Abstract
The invasion of erythrocytes by Plasmodium falciparum occurs through multiple pathways that can be studied in vitro by examining the invasion of erythrocytes treated with enzymes such as neuraminidase, trypsin, and chymotrypsin. We have studied the invasion pathways used by 31 Kenyan P. falciparum isolates from children with uncomplicated or severe malaria. Six distinct invasion profiles were detected, out of eight possible profiles. The majority of isolates (23 of 31) showed neuraminidase-resistant, trypsin-sensitive invasion, characteristic of the pathway mediated by an unknown parasite ligand and erythrocyte receptor “X.” The neuraminidase-sensitive, trypsin-sensitive phenotype consistent with invasion mediated by the binding of parasite ligand erythrocyte binding antigen 175 to glycophorin A, the most common invasion profile in a previous study of Gambian field isolates, was seen in only 3 of 31 Kenyan isolates. No particular invasion profile was associated with severe P. falciparum malaria, and there was no significant difference in the levels of inhibition by the various enzyme treatments between isolates from children with severe malaria and those from children with uncomplicated malaria (P, >0.1 for all enzymes; Mann-Whitney U test). These results do not support the hypothesis that differences in invasion phenotypes play an important role in malaria virulence and indicate that considerable gaps remain in our knowledge of the molecular basis of invasion pathways in natural P. falciparum infections.
Erythrocyte invasion by merozoites is a crucial step during the intraerythrocytic asexual cycle of Plasmodium falciparum. This stage of the parasite infection causes an estimated 500 million clinical malaria cases per year (29), and understanding the parasite's biology is important for the identification of new drugs and vaccines. Four key stages of invasion have been described: merozoite attachment to the erythrocyte, apical reorientation, tight-junction formation, and finally, erythrocyte entry that involves the formation of a parasitophorous vacuole (8). The molecular basis of these processes is not fully understood; however, several parasite molecules involved in invasion have been identified and are encoded by two families of genes: erythrocyte binding antigens (EBA-175, EBA-140, and EBA-181) and reticulocyte binding protein homologues (Rh1, Rh2a, Rh2b, and Rh4) (8). It is now clear that in contrast to P. vivax (which requires the Duffy antigen receptor), P. falciparum merozoites interact with multiple alternative receptors that are important for erythrocyte invasion (8). Invasion pathways have been identified by examining the entry of merozoites into erythrocytes that have mutant or null receptor phenotypes (11, 22) and by removing erythrocyte surface receptors with enzyme treatments (2, 6, 19), the commonly used enzymes being neuraminidase, trypsin, and chymotrypsin. Neuraminidase removes sialic acids from the erythrocyte surface. Testing the ability of a parasite isolate to invade neuraminidase-treated erythrocytes defines whether the isolate is sialic acid dependent or independent. Trypsin removes surface glycoproteins, including glycophorin A (GPA) and glycophorin C (GPC). Chymotrypsin has a different specificity from trypsin and removes other proteins, including glycophorin B (GPB) and Band 3.
The invasion profiles of various P. falciparum laboratory strains have been defined previously by studying the invasion of enzyme-treated erythrocytes. Presently, six different invasion profiles of laboratory strains have been described, as shown in Table 1. It has been found that the pathways described for particular laboratory strains are consistent across laboratories, indicating that the invasion phenotype is stable (2, 6), with an exception being a report showing different invasion profiles for two 3D7 lines (4).
TABLE 1.
Invasion profiles of P. falciparum laboratory strains
| Invasion profilea | Parasite ligandb | RBC receptor | Reference(s) |
|---|---|---|---|
| NmS TS CTR | EBA-175 | GPA | 22, 26 |
| BAEBL/EBA-140 | GPC | 16, 18 | |
| NmS TR CTS | ? | GPB | 6 |
| JESEBL/EBA-181 | Receptor E | 9 | |
| NmS TR CTR | PfRh1 | Receptor Y | 23, 31 |
| NmR TS CT? | ? | Receptor X | 6 |
| NmR TR CTS | PfRh2b | Receptor Z | 7 |
| MSP1 | Band 3 | 10 | |
| AMA1 | Kx | 12 | |
| NmR T? CTR | PfRh4 | ? | 30 |
Nm, neuraminidase; T, trypsin; CT, chymotrypsin; S, sensitive; R, resistant.
Pf, P. falciparum.
Following from the work on laboratory strains, there has been recent interest in studying the invasion phenotypes of field isolates to understand the erythrocyte invasion process in natural P. falciparum infections. This issue is of particular relevance for the design of vaccines against blood-stage antigens, which may target specific invasion pathways. To date, there have been three studies of the invasion profiles of field isolates, with isolates collected in India, The Gambia, and Brazil (1, 15, 21). These studies have reported the invasion profiles of field isolates invading neuraminidase- or trypsin-treated erythrocytes (1, 21) or neuraminidase-, trypsin-, or chymotrypsin-treated erythrocytes (15). Among isolates from each study site, several distinct invasion profiles were detected, although there was one preferred invasion profile employed by the majority of isolates from each site. Twelve out of 15 Indian isolates used a neuraminidase- and trypsin-resistant pathway (21), whereas among isolates from The Gambia and Brazil, the neuraminidase- and trypsin-sensitive invasion profile was predominant (30 of 38 isolates in The Gambia and 7 of 14 isolates in Brazil) (1, 15). The study in The Gambia indicated that the majority of isolates probably invaded through the EBA-175/GPA pathway (or the EBA-140/GPC pathway, as the GPC receptor is also neuraminidase and trypsin sensitive) (1), but the Brazilian study showed that 5 of the 7 Brazilian isolates with this enzyme sensitivity receptor phenotype could invade erythrocytes which lacked GPA (15). In addition, approximately a third (5 of 14) of the Brazilian isolates used trypsin-resistant receptors, which is an unusual phenotype elsewhere (15).
All of the above-cited studies were based on field isolates collected from patients with uncomplicated malaria (1, 15, 21). Parasite isolates from patients with severe P. falciparum malaria have not been studied previously, and it is unknown whether isolates from patients with severe malaria may use a specific invasion pathway or possibly a larger range of invasion pathways than isolates from patients with mild malaria. We therefore investigated the invasion profiles of Kenyan field isolates collected from children diagnosed with either uncomplicated or severe P. falciparum malaria and tested for an association between the invasion profile and disease severity.
MATERIALS AND METHODS
Parasite isolates.
Parasite isolates were collected from patients with P. falciparum infections attending Kilifi District Hospital, Kilifi, Kenya, from December 2003 to January 2004 and June to August 2004. At this site, malaria transmission is seasonal (June to August and December to February), with the average number of infective bites per individual estimated at approximately 10 to 30 per year (13). Children with cerebral malaria (those that could not be roused from a comatose state with a Blantyre score of ≤2), prostration (the inability to sit or, for babies, to breast feed), or respiratory distress (abnormally deep breathing) were considered to have severe malaria. This clinical definition identifies approximately the same group of children at risk for life-threatening malaria as the more comprehensive World Health Organization criteria (17). Uncomplicated cases of malaria were defined as those in children presenting at the outpatient department of the same hospital with no signs of severe disease who were treated successfully with oral antimalarial therapy. Blood samples were collected immediately prior to treatment after obtaining informed consent from the children's parents or guardians, and protocols were approved by ethical review boards in the United Kingdom and Kenya. The isolates were collected into heparinized tubes and processed within 12 h of collection. Lymphocytes and the buffy coat were removed, and the erythrocytes were washed three times in incomplete RPMI 1640 medium (incRPMI; RPMI 1640 medium plus 25 mM HEPES, 2 mM glutamine, 25 mM glucose, and 25 μg of gentamicin/ml, pH 7.2 to 7.4). The parasites were cultured as described below.
Parasite genotyping.
For DNA analysis, approximately 200 μl of each parasite culture (equivalent to 5 μl of packed erythrocytes) was sampled before growth and DNA was extracted by using the QIAamp DNA blood mini kit (QIAGEN, United Kingdom). To test for the presence of multiple clones of P. falciparum among the isolates, the highly polymorphic repeat sequences in two loci (msp1 block 2 and msp2 block 3) were amplified by using a nested PCR method with allele type-specific internal primers (28).
SI.
The selectivity index (SI) is a measure of whether invasion is a random process or whether more multiple invasions occur than would be expected if invasion proceeded by chance alone. An increase in the number of multiple invasions is thought to occur when only a subset of the total red blood cell (RBC) population is available for invasion (27). An SI of 1 indicates the random invasion of RBCs, whereas an SI of, for example, 2 indicates twofold more multiply infected RBCs than would be expected if invasion occurred by chance alone. To assess the SI, thin smears from the blood sample collected from each patient were prepared and stained with Giemsa. Three hundred ring-infected erythrocytes per slide were counted, and the number of rings in each erythrocyte was recorded. The SI was calculated as described in detail previously (27). Briefly, the number of observed multiply infected erythrocytes (i.e., those with two or more rings per erythrocyte) was divided by the number of expected multiply infected erythrocytes. The latter was calculated according to a Poisson distribution, where lambda (the mean number of parasites per erythrocyte) is −ln(1 − the level of parasitemia). The level of parasitemia was calculated as the number of infected erythrocytes per 1,000 examined.
Parasite cultures.
Field isolates were cultured in complete RPMI 1640 medium (incRPMI plus 10% pooled human AB serum). Cultures were set up at a hematocrit value of 1 to 2% with blood group O+ erythrocytes and incubated at 37°C with 3% CO2, 1% O2, and 96% N2. Cultures were monitored for 18 to 36 h by using Giemsa smears, and only those isolates with normal morphology that matured to the schizont (segmenter) stage were included in the study. Two laboratory strains, P. falciparum clone 7G8 (11) and strain Dd2 (6), were studied as controls for the invasion inhibition assays. The laboratory strains were cultured in complete RPMI 1640 medium as described above except with 10% pooled human O serum. Laboratory strains were synchronized by sorbitol lysis (14).
Donor erythrocytes.
Donor erythrocytes for the field isolate invasion assays were collected from a single Caucasian donor (blood group O+). Ten milliliters of whole blood was collected into 2 ml of acid citrate dextrose, mixed to prevent coagulation, and stored at 4°C for a maximum of 2 weeks. Before use, the white blood cells were removed by layering 6 ml of whole blood over 5 ml of Lymphoprep (Axis Shield) and centrifuging for 20 min at 400 × g. The erythrocyte pellet was washed twice with 10 ml of incRPMI, resuspended at a 50% hematocrit value, and used within 1 week. O+ erythrocytes for experiments with laboratory strains were obtained from the Scottish Blood Transfusion Service. They were treated as described above.
Enzyme treatment of erythrocytes.
Frozen aliquots of trypsin, chymotrypsin, and the soybean trypsin inhibitor were prepared. The enzymes were dissolved in incRPMI to the required concentrations, as follows: trypsin, 1 mg/ml; chymotrypsin, 1 mg/ml; and soybean trypsin inhibitor, 0.5 mg/ml. They were then frozen in 1-ml aliquots at −20°C. The neuraminidase (Vibrio cholerae neuraminidase; Calbiochem) was reconstituted in distilled H2O and stored at 4°C according to the manufacturer's instructions. The enzyme treatment was based on protocols described previously (1). An aliquot of each of the required enzymes was thawed. A tube with 50 mU of neuraminidase in 1 ml of incRPMI was prepared, and one control tube with 1 ml of incRPMI was prepared. To each tube containing 1 ml of an enzyme mixture (neuraminidase, chymotrypsin, or trypsin) or incRPMI alone (control), a volume of 100 μl of packed donor erythrocytes was added. The tubes were incubated on a rotating wheel for 1 h at 37°C. After that time, all samples were washed once with 1 ml of incRPMI and then soybean trypsin inhibitor was added to the trypsin and chymotrypsin samples to inhibit the enzymes. IncRPMI was added to the control and neuraminidase samples. The samples were incubated on a rotating wheel for 10 min at room temperature and then washed three times before the erythrocytes were resuspended at a 50% hematocrit value in incRPMI. The enzyme-treated erythrocytes and controls were stored at 4°C and used for a maximum of 1 week, after which samples were freshly prepared.
Agglutination of enzyme-treated erythrocytes.
Agglutination assays of enzyme-treated erythrocytes were carried out to ensure that the specific epitopes were being removed successfully. A 20-μl sample of each of the different erythrocyte suspensions was diluted in 200 μl of 0.9% NaCl. Forty microliters of a 5% hematocrit sample of each aliquot was added to the same volume of anti-M antibody (Biotest [UK] Ltd.), anti-S antibody (Biotest [UK] Ltd.), or peanut (Arachis hypogaea) lectin (Sigma; reconstituted in phosphate-buffered saline and stored in aliquots at −20°C) in a Pyrex tube. The samples in the tubes were mixed, and the tubes were covered with Parafilm. The samples were incubated for 5 min (anti-S antibody) or 30 min (anti-M antibody and peanut lectin) at room temperature. The anti-S antibody samples were centrifuged for 1 min at 150 × g before the agglutination was assessed. To check for agglutination, the pellets were gently dislodged and the agglutination was assessed by eye and scored (one large agglutinate, ++; small agglutinates, +; and no agglutinates, −). Trypsin activity was shown by the complete loss of anti-M-mediated agglutination, chymotrypsin activity by the complete loss of anti-S-mediated agglutination, and neuraminidase activity by the gain of peanut lectin-mediated agglutination after enzyme treatment.
Invasion assays.
The assay to determine the parasite multiplication rate (PMR) in the first cycle of in vitro culture was set up as described previously (3, 5). Invasion inhibition assays were carried out with two laboratory strains (7G8 and Dd2) in three independent experiments and with the Kenyan field isolates in a single experiment. All parasites were cultured as described above until the mature schizont stage. The schizont samples were enriched by centrifugation through 55% Percoll. The schizonts from the enriched samples were washed twice, and the schizont concentrations were adjusted to around 1% parasitemia with normal group O+ RBCs or the matching enzyme-treated RBCs. The mixtures were incubated in 96-well plates at a volume of 50 μl and a hematocrit value of 2%, with duplicate wells for each treatment. The 96-well plate was placed within a gas modulator chamber to allow culturing in 3%CO2, 1% O2, and 96% N2 for 24 h at 37°C. Thin blood films from the preinvasion sample and the samples of ring-invaded erythrocytes after 24 h were made and stained with Giemsa to determine the level of parasitemia. At least 1,000 erythrocytes from each slide of the duplicate wells per sample were counted. The PMR is determined by dividing the number of ring-infected erythrocytes after invasion by the preinvasion level of parasitemia. Invasion inhibition was measured as follows: 100 − [(PMR after enzyme treatment/control PMR) × 100)]. In a few cases, no ring-infected erythrocytes were seen and the invasion inhibition was measured as 100%. Where the PMR in enzyme-treated erythrocytes was higher than that in control erythrocytes, the invasion inhibition is given as a negative value due to an increase in invasion.
Rosette frequency.
Mature, pigmented trophozoite-stage parasites were stained with 25 μg of ethidium bromide/ml for 5 min at 37°C, and a wet preparation was assessed for rosetting by microscopy. The rosette frequency is the percentage of mature infected erythrocytes binding two or more uninfected erythrocytes.
Statistical analysis.
Statview software (version 5; SAS Institute Inc.) was used for statistical analysis. All statistical tests of the Kenyan field isolate data were nonparametric tests as the data were not normally distributed. For statistical analyses, any negative invasion inhibition results were counted as 0. The Mann-Whitney U test or Kruskal-Wallis test was used to look for differences between two or more groups, respectively. Correlations between the SI, the PMR, the rosette frequency, and the invasion inhibition data were determined with Spearman's rank correlation.
RESULTS
Diversity in invasion profiles of Kenyan P. falciparum isolates.
The invasion profiles of 31 Kenyan field isolates in the first cycle of in vitro growth were examined. These isolates were a subset of those studied previously to examine the relationships among the PMR, the erythrocyte SI, and malaria severity (5). The inhibition of the invasion of neuraminidase-treated erythrocytes by Kenyan field isolates varied from −32.4% (indicating an increase in invasion) to 100% (i.e., no ring-infected erythrocytes were counted), with a median of 12.8%. The inhibition of the invasion of trypsin-treated erythrocytes varied from 28 to 100%, with a median of 89.1%. The inhibition of the invasion of chymotrypsin-treated erythrocytes ranged from −66.7 to 100%, with a median of 62.8%.
The invasion profiles of the field isolates were defined by the sensitivity of the isolates to invasion inhibition by neuraminidase, trypsin, and chymotrypsin. Isolates exhibiting levels of inhibition above 50% compared to the control (no enzyme treatment) were defined as sensitive, and those with levels below 50% were defined as resistant according to the common definition criteria (1, 15). The Kenyan isolates showed six out of a possible eight invasion profiles (Table 2). The most common profile, seen in 18 of 31 isolates, was neuraminidase-resistant and trypsin- and chymotrypsin-sensitive invasion, which matches the profile of the laboratory strain 7G8 (Table 2) (6). Five of the Kenyan isolates also showed neuraminidase resistance and trypsin sensitivity but were chymotrypsin resistant. Three of the Kenyan isolates were sensitive to all three enzymes, indicating a pathway whose molecular basis is completely unknown and that has been identified previously in only one Brazilian field isolate (15). Three of the isolates showed no sensitivity to chymotrypsin and were neuraminidase and trypsin sensitive, exhibiting the invasion profile predominant among samples of Brazilian isolates and Gambian isolates (although the chymotrypsin sensitivity of the Gambian isolates is unknown) (1, 15). One isolate showed the same profile as the laboratory strain Dd2 (6) and was neuraminidase sensitive and chymotrypsin and trypsin resistant. Finally, one isolate was resistant to both neuraminidase and trypsin treatment but was sensitive to chymotrypsin treatment. Two more profiles are theoretically possible but were not seen in this study (resistance to all three enzymes and a neuraminidase-sensitive, trypsin-resistant, and chymotrypsin-sensitive profile).
TABLE 2.
Invasion profiles of Kenyan field isolates
| Invasion profilea | Isolateb | Symptoms of clinical severityc | % Inhibition byd:
|
PMRe | SIe | ABO blood group typef | ||
|---|---|---|---|---|---|---|---|---|
| Neuraminidase (50 mU/ml) | Trypsin (1 mg/ml) | Chymotrypsin (1 mg/ml) | ||||||
| NmRTSCTS | 7G8 | 33.3 (19.2) | 80.7 (5.5) | 58.2 (7.5) | ||||
| 6 U | NA | 9.3 | 86.1 | 62.8 | 2.91 | 2.35 | O | |
| 7 U | NA | 12.8 | 83.0 | 76.6 | 4.70 | 1.28 | A | |
| 8 S | Pr, RD, An | 7.1 | 78.6 | 78.6 | 1.75 | 2.54 | A | |
| 9 S | C | −32.4 | 97.2 | 90.1 | 2.73 | 1.85 | O | |
| 10 S | Pr, RD | 0 | 87.8 | 73.2 | 2.05 | 2.06 | A | |
| 11 U | NA | 49.1 | 75.4 | 59.7 | 2.64 | 1.55 | O | |
| 12 S | Pr, RD | −22.8 | 85.7 | 62.9 | 2.50 | 1.96 | O | |
| 13 U | NA | 8.5 | 95.8 | 56.3 | 5.92 | 0.37 | O | |
| 14 U | NA | 1.1 | 89.1 | 56.5 | 3.29 | 1.32 | O | |
| 15 U | NA | 12.5 | 79.2 | 58.3 | 4.00 | ND | O | |
| 16 S | C, RD | 0 | 92.9 | 92.9 | 3.50 | 1.57 | O | |
| 17 U | NA | 7.5 | 79.1 | 64.2 | 6.09 | 0 | O | |
| 18 U | NA | 42.0 | 88.0 | 94.0 | 4.17 | 1.91 | O | |
| 19 U | NA | 17.0 | 85.1 | 59.6 | 1.74 | 4.97 | O | |
| 20 U | NA | 10.3 | 92.3 | 92.3 | 2.29 | 1.13 | O | |
| 21 S | Pr | 23.5 | 94.1 | 70.6 | 0.71 | 1.31 | B | |
| 22 S | Pr, RD, An | 33.3 | 92.6 | 55.6 | 1.13 | 2.33 | B | |
| 23 U | NA | 29.6 | 77.8 | 77.8 | 1.50 | 3.74 | O | |
| NmRTSCTR | 1 U | NA | 13.8 | 100 | 43.1 | 0.58 | 2.34 | O |
| 2 U | NA | 8.9 | 91.1 | 48.2 | 3.15 | 1.76 | ND | |
| 3 S | RD | −13.2 | 89.5 | 42.1 | 1.58 | 1.71 | O | |
| 4 S | C, RD | 6.3 | 100 | −66.7 | 0.48 | 1.89 | A | |
| 5 U | NA | −23.6 | 94.4 | 28.3 | 2.33 | 1.31 | O | |
| NmSTSCTS | 29 S | Pr, An | 100 | 82.5 | 90.2 | 1.43 | 2.45 | B |
| 30 U | NA | 56.0 | 100 | 100 | 0.69 | ND | A | |
| 31 U | NA | 56.1 | 100 | 63.4 | 0.82 | 1.51 | B | |
| NmSTSCTR | 26 S | Pr | 75.2 | 96.0 | 30.2 | 3.51 | 1.63 | O |
| 27 U | NA | 100 | 100 | 31.5 | 0.73 | 2.35 | AB | |
| 28 U | NA | 58.0 | 81.3 | 38.6 | 1.76 | 0.86 | B | |
| NmSTRCTR | Dd2 | 89.2 (10.2) | 47.6 (16.9) | 11.1 (30.5) | ||||
| 25 U | NA | 58.5 | 28.6 | 8.3 | 2.17 | 3.27 | ND | |
| NmR TR CTS | 24 S | Pr | 16.0 | 28.0 | 100 | 0.25 | 1.76 | B |
Nm, neuraminidase; T, trypsin; CT, chymotrypsin; S, sensitive; R, resistant.
Each field isolate designation includes a label to indicate whether the isolate was derived from a child with uncomplicated (U) or severe (S) malaria. Laboratory strains 7G8 and Dd2 are indicated in italics. For the laboratory strains, the means (and standard deviations) of results from three experiments are shown. Field isolate data are from a single experiment for each isolate. Field isolates corresponding to infections with the indicated number of genotypes are as follows: one genotype, isolates 8, 21, 22, and 26; two genotypes, isolates 2, 11, 12, and 13; three genotypes, isolates 9 and 23; four genotypes, isolates 5 and 14. Other isolates were not genotyped.
Severe malaria was indicated by a comatose state (C), respiratory distress (RD), prostration (Pr), and anemia (An). For the uncomplicated malaria cases, NA indicates not applicable.
Percentages of inhibition of invasion are relative to the level of invasion of non-enzyme-treated control erythrocytes. A result of 100% inhibition indicates that no rings of infected erythrocytes were counted, whereas a negative percentage of inhibition (e.g., −32%) indicates an increase in invasion compared to the control.
These results have been published previously in more detail (5). PMR, parasite multiplication rate; SI, selectivity index.
For two samples, the corresponding ABO type was not determined (ND).
The minimum number of parasite genotypes per isolate from each patient was estimated for isolates from 12 patients, and as expected for an area with moderately high levels of malaria transmission, multiple infections were common (mean number of genotypes overall, 2.2; mean number of genotypes in isolates from patients with severe malaria [n = 6], 1.5; and mean number of genotypes in isolates from patients with uncomplicated malaria [n = 6], 2.8). Four of six patients with severe malaria had single infections, whereas none of the patients with uncomplicated malaria had single infections. The presence of multiple genotypes within an isolate from a single patient has the potential to complicate the interpretation of invasion profile data, because the profile seen may be a composite of different parasite types. In a mixed-clone infection, it is unclear whether a single genotype tends to dominate the infection or whether the isolate consists of roughly equal proportions of the different clones. A previous study found no significant difference in the invasion profiles corresponding to single-clone infections and multiple-clone infections, suggesting that the presence of mixed-clone infections does not necessarily confound the data (1). In addition, for practical reasons, the examination of only single-clone infections in areas with moderate to high levels of transmission would severely restrict the number of samples available and would exclude potentially important data.
Invasion profiles of Kenyan isolates and malaria severity.
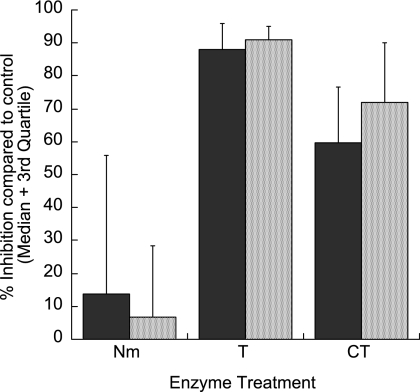
Previous studies of the invasion pathways used by field isolates have been carried out with isolates taken from patients with uncomplicated malaria. It is unknown whether isolates from patients with severe P. falciparum malaria may invade by a specific invasion pathway. We therefore compared the levels of inhibition of the invasion of enzyme-treated erythrocytes by isolates collected from patients with severe and uncomplicated malaria. Nineteen of the isolates were from children with uncomplicated malaria (mean age, 46.3 months [standard deviation, 34.9 months]; mean hemoglobin level, 9.8 g/dl [standard deviation, 1.3 g/dl[), and 12 isolates were collected from children with severe malaria (mean age, 19.6 months [standard deviation, 11.2 months]; mean hemoglobin level, 7.7 g/dl [standard deviation, 3.1 g/dl]). Of the patients with severe malaria, three had cerebral malaria, four showed prostration and respiratory distress, one had respiratory distress only, one exhibited prostration and severe anemia, and three presented with prostration only. As seen in Table 2, the isolates from both clinical groups showed similar distributions of recorded invasion profiles. Figure 1 shows the median levels of the inhibition of invasion by isolates from patients with uncomplicated or severe malaria in association with each of the enzyme treatments, compared to the invasion of non-enzyme-treated control erythrocytes. The levels of inhibition did not differ between the isolates from patients with uncomplicated malaria and those from patients with severe malaria in association with any of the enzyme treatments (P, >0.1 for all enzyme treatments; Mann-Whitney U test).
FIG. 1.
Percentages of inhibition of erythrocyte invasion by Kenyan field isolates from children with uncomplicated (black bars) or severe (gray bars) P. falciparum malaria. The erythrocytes were treated with neuraminidase (Nm; 50 mU/ml), trypsin (T; 1 mg/ml), or chymotrypsin (CT; 1 mg/ml). The median values are shown; error bars represent third quartile values. No significant differences between the isolates from patients with uncomplicated malaria and those from patients with severe malaria were associated with any of the enzyme treatments (P, >0.1 for all enzyme treatments; Mann-Whitney U test).
Invasion profiles and other parasite variables.
We determined the SIs (a measure of whether invasion occurs randomly in any available erythrocyte or is selective for a subset of erythrocytes) (27), PMRs (5), and rosette frequencies (a parasite virulence-associated factor) (24, 25) for the isolates included in this study as part of a larger study of PMRs and clinical malaria isolate phenotypes described elsewhere (5). None of these variables correlated significantly with the level of inhibition by any of the enzyme treatments (P > 0.1; Spearman's rank correlation).
ABO blood group and invasion phenotype.
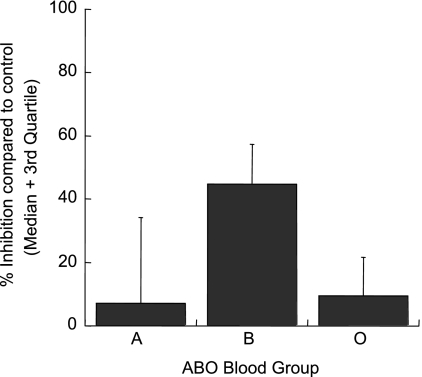
We found a significant difference in neuraminidase sensitivity among isolates taken from patients of different ABO blood group types (Fig. 2). Seventeen isolates were collected from patients with blood group O, six were from patients with blood group B, and five were from patients with blood group A. The single sample from a patient with blood group AB was excluded from this analysis, and for two samples the corresponding blood group data were missing. It was found that isolates from patients with blood group B were more inhibited by the neuraminidase treatment of erythrocytes, with a median level of inhibition of 44.7% (interquartile range [IQR], 23.5 to 58.0%; n = 6) compared to 7.1% (IQR, 3.1 to 34.4%; n = 5) and 9.3% (IQR, 0 to 23.3%; n = 17) for blood groups A and O, respectively (P = 0.02; Kruskal-Wallis test). This finding indicates that isolates growing in blood group type B are more dependent on sialic acid for the invasion of type O erythrocytes (the donor of the erythrocytes used for all invasion assays was type O). Trypsin or chymotrypsin sensitivity was not related to the ABO blood group. No other patient variable (age, level of parasitemia, hemoglobin level, or white blood cell count) was associated with the level of inhibition of the invasion of enzyme-treated erythrocytes.
FIG. 2.
Percentages of inhibition of the invasion of neuraminidase-treated erythrocytes by Kenyan field isolates from children with the ABO blood group A (n = 5), B (n = 6), or O (n = 17). The median levels of inhibition are shown; error bars represent third quartile values. Isolates from patients with blood group B were more inhibited by the treatment of erythrocytes with neuraminidase than those from patients with blood group A or O (P = 0.02; Kruskal-Wallis test).
DISCUSSION
We examined the abilities of Kenyan field isolates to invade erythrocytes treated with three different enzymes in order to assess the invasion pathways used by parasites isolated from patients with clinical P. falciparum malaria infections. In contrast to the results of previous studies from The Gambia and Brazil (1, 15), but similar to those of a study from India (21), it was found that the neuraminidase- and trypsin-sensitive, chymotrypsin-resistant receptors (characteristic of EBA-175/GPA- and EBA-140/GPC-mediated invasion) were rarely used by Kenyan field isolates (Table 2). Geographical variation in invasion profiles may have important implications for blood-stage vaccines which target antigens involved in invasion pathways. For example, a vaccine based on EBA-175 and/or EBA-140 may not be as effective in Kenya as it would be in The Gambia. Furthermore, the redundancy of invasion pathways used by field isolates indicates that a vaccine based on only one invasion ligand may not greatly inhibit P. falciparum invasion and growth but may instead select for parasites which invade by different mechanisms.
The invasion patterns of the majority of the Kenyan isolates (23 of 31) could be broadly described as neuraminidase resistant and trypsin sensitive. This profile was originally described for the laboratory strain 7G8, and it was inferred that this profile indicated the existence of an erythrocyte receptor “X” (6, 11). The finding that this pathway occurs commonly in clinical malaria isolates in Kenya argues that further research to delineate the molecular interactions underlying this pathway is of major importance. Indeed, it is likely that X is not only one receptor, as invasion by this pathway may be either sensitive or resistant to the treatment of erythrocytes with chymotrypsin. The parasite ligand(s) involved in the neuraminidase-resistant, trypsin-sensitive pathway is also unclear, although one possible candidate is the Rh4 protein that mediates neuraminidase-resistant, chymotrypsin-resistant invasion (30) (the trypsin sensitivity of Rh4-mediated invasion has not yet been described).
The work described here constitutes the first study to examine the invasion profiles of field isolates collected from children with severe P. falciparum malaria. Although no significant association between the invasion profiles and disease severity was seen in this study, the sample size was small (only 12 isolates were from children with assorted severe malaria syndromes), so we cannot rule out that a difference may be seen in a larger study, and further work is required. However, the lack of an observed association between invasion profiles and disease severity is consistent with the findings of a previous study of a larger group of isolates collected at the same study site. No significant differences in the multiplication rates or the red cell SIs between isolates from children with uncomplicated malaria and those with severe malaria were observed (5), in contrast with previous results from Thailand (3). Due to the strong association between the PMR and disease severity in Thailand, a study of the invasion profiles of Thai field isolates, examining whether the high multiplication rates displayed by isolates from patients with severe malaria can be associated with overall less sensitivity to the enzyme treatment of erythrocytes or with a specific preferred invasion profile, may provide further insight.
As knowledge of the P. falciparum ligands involved in invasion increases and technological advances allow their study in field samples, it is important to characterize the expression levels of putative ligands and correlate these findings with invasion phenotypes. Thirteen of the samples analyzed in this study were also included in a recent study of the expression profiles of four Rh genes (those encoding Rh1, Rh2a, Rh2b, and Rh4) and the EBA-175 gene (20). Although the numbers are small and, therefore, this analysis should be regarded as preliminary, a positive correlation between the relative levels of EBA-175 and sensitivity to the treatment of erythrocytes with neuraminidase was found, consistent with the known neuraminidase sensitivity of the EBA-175 receptor GPA (26).
One unexpected finding from this study was a significant difference in the levels of inhibition by neuraminidase treatment depending on the ABO blood group of the host from which the parasites were isolated. Isolates from patients with blood group B were the most inhibited by the treatment of erythrocytes with neuraminidase compared to isolates from patients with blood group A or O (Fig. 2) (P = 0.02; Kruskal-Wallis test). This finding suggests that the host ABO type may influence the invasion phenotype of the parasite. It remains unclear whether these results occurred by chance, as the sample size was small and the study was not designed to look at the invasion inhibition in relation to the host ABO blood group type, but these findings may merit further investigation.
In summary, this work shows that a variety of invasion pathways are used by Kenyan field isolates and that there is no single discernible pathway specifically associated with severe P. falciparum malaria. Further studies in a variety of geographical areas with isolates associated with various disease syndromes, along with parallel studies examining the transcription of parasite invasion ligands (20), will be required to fully characterize the diversity in invasion pathways and indicate if these pathways may be effectively targeted by vaccines based on the relevant ligands.
Acknowledgments
We are grateful to the clinical, nursing, and laboratory staff at the KEMRI Unit in Kilifi, Kenya, for their assistance with sample collection for this study and to the patients and their parents for their participation. This work is published with the permission of the director of KEMRI.
This work was supported by the Wellcome Trust (PhD studentship to A.-M.D. and senior research fellowship to J.A.R., grant number 067431).
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Baum, J., M. Pinder, and D. J. Conway. 2003. Erythrocyte invasion phenotypes of Plasmodium falciparum in The Gambia. Infect. Immun. 71:1856-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binks, R. H., and D. J. Conway. 1999. The major allelic dimorphisms in four Plasmodium falciparum merozoite proteins are not associated with alternative pathways of erythrocyte invasion. Mol. Biochem. Parasitol. 103:123-127. [DOI] [PubMed] [Google Scholar]
- 3.Chotivanich, K., R. Udomsangpetch, J. A. Simpson, P. Newton, S. Pukrittayakamee, S. Looareesuwan, and N. J. White. 2000. Parasite multiplication potential and the severity of Falciparum malaria. J. Infect. Dis. 181:1206-1209. [DOI] [PubMed] [Google Scholar]
- 4.Cortes, A., A. Benet, B. M. Cooke, J. W. Barnwell, and J. C. Reeder. 2004. Ability of Plasmodium falciparum to invade Southeast Asian ovalocytes varies between parasite lines. Blood 104:2961-2966. [DOI] [PubMed] [Google Scholar]
- 5.Deans, A. M., K. E. Lyke, M. A. Thera, C. V. Plowe, A. Kone, O. K. Doumbo, O. Kai, K. Marsh, M. J. Mackinnon, A. Raza, and J. A. Rowe. 2006. Low multiplication rates of African Plasmodium falciparum isolates and lack of association of multiplication rate and red blood cell selectivity with malaria virulence. Am. J. Trop. Med. Hyg. 74:554-563. [PMC free article] [PubMed] [Google Scholar]
- 6.Dolan, S. A., J. L. Proctor, D. W. Alling, Y. Okubo, T. E. Wellems, and L. H. Miller. 1994. Glycophorin B as an EBA-175 independent Plasmodium falciparum receptor of human erythrocytes. Mol. Biochem. Parasitol. 64:55-63. [DOI] [PubMed] [Google Scholar]
- 7.Duraisingh, M. T., T. Triglia, S. A. Ralph, J. C. Rayner, J. W. Barnwell, G. I. McFadden, and A. F. Cowman. 2003. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 22:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaur, D., D. C. Mayer, and L. H. Miller. 2004. Parasite ligand-host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int. J. Parasitol. 34:1413-1429. [DOI] [PubMed] [Google Scholar]
- 9.Gilberger, T. W., J. K. Thompson, T. Triglia, R. T. Good, M. T. Duraisingh, and A. F. Cowman. 2003. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J. Biol. Chem. 278:14480-14486. [DOI] [PubMed] [Google Scholar]
- 10.Goel, V. K., X. Li, H. Chen, S. C. Liu, A. H. Chishti, and S. S. Oh. 2003. Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc. Natl. Acad. Sci. USA 100:5164-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadley, T. J., F. W. Klotz, G. Pasvol, J. D. Haynes, M. H. McGinniss, Y. Okubo, and L. H. Miller. 1987. Falciparum malaria parasites invade erythrocytes that lack glycophorin A and B (MkMk). Strain differences indicate receptor heterogeneity and two pathways for invasion. J. Clin. Investig. 80:1190-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, K., D. C. Mayer, S. Singh, M. Reid, and L. H. Miller. 2005. Domain III of Plasmodium falciparum apical membrane antigen 1 binds to the erythrocyte membrane protein Kx. Proc. Natl. Acad. Sci. USA 102:5552-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinyanjui, S. M., T. Howard, T. N. Williams, P. C. Bull, C. I. Newbold, and K. Marsh. 2004. The use of cryopreserved mature trophozoites in assessing antibody recognition of variant surface antigens of Plasmodium falciparum-infected erythrocytes. J. Immunol. Methods 288:9-18. [DOI] [PubMed] [Google Scholar]
- 14.Lambros, C., and J. P. Vanderberg. 1979. Synchronisation of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418-420. [PubMed] [Google Scholar]
- 15.Lobo, C. A., K. de Frazao, M. Rodriguez, M. Reid, M. Zalis, and S. Lustigman. 2004. Invasion profiles of Brazilian field isolates of Plasmodium falciparum: phenotypic and genotypic analyses. Infect. Immun. 72:5886-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maier, A. G., M. T. Duraisingh, J. C. Reeder, S. S. Patel, J. W. Kazura, P. A. Zimmerman, and A. F. Cowman. 2003. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 9:87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh, K., D. Forster, C. Waruiru, I. Mwangi, M. Winstanley, V. Marsh, C. Newton, P. Winstanley, P. Warn, N. Peshu, G. Pasvol, and R. Snow. 1995. Indicators of life-threatening malaria in African children. N. Engl. J. Med. 332:1399-1404. [DOI] [PubMed] [Google Scholar]
- 18.Mayer, D. C., J. B. Mu, X. Feng, X. Z. Su, and L. H. Miller. 2002. Polymorphism in a Plasmodium falciparum erythrocyte-binding ligand changes its receptor specificity. J. Exp. Med. 196:1523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell, G. H., T. J. Hadley, M. H. McGinniss, F. W. Klotz, and L. H. Miller. 1986. Invasion of erythrocytes by Plasmodium falciparum malaria parasites: evidence for receptor heterogeneity and two receptors. Blood 67:1519-1521. [PubMed] [Google Scholar]
- 20.Nery, S., A. M. Deans, M. Mosobo, K. Marsh, J. A. Rowe, and D. J. Conway. 2006. Expression of Plasmodium falciparum genes involved in erythrocyte invasion varies among isolates cultured directly from patients. Mol. Biochem. Parasitol. 149:208-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okoyeh, J. N., C. R. Pillai, and C. E. Chitnis. 1999. Plasmodium falciparum field isolates commonly use erythrocyte invasion pathways that are independent of sialic acid residues of glycophorin A. Infect. Immun. 67:5784-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasvol, G., M. Jungery, D. J. Weatherall, S. F. Parsons, D. J. Anstee, and M. J. Tanner. 1982. Glycophorin as a possible receptor for Plasmodium falciparum. Lancet ii:947-950. [DOI] [PubMed] [Google Scholar]
- 23.Rayner, J. C., E. Vargas-Serrato, C. S. Huber, M. R. Galinski, and J. W. Barnwell. 2001. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J. Exp. Med. 194:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowe, J. A., J. Obiero, K. Marsh, and A. Raza. 2002. Positive correlation between rosetting and parasitemia in Plasmodium falciparum clinical isolates. Am. J. Trop. Med. Hyg. 66:458-460. [DOI] [PubMed] [Google Scholar]
- 25.Rowe, J. A., J. Shafi, O. K. Kai, K. Marsh, and A. Raza. 2002. Nonimmune IgM, but not IgG, binds to the surface of Plasmodium falciparum-infected erythrocytes and correlates with rosetting and severe malaria. Am. J. Trop. Med. Hyg. 66:692-699. [DOI] [PubMed] [Google Scholar]
- 26.Sim, K. L., C. E. Chitnis, K. Wasniowska, T. J. Hadley, and L. H. Miller. 1994. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264:1941-1944. [DOI] [PubMed] [Google Scholar]
- 27.Simpson, J. A., K. Silamut, K. Chotivanich, S. Pukrittayakamee, and N. J. White. 1999. Red cell selectivity in malaria: a study of multiple-infected erythrocytes. Trans. R. Soc. Trop. Med. Hyg. 93:165-168. [DOI] [PubMed] [Google Scholar]
- 28.Snounou, G., X. Zhu, N. Siripoon, W. Jarra, S. Thaithong, K. N. Brown, and S. Viriyakosol. 1999. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans. R. Soc. Trop. Med. Hyg. 93:369-374. [DOI] [PubMed] [Google Scholar]
- 29.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434:214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stubbs, J., K. M. Simpson, T. Triglia, D. Plouffe, C. J. Tonkin, M. T. Duraisingh, A. G. Maier, E. A. Winzeler, and A. F. Cowman. 2005. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science 309:1384-1387. [DOI] [PubMed] [Google Scholar]
- 31.Triglia, T., M. T. Duraisingh, R. T. Good, and A. F. Cowman. 2005. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol. Microbiol. 55:162-174. [DOI] [PubMed] [Google Scholar]