
| FliI-RC | ACCAGTACTTGTTGTCTCGCCTGCTAATG |
| FlhDC-FH | ACATCACGGGGTGCGGTGAGACCGCATAAAAATAAAGTTGGTTATTCTGGAGTGTAGGCTGGAGCTGCTTC |
| FlhDC-RH | GGGCAAAAGAAAGCAGCGGTACATCGTTACCGCTGCTGGAATGTTTCGCCCATATGAATATCCTCCTTA |
| FlhDC-FV | GGTGGGTCTGCTTATTGCAG |
| FlhDC-RV | AGGCTTCCACCGGTCATCA |
| FlhDC-FC | GCCGAATTCCAGTTGCGTCGATTTAGG |
| FlhDC-RC | ACCAGTACTAGGCTTCCACCGGTCATCA |
| FliC-FH | CAGTCTGCGCTGTCGAGTTCTATCGAGCGTCTGTCTTCTGGCTTGCGTAGTGTAGGCTGGAGCTGCTTC |
| FliC-RH | ACCGGCCTGCTGGATAATCTGCGCTTTCGACATGTTGGACACTTCGGTCGCATATGAATATCCTCCTTAG |
| FliC-FV | TGGAAACCCAAAACGTAATC |
| FliC-RV | GCCGTCAGTCTCAGTTAATC |
| FliC-FC | GCCGAATTCCGGGGTTATCGGCCTGAA |
| FliC-RC | ACCAGTACTATGTTGCCGGATGCGGCG |
| CheW-FH | GCCGCCTGAATGAGTAAAAAGGTAACAATATGACCGGTATGACGAATGTAGTGTAGGCTGGAGCTGCTTC |
| CheW-RH | GATGCGCAGATCATCGGGTTCATTTCAATTGAGGAAATCGGGAGAATTACCATATGAATATCCTCCTTAG |
| CheW-FV | TGGCGACGGCAGCGTGGC |
| CheW-RV | TGACTACGCGGATACGGTT |
| FliC EX F | ATGGCACAAGTCATTAATACCAAC |
| FliC EX R | TATAGGCCGGCCTACCCTGCAGCAGAGACAGAACCTG |
Complementation of isogenic mutants.
Complementation of individual flagellum genes was performed using the pACYC184 vector (6, 25). The gene to be complemented was amplified, cut with the EcoRI and ScaI restriction enzymes built into the primers (the FC and RC primers [Table 1]), and ligated into the pACYC184 vector previously cut with the same enzymes. The resulting construct was chloramphenicol sensitive and tetracycline resistant. The ligation mixture was electroporated into electrocompetent E. coli DH5α or E.cloni Elite cells, plated onto agar containing tetracycline (20 μg/ml), and incubated at 37°C overnight. The presence of the correct insert in the resulting colonies was verified using pACYC184-specific primers, gene-specific primers (the FC and RC primers), chloramphenicol sensitivity, and EcoRI/ScaI restriction enzyme digestion. The recombinant plasmids were then introduced into the appropriate electrocompetent flagellar mutants.
Motility assays.
To assess the swimming motility of the wild type, isogenic mutants of the wild type, or the complemented strains, the bacteria were initially grown on 1.5% tryptone agar plates. After overnight incubation, isolated colonies were stab inoculated onto 0.3% soft tryptone agar plates containing 0.3% glucose and incubated at 37°C for 24 h. The diameter of each zone of growth was determined to quantify motility.
HBMEC association and invasion assays.
HBMEC association and invasion assays were carried out as described previously (36). Briefly, HBMEC grown to confluence on collagen-coated 24-well plates were infected with bacteria grown overnight in LB broth. The multiplicity of infection of bacteria in cells was ∼100, and the plates were incubated for 90 min at 37°C in the presence of 5% CO2. When centrifugation was required, the bacteria were centrifuged onto HBMEC at 800 × g for 10 min. After incubation, the wells were washed four times with wash medium (medium 199-Ham F-12 medium [1:1] with 2 mM glutamine), and the HBMEC were lysed with 0.025% Triton X-100 for 10 to 15 min. The associated bacteria were enumerated by plating on sheep blood agar plates. For invasion assays, following the 90-min incubation, the cells were washed once and incubated with medium containing 100 μg/ml of gentamicin for 1 h to kill the extracellular bacteria. The cells were then washed and lysed as described above for the association assay. All assays were performed in triplicate. To calculate association or invasion frequencies, the ratio of the number of adherent or invaded bacteria to the number of bacteria in the original inoculum was used. The association and invasion frequencies were calculated relative to the frequencies of the wild type and were expressed as relative association and invasion rates. In addition, in order to determine the sensitivity of the mutants to assay procedures, colony counts were obtained for the wild type and flagellar mutants after incubation with 0.025% Triton X-100 for 30 min.
Effect of flagellin on E. coli K1 association with HBMEC.
Recombinant flagellin was obtained by cloning the fliC gene into a pQE80-based vector (QIAGEN, Valencia, CA), pQE80-XY1. Briefly, the gene was amplified with PfuUltra DNA polymerase (Stratagene, La Jolla, CA) using primers FliC EX F and FliC EX R (Table 1) and was digested with FseI. The PCR product was then ligated into pQE80-XY1 previously cut with AfeI and FseI and electroporated into E. coli DH10B. Colonies on plates containing ampicillin (100 μg/ml) were screened by PCR for the insert and confirmed by sequencing. The vector containing the fliC insert was subsequently transformed into E. coli BL21(DE3) for protein expression and purification as described by Wilson et al. (37). Briefly, overnight cultures grown in LB broth were diluted 1:100 in 1.5 liters of LB broth and grown at 37°C with agitation until the OD600 was 0.5. The cultures were cooled to 25°C, induced with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and grown with agitation for an additional 4 h at 25°C. Cells were harvested and resuspended in 20 ml buffer A (50 mM Tris-HCl [pH 8.0], 300 mM NaCl) supplemented with 0.1 mg/ml lysozyme, incubated for 30 min on ice, and lysed by sonication. Insoluble material was removed by centrifugation at 14,000 rpm. His6-tagged FliC protein was affinity purified from the soluble lysate by incubation with agitation with 5 ml Ni-nitrilotriacetic acid agarose (QIAGEN, Valencia, CA) overnight at 4°C. The lysate was loaded onto two disposable standing columns; each column was washed with 12 ml buffer A and 12 ml buffer B (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 5 mM imidazole). Bound protein was eluted with buffer C (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 300 mM imidazole), and 1-ml fractions were collected. Twenty-five microliters of each fraction was electrophoresed in a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel, and fractions containing His6-FliC were pooled, dialyzed against buffer E (10 mM Tris-HCl [pH 8.0], 10 mM NaCl) using Slide-A-Lyzer dialysis cassettes (Pierce, Rockford, IL), and concentrated to 2 mg/ml with a Centricon YM-10 centrifugal concentrator (Millipore, Billerica, MA). The dialyzed protein (0.5 μg) was then electrophoresed on a 4 to 12% bis-Tris SDS-PAGE gel (Invitrogen, Carlsbad, CA) to assess its purity.
To investigate the role of flagella as a putative adhesin, 100 μg of the purified recombinant flagellin or 10 mM Tris-HCl was preincubated with HBMEC at 37°C for 1 h, and association assays with centrifugation were carried out as described above. His6-AslA (100 μg) was used as a nonspecific protein control. This protein contributes to E. coli K1 invasion of HBMEC but not to association (14; K. S. Kim, unpublished data).
Western blot analysis of flagellin.
The purified flagellin obtained as described above was subjected to Western blot analysis using anti-flagellin and anti-His antibodies. To do this, 0.5 μg of the total protein was analyzed for flagellin or the His tag after electrophoresis on a 4 to 12% bis-Tris gel (Invitrogen, Carlsbad, CA) and transfer to an NCP membrane (Bio-Rad, Hercules, CA). A rabbit polyclonal antibody against flagellin (anti-H7) was used as the primary antibody (1:500 dilution; Becton Dickinson, Sparks, MD). An anti-rabbit antibody (Cell Signaling Technology, Danvers, MA) labeled with horseradish peroxidase was used as the secondary antibody (1:5,000 dilution). To identify the His tag, an anti-His mouse monoclonal antibody (1:1,000 dilution) and a secondary anti-mouse antibody (1:5,000 dilution) conjugated with horseradish peroxidase were used. The membrane was then incubated with a chemiluminescent substrate (ECL; Amersham Biosciences, Piscataway NJ) according to the manufacturer's recommendations and visualized.
Data analysis.
Student's t test was employed to determine the statistical significance of association or invasion frequencies for experimental groups. A P value of <0.05 was considered statistically significant.
RESULTS
Flagellar mutants had similar growth rates but impaired swimming motility.
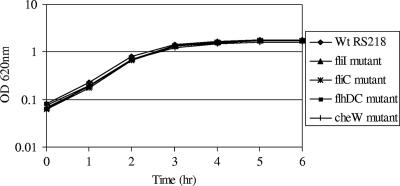
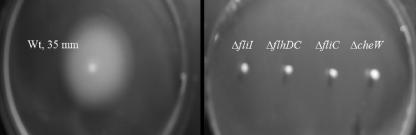
Since flagella are locomotive organelles of bacteria, deletion of genes affecting the flagellar machinery is expected to impair the motility of mutants. Since flagella are produced using more than 50 genes which are organized into hierarchal classes (7), defined deletion mutants with mutations in flhDC, fliI, fliC, and cheW were constructed to obtain representative of each class. As shown in Fig. 1, all the flagellar mutants had similar growth rates when they were grown for 6 h in LB broth with agitation. As expected, all the mutants exhibited defects in motility (Fig. 2). The motility of the wild-type bacteria was ∼35 mm, while the motilities of the flagellar mutants ranged from 2 mm (ΔfliI, ΔflhDC, and ΔfliC mutants) to 4 mm (ΔcheW mutant). Similar results were obtained in the absence of glucose as well as sodium chloride (data not shown). Also, none of the mutants had a motility phenotype when it was suspended in a film of liquid and examined with a microscope (data not shown).
FIG. 1.
Growth curves for wild-type strain RS218 and isogenic flagellar mutants of this strain. Bacteria were grown in LB broth at 37°C for 6 h with agitation, and the OD620 was determined at hourly intervals. The data are the means of two independent experiments. Wt, wild type.
FIG. 2.
Motility of the wild type and different flagellar mutants in 0.3% tryptone agar containing 0.3% glucose. Bacteria were grown on 1.5% tryptone agar plates, and isolated colonies were stab inoculated and grown at 37°C for 24 h. Wt, wild type.
Flagellum-deficient mutants exhibited defects in association with and invasion of HBMEC.
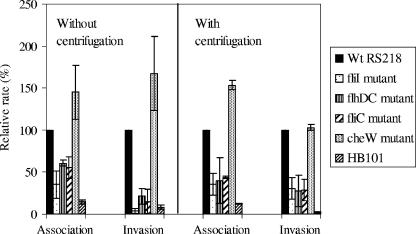
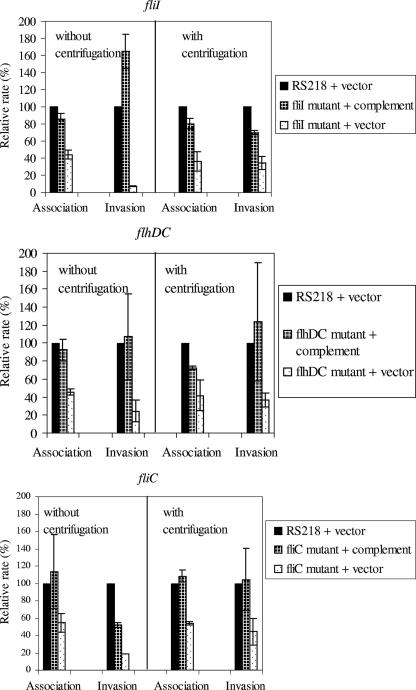
We next examined whether the mutants mentioned above are defective for association with and invasion of HBMEC and whether their defects are related to the defects in motility. As shown in Fig. 3, the fliI, flhDC, and fliC mutants exhibited defects in association and invasion frequencies compared to the parent wild-type strain, while the cheW mutant did not exhibit such defects. The mean relative HBMEC association rates of the fliI, flhDC and fliC mutants without centrifugation were 35, 61, and 56%, respectively, which were significantly (P < 0.05) less than that of the wild type. Similar statistically significant defects in association were observed even after the mutants were centrifuged onto HBMEC, and the mean association rates were 36, 40, and 44% for the fliI, flhDC, and fliC mutants, respectively. Thus, the use of a centrifugation step to establish close contact between bacteria and HBMEC did not restore the association of the fliI, flhDC, and fliC mutants with HBMEC, suggesting that motility is probably not involved in the E. coli K1 association with HBMEC. This conclusion was supported by our demonstration that the cheW mutant exhibited no defects in HBMEC association with and without centrifugation. Similar results were obtained with the motAB deletion mutant (data not shown). Deletion of motAB results in paralyzed flagella and no motility. No significant defects in HBMEC association were observed with and without centrifugation of this mutant onto HBMEC.
FIG. 3.
Relative association and invasion frequencies of different flagellar mutants with HBMEC compared to those of the wild-type strain. The assays were carried out with and without centrifugation of bacteria. The fliI, flhDC, and fliC mutants exhibited statistically significant (P < 0.05) decreases in the relative association and invasion frequencies, while the cheW mutant displayed no defects in association and invasion. The bars indicate the means of two or three independent experiments, and the error bars indicate the standard deviations. Wt, wild type.
Similar to the decreased association rates, the invasion frequencies of the fliI, flhDC, and fliC mutants were also decreased (Fig. 3). The mean relative invasion frequencies without centrifugation of the bacteria onto HBMEC were 4, 22, and 15% for the fliI, flhDC, and fliC mutants, respectively, while their relative invasion frequencies after centrifugation were 31, 28, and 29%, respectively. Irrespective of whether the assays were carried out with or without centrifugation, the relative invasion defects of the fliI, flhDC, and fliC mutants were statistically significant (P < 0.05) compared to the invasion of the wild type. On the other hand, similar to the association rate, the invasion rate of the cheW mutant was not significantly less than that of the wild type. Also, no increase in sensitivity to Triton X-100 was observed for the ΔfliI, ΔflhDC, and ΔfliC flagellar mutants, as indicated by the lack of significant decreases in colony counts for the mutants after incubation with 0.025% Triton X-100 for 30 min compared to the colony counts for the wild type (data not shown).
While defects in the HBMEC association rates of the fliI, flhDC, and fliC mutants were observed irrespective of centrifugation, no statistically significant differences were observed when the association rates obtained in the two sets of experiments (i.e., with and without centrifugation) were compared. Also, while the relative invasion frequencies of the fliI, flhDC, and fliC mutants without centrifugation were lower than the relative invasion frequencies with centrifugation, the differences were not statistically significant for the flhDC and fliC mutants.
Complementation restored motility, association, and invasion.
To verify that the defects associated with the fliI, flhDC, and fliC mutants were indeed due to the mutant genes and not to any polar effects on downstream genes, wild-type copies of each gene were introduced into the mutants in trans. The results are shown in Fig. 4 and 5.
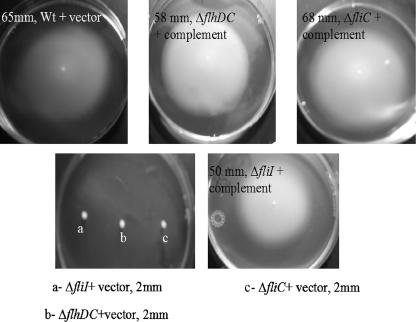
FIG. 4.
Motility of the flagellum mutant strains transcomplemented with the corresponding genes or of the vector alone compared to the motility of the wild-type strain with the vector. Soft agar plates were stab inoculated with isolated colonies of individual strains and incubated at 37°C for 24 h. Wt, wild type.
FIG. 5.
Relative association and invasion frequencies of the flagellum mutants transcomplemented with the corresponding genes. The bars indicate the means of two or three independent experiments, and the error bars indicate the standard deviations.
Motility defects were observed after deletion of fliI, flhDC, and fliC, but the motility was completely restored to the levels of the wild type after complementation, while introduction of the vector alone had no effect on motility (Fig. 4).
Transcomplementation also restored the association and invasion defects associated with the fliI, flhDC, and fliC mutations (Fig. 5). The association and invasion frequencies of the complemented strains were significantly higher than those of the mutants containing the vector alone, and the levels were as high as (or almost as high as) the levels of the parent strain with the vector alone. Similar to the results shown in Fig. 3, for each mutant containing the vector alone no overall statistically significant differences were observed between the HBMEC association and invasion frequencies with or without centrifugation.
Flagellum mutation does not affect type 1 fimbriation in E. coli RS218.
It has been suggested that type 1 fimbriae are regulated by flagella, and we have previously shown that type 1 fimbriae are involved in E. coli K1 association with HBMEC (36). Hence, in our next experiment we examined whether the HBMEC association and invasion defects of the flagellum mutants were due to any reduction in type 1 fimbria expression. The type 1 fimbria expression in the fliC mutant was examined by the yeast aggregation assay (36) and the PCR-based invertible element orientation assay, as described previously (1, 36). We did not observe any defects in type 1 fimbria expression in the fliC mutant with either of these methods (data not shown); e.g., the fliC mutant and the wild type exhibited the same yeast aggregation titer, 1: 64.
Purified flagellin blocks association of wild-type strain RS218 with HBMEC.
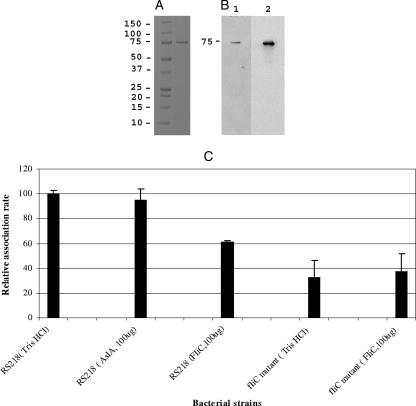
We next tried to obtain more information concerning the role of E. coli K1 flagella in promoting the association with HBMEC by using purified flagellin. Recombinant flagellin obtained with the cloned fliC gene was electrophoresed on a 4 to 12% bis-Tris gel (Invitrogen, Carlsbad, CA). A single band corresponding to a ∼75-kDa product was observed (Fig. 6A). Since the molecular mass of FliC is ∼60 kDa, this protein may represent an aggregated product; no other bands were observed, suggesting that the FliC represented the purified flagellin in the preparation. Our Western blot analysis with antibodies to histidine and H7 recognized the ∼75-kDa band (Fig. 6B, lanes 1 and 2, respectively). Next, we performed blocking assays with the purified flagellin. HBMEC were preincubated with 100 μg flagellin for 1 h at 37°C, and this was followed by association assays with the wild type or the fliC mutant with centrifugation. As shown in Fig. 6C, the association rate of the wild type was ∼40% (P < 0.05) lower than that of the 10 mM Tris-HCl control. On the other hand, addition of purified flagellin had no effect on the association rate of the fliC mutant (Fig. 6C). In addition, the nonspecific His-tagged AslA protein had no effect on E. coli K1 association when it was used at a similar concentration. These findings suggest that flagellin (FliC) contributes to E. coli K1 association with HBMEC.
FIG. 6.
(A and B) SDS-PAGE analysis of purified FliC obtained from the wild-type strain (panel A) and Western blot with anti-His and anti-flagellin antibodies (panel B, lanes 1 and 2, respectively). (C) HBMEC were preincubated with 100 μg of the preparation or 10 mM Tris-HCl for 1 h, followed by an association assay with the wild type or the fliC mutant with centrifugation. Preincubation treatments are indicated in parentheses. One hundred micrograms of His-tagged AslA was used as a nonspecific His-tagged protein control. Preincubation with FliC significantly (P < 0.05) blocked association of the wild type but had no effect on the fliC mutant. The bars indicate the means of two independent experiments, and the error bars indicate the standard deviations.
DISCUSSION
Flagella are locomotive organelles of bacteria, enabling the organisms to reach tissues, obtain nourishment, and/or colonize (3, 25, 26). On some occasions they have also been shown to be virulence factors enabling pathogenic bacteria to adhere to or to secrete effectors into eukaryotic cells for subsequent invasion (11, 23). However, this paradigm has been tested in multiple organisms only on mucosal surfaces, such as the respiratory, intestinal, or urogenital tract (25, 30, 31, 38). Crossing of the BBB by E. coli K1 is required for meningitis, and this has been shown to be a multistep process, involving binding to and invasion of HBMEC, which constitute the BBB. The role of bacterial flagella in crossing the BBB to cause meningitis, however, is unknown.
In recent studies using E. coli DNA microarrays (10), we observed that the level of expression of fliI was markedly greater (threefold) in E. coli K1 associated with HBMEC than in nonassociated bacteria (Xie et al., unpublished data). Since fliI encodes the ATPase involved in the flagellar secretion machinery, we investigated whether flagella are involved in the E. coli K1 association with and invasion of HBMEC in vitro by virtue of the flagellar structure and/or flagellum-mediated motility. For this purpose defined mutants were constructed for each class of genes in order to affect flagella (ΔfliI, ΔflhDC, and ΔfliC) or motility alone (ΔcheW), and their abilities to associate with and invade HBMEC were examined. Since flagella mediate motility, HBMEC association and invasion assays were conducted with and without centrifugation. A centrifugation step was used to promote close contact between bacteria and HBMEC and was expected to minimize any motility-mediated effect on bacterial association and invasion. As expected, mutants with all the mutations that affected flagellar structure (ΔfliI, ΔflhDC, and ΔfliC) exhibited significant defects in motility, like cheW (and motAB) deletion mutants. However, only fliI, flhDC, and fliC mutants exhibited significant defects in HBMEC association and invasion (Fig. 3). These defects were observed even after centrifugation of flagellar mutants onto HBMEC. Also, the defects were not due to increased sensitivity of the mutants to Triton X-100 since no significant differences in colony counts were observed when preparations were incubated with 0.025% Triton X-100 for 30 min. These findings suggest that flagella play an important role in E. coli K1 association with and invasion of HBMEC and that flagellum-mediated motility is less likely to be a factor contributing to HBMEC association and invasion. Additionally, since the fliI mutant still synthesized FliC but was defective for FliC secretion and flagellum formation, these findings suggest that the presence of flagellin outside the bacterial cell in the form of intact flagella is important for HBMEC association and invasion.
It is interesting that significant invasion defects were observed with the fliI, flhDC, and fliC mutants with and without centrifugation of bacteria compared to the characteristics of the wild-type. However, when invasion frequencies were compared as a measure of association rates, no significant defects in invasion were observed after the bacteria were centrifuged (Fig. 3). Thus, it is more likely that the flagellum contributes to association than that it contributes to invasion in the E. coli K1 interaction with HBMEC. Also, the defects in the association with and invasion of HBMEC of the fliI, flhDC, and fliC mutants (Fig. 3) were not due to their growth defects since none of the mutants exhibited a significant change in growth over a 6-h period in LB broth (Fig. 1), as well as in a 90-min experiment in tissue culture medium for HBMEC association and invasion assays (data not shown). In addition, complementation of the fliI, flhDC, and fliC mutants with the corresponding genes restored the association and invasion frequencies to the levels of the wild type (Fig. 5). These findings indicate that the HBMEC association and invasion defects seen with the fliI, flhDC, and fliC mutants were not related to any polar effect of downstream genes. It is noteworthy that complementation also restored the motility defects of these mutants (Fig. 4), but as indicated above, flagellum-mediated motility was not a major factor contributing to the E. coli K1 association with and invasion of HBMEC. This conclusion was also supported by our demonstration that the cheW and motAB mutants were defective for motility but retained the ability to associate with HBMEC (Fig. 2 and 3 and data not shown).
We have previously shown that type 1 fimbriae are important in the E. coli K1 association with HBMEC (36), and we examined whether the HBMEC association and invasion defects observed with the flagellum mutants were related to any reduction in type 1 fimbria expression. Barnich et al. (4) showed that type 1 fimbriation was significantly reduced in a fliC mutant of adherent invasive E. coli strain LF82 that was isolated from a patient with Crohn's disease. However, the archetypal ΔfliC flagellum mutant did not exhibit any reduction in type 1 fimbria expression compared to the expression in the wild-type strain. Our results are similar to those of Wright et al. (38), who showed that the levels of type 1 fimbria expression were the same in wild-type uropathogenic E. coli strain UTI89 and its isogenic fliC mutant.
Since we demonstrated that flagella are likely to be an association factor, we examined this conclusion using purified recombinant flagellin. Purified recombinant flagellin was prepared using the cloned fliC gene, and the preparation produced a single band corresponding to aggregated FliC, which cross-reacted with anti-His and anti-flagellin antibodies (Fig. 6A and B). It was interesting that preincubation of HBMEC with this purified flagellin preparation resulted in blocking of 40% of the wild-type E. coli K1 association with HBMEC, while preincubation with recombinant flagellin had no effect on the association of the fliC mutant with HBMEC (Fig. 6C). These findings suggest that the blocking effect of the exogenous flagellin preparation was specific to flagellin (FliC). Since flagella were demonstrated to be an association factor rather than an invasion factor in our study (Fig. 3 and 5), the effect of purified flagellin on invasion was not determined. Studies are in progress to further examine and characterize the role of flagellin in the E. coli K1 association with and subsequent invasion of HBMEC.
This is the first study demonstrating that bacterial flagella play a role in the E. coli K1 association with and invasion of HBMEC. Previously, there has been only one study in which researchers examined the role of flagella in binding to endothelial cells, which was a study of the binding of the protozoan parasite Trypanosoma congolese to bovine aortal endothelial cells (13). Incidentally, the human counterpart of this organism, Trypanosoma brucei, causes meningitis (12, 27). Thus, it is likely that flagella play an important role in the pathogenesis of E. coli K1 meningitis.
Acknowledgments
This work was supported by NIH grants.
We thank Donna Pearce for help with the Western blots.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Aberg, A., V. Shingler, and C. Balsalobre. 2006. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 60:1520-1533. [DOI] [PubMed] [Google Scholar]
- 2.Achtman, M., A. Mercer, B. Kusecek, A. Pohl, M. Heuzenroeder, W. Aaronson, A. Sutton, and R. P. Silver. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. [DOI] [PubMed] [Google Scholar]
- 4.Barnich, N., J. Boudeau, L. Claret, and A. Darfeuille-Michaud. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol. Microbiol. 48:781-794. [DOI] [PubMed] [Google Scholar]
- 5.Bortolussi, R., P. Ferrieri, B. Bjorksten, and P. G. Quie. 1979. Capsular K1 polysaccharide of Escherichia coli: relationship to virulence in newborn rats and resistance to phagocytosis. Infect. Immun. 25:293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collazo, C. M., and J. E. Galan. 1997. The invasion-associated type-III protein secretion system in Salmonella--a review. Gene 192:51-59. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cello, F., Y. Xie, M. Paul-Satyaseela, and K. S. Kim. 2005. Approaches to bacterial RNA isolation and purification for microarray analysis of Escherichia coli K1 interaction with human brain microvascular endothelial cells. J. Clin. Microbiol. 43:4197-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 12.Grab, D. J., O. Nikolskaia, Y. V. Kim, J. D. Lonsdale-Eccles, S. Ito, T. Hara, T. Fukuma, E. Nyarko, K. J. Kim, M. F. Stins, M. J. Delannoy, J. Rodgers, and K. S. Kim. 2004. African trypanosome interactions with an in vitro model of the human blood-brain barrier. J. Parasitol. 90:970-979. [DOI] [PubMed] [Google Scholar]
- 13.Hemphill, A., and C. A. Ross. 1995. Flagellum-mediated adhesion of Trypanosoma congolense to bovine aorta endothelial cells. Parasitol. Res. 81:412-420. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman, J. A., J. L. Badger, Y. Zhang, S. H. Huang, and K. S. Kim. 2000. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect. Immun. 68:5062-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, S. H., Y. H. Chen, Q. Fu, M. Stins, Y. Wang, C. Wass, and K. S. Kim. 1999. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect. Immun. 67:2103-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, S. H., Z. S. Wan, Y. H. Chen, A. Y. Jong, and K. S. Kim. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 183:1071-1078. [DOI] [PubMed] [Google Scholar]
- 17.Khan, N. A., S. Shin, J. W. Chung, K. J. Kim, S. Elliott, Y. Wang, and K. S. Kim. 2003. Outer membrane protein A and cytotoxic necrotizing factor-1 use diverse signaling mechanisms for Escherichia coli K1 invasion of human brain microvascular endothelial cells. Microb. Pathog. 35:35-42. [DOI] [PubMed] [Google Scholar]
- 18.Khan, N. A., Y. Wang, K. J. Kim, J. W. Chung, C. A. Wass, and K. S. Kim. 2002. Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J. Biol. Chem. 277:15607-15612. [DOI] [PubMed] [Google Scholar]
- 19.Kim, K. J., S. J. Elliott, F. Di Cello, M. F. Stins, and K. S. Kim. 2003. The K1 capsule modulates trafficking of E. coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell. Microbiol. 5:245-252. [DOI] [PubMed] [Google Scholar]
- 20.Kim, K. S. 2006. Microbial translocation of the blood-brain barrier. Int. J. Parasitol. 36:607-614. [DOI] [PubMed] [Google Scholar]
- 21.Kim, K. S. 2003. Pathogenesis of bacterial meningitis: from bacteraemia to neuronal injury. Nat. Rev. Neurosci. 4:376-385. [DOI] [PubMed] [Google Scholar]
- 22.Kim, K. S., H. Itabashi, P. Gemski, J. Sadoff, R. L. Warren, and A. S. Cross. 1992. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J. Clin. Investig. 90:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konkel, M. E., J. D. Klena, V. Rivera-Amill, M. R. Monteville, D. Biswas, B. Raphael, and J. Mickelson. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korhonen, T. K., M. V. Valtonen, J. Parkkinen, V. Vaisanen-Rhen, J. Finne, F. Orskov, I. Orskov, S. B. Svenson, and P. H. Makela. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48:486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane, M. C., V. Lockatell, G. Monterosso, D. Lamphier, J. Weinert, J. R. Hebel, D. E. Johnson, and H. L. Mobley. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73:7644-7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millikan, D. S., and E. G. Ruby. 2004. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 186:4315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olowe, S. A. 1975. A case of congenital trypanosomiasis in Lagos. Trans. R. Soc. Trop. Med. Hyg. 69:57-59. [DOI] [PubMed] [Google Scholar]
- 28.Prasadarao, N. V., C. A. Wass, S. H. Huang, and K. S. Kim. 1999. Identification and characterization of a novel Ibe10 binding protein that contributes to Escherichia coli invasion of brain microvascular endothelial cells. Infect. Immun. 67:1131-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasadarao, N. V., C. A. Wass, and K. S. Kim. 1996. Endothelial cell GlcNAc beta 1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect. Immun. 64:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prince, A. 2006. Flagellar activation of epithelial signaling. Am. J. Respir. Cell Mol. Biol. 34:548-551. [DOI] [PubMed] [Google Scholar]
- 31.Ramos, H. C., M. Rumbo, and J. C. Sirard. 2004. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12:509-517. [DOI] [PubMed] [Google Scholar]
- 32.Robbins, J. B., G. H. McCracken, Jr., E. C. Gotschlich, F. Orskov, I. Orskov, and L. A. Hanson. 1974. Escherichia coli K1 capsular polysaccharide associated with neonatal meningitis. N. Engl. J. Med. 290:1216-1220. [DOI] [PubMed] [Google Scholar]
- 33.Schmiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin, S., G. Lu, M. Cai, and K. S. Kim. 2005. Escherichia coli outer membrane protein A adheres to human brain microvascular endothelial cells. Biochem. Biophys. Res. Commun. 330:1199-1204. [DOI] [PubMed] [Google Scholar]
- 35.Stins, M. F., F. Gilles, and K. S. Kim. 1997. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J. Neuroimmunol. 76:81-90. [DOI] [PubMed] [Google Scholar]
- 36.Teng, C. H., M. Cai, S. Shin, Y. Xie, K. J. Kim, N. A. Khan, F. Di Cello, and K. S. Kim. 2005. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect. Immun. 73:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson, C. G., T. Kajander, and L. Regan. 2005. The crystal structure of NlpI. A prokaryotic tetratricopeptide repeat protein with a globular fold. FEBS J. 272:166-179. [DOI] [PubMed] [Google Scholar]
- 38.Wright, K. J., P. C. Seed, and S. J. Hultgren. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 73:7657-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie, Y., K. J. Kim, and K. S. Kim. 2004. Current concepts on Escherichia coli K1 translocation of the blood-brain barrier. FEMS Immunol. Med. Microbiol. 42:271-279. [DOI] [PubMed] [Google Scholar]