Abstract
Group A Streptococcus (GAS) genes that encode proteins putatively involved in polysaccharide utilization show growth phase-dependent expression in human saliva. We sought to determine whether the putative polysaccharide transcriptional regulator MalR influences the expression of such genes and whether MalR helps GAS infect the oropharynx. Analysis of 32 strains of 17 distinct M protein serotypes revealed that MalR is highly conserved across GAS strains. malR transcripts were detectable in patients with GAS pharyngitis, and the levels increased significantly during growth in human saliva compared to the levels during growth in glucose-containing or nutrient-rich media. To determine if MalR influenced the expression of polysaccharide utilization genes, we compared the transcript levels of eight genes encoding putative polysaccharide utilization proteins in the parental serotype M1 strain MGAS5005 and its ΔmalR isogenic mutant derivative. The transcript levels of all eight genes were significantly increased in the ΔmalR strain compared to the parental strain, especially during growth in human saliva. Following experimental infection, the ΔmalR strain persistently colonized the oropharynx in significantly fewer mice than the parental strain colonized, and the numbers of ΔmalR strain CFU recovered were significantly lower than the numbers of the parental strain CFU recovered. These data led us to conclude that MalR influences the expression of genes putatively involved in polysaccharide utilization and that MalR contributes to the persistence of GAS in the oropharynx.
Genetic studies have shown that some of the microbial genes that are most widely and differentially expressed are those that encode proteins putatively involved in carbohydrate transport and metabolism (1, 3, 32, 37, 44). Changes in the transcript levels of these genes are often accompanied by alterations in genes encoding classical virulence factors, such as secreted toxins, proteases, immune effector molecules, and capsule synthesis proteins (17, 25, 28, 35, 46). Thus, genome-wide investigations of major bacterial pathogens have suggested that there is a close link between basic metabolic processes and pathogenesis (25, 32, 35). These findings in turn have led to recent investigations attempting to more clearly elucidate the relationship between bacterial nutrition and infection (7, 17, 34, 41).
In humans, group A Streptococcus (GAS) causes diverse infections ranging from innocuous (e.g., simple colonization and uncomplicated pharyngeal and skin infections) to life threatening (e.g., necrotizing fasciitis and toxic shock syndrome) (10). This diversity implies that GAS infection involves complex regulatory networks that are different in different environments (21). When faced with new surroundings, GAS not only alters the transcription of genes involved in meeting basic metabolic demands but also alters the transcription of genes encoding major virulence factors (15, 25, 35, 38). Therefore, gene regulation in GAS forms a link between basic nutrition and pathogenesis.
The major site of GAS infection and colonization in humans is the oropharynx (29). A key mediator of acquired and innate immunity in the human oropharynx is saliva, and we have recently been exploring molecular interactions between GAS and human saliva (33). Using expression microarrays, we analyzed GAS transcription during the logarithmic and stationary growth phases of GAS organisms in human saliva (35). Of the nearly 1,700 genes in the GAS genome, we found that 14 of the 15 genes that had the largest growth phase-dependent changes in transcript levels encode proteins that are thought to be involved in carbohydrate transport and metabolism (35). Moreover, 8 of these 14 genes encode proteins that are either putatively or known to be involved in the uptake and processing of polysaccharides (35). In studies of GAS growth in a nonhuman primate model of pharyngitis, in human blood ex vivo, and in soft tissue infection in mice, we found that the transcription of genes encoding polysaccharide utilization proteins was highly dynamic (14, 15, 44). Taken together, these data suggest that the regulation of genes involved in polysaccharide degradation, transport, and utilization likely contributes to GAS host-pathogen interactions.
Recently, we demonstrated that the GAS putative maltodextrin binding protein MalE was important for colonization of the oropharynx (34). The transcript level of malE was increased nearly 100-fold during growth in human saliva compared to growth in a standard laboratory medium (34). In this work we sought to determine whether the transcript levels of seven other genes encoding proteins putatively involved in polysaccharide digestion, transport, and metabolism were elevated in human saliva in a fashion similar to malE. We also investigated whether the transcript levels of putative polysaccharide utilization genes were influenced by MalR, a putative transcriptional repressor not previously studied in GAS. We discovered that MalR influences the expression of at least eight genes residing in two distinct locations on the GAS chromosome. Moreover, we found that MalR is essential for high-level persistence of GAS in human saliva and in the mouse oropharynx.
MATERIALS AND METHODS
Bacterial strains and culture media.
The genome of serotype M1 strain MGAS5005 has been completely sequenced (39). Strain MGAS5005 has been studied extensively in animal models of GAS infection and in vitro, including transcriptome analysis during growth in human saliva (34, 35, 40, 44). The other bacterial strains used in this study are listed in Table 1. GAS was grown on Trypticase soy agar containing 5% sheep blood (BSA) (Becton Dickinson) or in Todd-Hewitt broth containing 0.2% (wt/vol) yeast extract (THY) (Difco) at 37°C without shaking in 5% CO2. Spectinomycin (Sigma) was added to THY or THY agar at a concentration of 150 μg/ml when appropriate. Carbohydrates were added at a concentration of 1.0% (wt/vol) to a carbohydrate-free preparation of a commercially available chemically defined medium (CDM) (JR Biosciences) (45). In this paper, we use the terms glucose-medium, maltose-medium, maltotriose-medium, etc., to refer to the carbohydrate-free CDM with the carbohydrate indicated added at a concentration of 1.0% (wt/vol).
TABLE 1.
Homology of MalR in various GAS strains
| Strain | M serotype | Year isolated | Disease or source | Nucleotide sequence changes compared with strain MGAS5005 | % MalR amino acid sequence identity with strain MGAS5005 |
|---|---|---|---|---|---|
| SF370 | 1 | 1985 | Wound infection | None | 100 |
| MGAS2221 | 1 | 1988 | Scarlet fever | None | 100 |
| MGAS9506 | 1 | 2001 | Pharyngeal | None | 100 |
| MGAS10606 | 1 | 2002 | Pharyngeal | None | 100 |
| MGAS9490 | 2 | 2001 | Pharyngeal | G93A, A648G | 100 |
| MGAS10270 | 2 | 2002 | Pharyngeal | G93A, A648G | 100 |
| MGAS10599 | 2 | 2002 | Pharyngeal | G93A, A648G | 100 |
| MGAS315 | 3 | 1988 | Toxic shock | G93A, C843T | 100 |
| SSI-1 | 3 | 1994 | Toxic shock | G93A, C843T | 100 |
| MGAS10652 | 3 | 2002 | Pharyngeal | G93A, C843T | 100 |
| MGAS11787 | 3 | 2003 | Pharyngeal | G93A, C843T | 100 |
| MGAS9503 | 4 | 2001 | Pharyngeal | G93A, A648G | 100 |
| MGAS10750 | 4 | 2002 | Pharyngeal | G93A, A648G | 100 |
| MGAS9512 | 6 | 2001 | Pharyngeal | G93A, A320T, C324T, T369C, T513C, A648Ga | 99.7 |
| MGAS10394 | 6 | 2001 | Pharyngeal | G93A, A320T, C324T, T369C, T513C, A648Ga | 99.7 |
| MGAS11802 | 6 | 2003 | Pharyngeal | G93A, A320T, C324T, T369C, T513C, A648Ga | 99.7 |
| MGAS11791 | 6 | 2003 | Pharyngeal | G93A, A320T, C324T, T369C, T513C, A648Ga | 99.7 |
| MGAS11847 | 11 | 2003 | Pharyngeal | G93A, A320T, C324T, T369C, T513C, A648Ga | 99.7 |
| MGAS2096 | 12 | 1980 | APSGNb | T168C, A648G | 100 |
| MGAS9429 | 12 | 1998 | Pharyngeal | T168C, A648G | 100 |
| MGAS9511 | 12 | 2001 | Pharyngeal | T168C, A648G | 100 |
| MGAS10604 | 12 | 2002 | Pharyngeal | T168C, A648G | 100 |
| MGAS11785 | 12 | 2003 | Pharyngeal | T168C, A648G | 100 |
| MGAS8232 | 18 | 1987 | Pharyngeal | G93A, T624G, A648G | 100 |
| MGAS11789 | 18 | 2003 | Pharyngeal | G93A, T624G, A648G | 100 |
| MGAS10603 | 22 | 2002 | Pharyngeal | G93A, A648G | 100 |
| MGAS6180 | 28 | 1998 | Blood | G93A, A648G | 100 |
| MGAS9513 | 28 | 2001 | Pharyngeal | G93A, A648G | 100 |
| MGAS10602 | 28 | 2002 | Pharyngeal | G93A, A648G | 100 |
| MGAS10600 | 75 | 2002 | Pharyngeal | G93A, T624G, A648G | 100 |
| MGAS9516 | 77 | 2001 | Pharyngeal | G93A, A648G | 100 |
| MGAS10601 | 77 | 2002 | Pharyngeal | G93A, A648G | 100 |
| MGAS9502 | 89 | 2001 | Pharyngeal | G93A, T624G, A648G | 100 |
| MGAS11790 | 102 | 2003 | Pharyngeal | G93A, A648G | 100 |
| MGAS11861 | 124 | 2003 | Pharyngeal | G93A, A648G | 100 |
The A320T nucleotide substitution in serotype M6 and M11 strains results in an Asp107Val replacement at the amino acid level.
APSGN, acute poststreptococcal glomerulonephritis.
Growth of GAS in human saliva.
GAS was grown in human saliva as previously described (33). Briefly, saliva was collected, briefly centrifuged, and passed through a 0.20-μm filter. GAS was grown in saliva without shaking at 37°C with 5% CO2. Healthy volunteers donated saliva according to a protocol for human subjects approved by the Baylor College of Medicine Institutional Review Board (33). Saliva pooled from at least four donors was used to minimize the effects of donor variation on the study results. All comparative growth experiments were done in duplicate on five separate occasions for a total of 10 replicates. To determine whether any observed growth differences might be related to differences in clumping induced by saliva, Gram staining was performed on culture aliquots. The average numbers of GAS in aggregates were determined by counting 10 high-power fields for each aliquot.
DNA sequence analysis.
Chromosomal DNA was isolated using a DNeasy kit (QIAGEN). DNA sequencing primers were designed on the basis of the serotype M1 strain MGAS5005 genome (39). The malR open reading frame M5005_Spy_1057 corresponds to open reading frame SPy1293 in serotype M1 strain SF370 (13). Sequence data obtained from both DNA strands with an Applied Biosystems 3730XL DNA analyzer were assembled with Sequencher 4.5. The inferred amino acid sequences were aligned and compared using the software program CLUSTALW. All sequences that differed from the strain MGAS5005 sequence were confirmed by repeating the entire DNA sequence analysis.
RNA isolation and transcript level analysis.
Serotype M1 strain MGAS5005 was grown to the mid- and late-logarithmic phases. RNA was isolated with a Fast Prep Blue kit (Q/BioGene) and was purified using an RNeasy kit (QIAGEN) (35). The quality and the concentration of RNA were assessed with an Agilent 2100 Bioanalyzer and by analysis of the A260/A280 ratio. cDNAs were created from the RNA using Superscript III (Invitrogen) by following the manufacturer's instructions. For comparison of gene transcript levels during growth in various media, TaqMan real-time quantitative reverse transcription-PCR (QRT-PCR) was performed in quadruplicate with an ABI Thermocycler 7700 (Applied Biosystems) using the ΔCT method of analysis (8). To compare gene transcript levels of the parental and ΔmalR and ΔmalRv2 isogenic mutant strains, the ΔΔCT method was employed (user bulletin no. 2, ABI PRISM 7700 sequence detection system; Applied Biosystems). TaqMan primers and probes for genes of interest and the internal control gene proS are listed in Table 2. All experiments were performed in quadruplicate on two separate occasions.
TABLE 2.
Primers and probes used in this study
| Primer | Sequence (5′→3′) | Target |
|---|---|---|
| malR-A | GTT AGC TTG TTT AAA GGT ACC ACC | 5′ primer for 5′ region of malR |
| malR-B | GTT ATA GTT ATT ATA ACA TGT ATT TGC CTT TTT GAG CGA CAT CTT | 3′ primer for 5′ region of malR with spc sequence overlap |
| malR-C | CTA TTT AAA TAA CAG ATT AAA AAA ATT ATA AGA AAC AAA GAG AAA GCG TCC GT | 5′ primer for 3′ region of malR with spc sequence overlap |
| malR-D | AAC TTC TAC ACT GTC CGC AGA AAC | 3′ primer for 3′ region of malR |
| malR-spcF | AAG ATG TCG CTC AAA AGG CAA ATA CAT GTT ATA ATA ACT ATA AC | 5′ primer for spc along with malR overlap sequence |
| malR-spcR | GGA CGC TTT CTC TTT GTT TTA AAC CTT ATA ATT TTT TTT AAT CTG TTA TTT AAA TAG | 3′ primer for spc along with malR overlap sequence |
| malR-SouthF | CGT CGT TGG CTA GAA TTT GCT | 5′ primer for Southern blotting |
| malR-SouthR | AGA ATC CAG CTT TTG CGA TG | 3′ primer for Southern blotting |
| malR-F | AAC CCT TCG ACG GTG AGT AG | 5′ malR primer for QRT-PCR |
| malR-R | AAT CTG TGC TGC GAC ATT TG | 3′ malR primer for QRT-PCR |
| malR-P | TCC AAA TCT GCC ATC GCT TTT C | malR TaqMan probe |
| malP-F | TGA TGA AGC GGT AGC TGT TG | 5′ malP primer for QRT-PCR |
| malP-R | GCA GTT GGC CAT TTT TCA AG | 3′ malP primer for QRT-PCR |
| malP-P | TGG TTG GTT ACA CTA ACC ACA CTA TTC TTG CA | malP TaqMan probe |
| malQ-F | CTC AGC CCC AGG TAT AGC AT | 5′ malQ primer for QRT-PCR |
| malQ-R | GG TTT TCT GCG ATG ATA GGG | 3′ malQ primer for QRT-PCR |
| malQ-P | TCC TAA GGC CTC ACG AAC CGC | malQ TaqMan probe |
| malE-F | TGA AAT CAT GGC AAA AAG TTA TCG | 5′ malE primer for QRT-PCR |
| malE-R | GAT CCA CAT CCC ACT AAC AAG GTA CT | 3′ malE primer for QRT-PCR |
| malE-P | GCA AGT GTC AAA CTT GCT CCG CCG | malE TaqMan probe |
| malF-F | GAT TGG GTG GGA CTT GCT AA | 5′ malF primer for QRT-PCR |
| malF-R | CAG CCC AAA TCA AAG TCC AT | 3′ malF primer for QRT-PCR |
| malF-P | TTC CTGCCA TAC GGC CGC TT | malF TaqMan probe |
| malG-F | TCA ATG CTT CAG CCA AGA TAC C | 5′ malG primer for QRT-PCR |
| malG-R | AGT GAT TGG GAC GGC AAT TA | 3′ malG primer for QRT-PCR |
| malG-P | ATG GCC TTC ACT GCT GGT TCT GTC | malG TaqMan probe |
| pulA-F | GGA CTA GCC CGC GAT GAA | 5′ pulA primer for QRT-PCR |
| pulA-R | CCT TGC TAG ACG ATT GAA GAC CAT | 3′ pulA primer for QRT-PCR |
| pulA-P | CTC AAC AAG GAG ATG GCA ATG CTA AAT TCT GG | pulA TaqMan probe |
| dexB-F | AGC GTC AGC TTG GAG AAG AG | 5′ dexB primer for QRT-PCR |
| dexB-R | CCA CAT CTG TAT TGG CGA TG | 3′ dexB primer for QRT-PCR |
| dexB-P | TGC CAC AGA CTT AGC AGG TGC ACA | dexB TaqMan probe |
| msmK-F | GTT AAA GAT GGC CGC ATT GT | 5′ msmK primer for QRT-PCR |
| msmK-R | ACC AGC AGC TTC AAG CAT TT | 3′ msmK primer for QRT-PCR |
| msmK-P | TGA CAT TGC TAT TCC AGA AGG ACA GCA | msmK TaqMan probe |
| proS-F | TGA GTT TAT TAT GAA AGA GGC TAT AGT TTC | 5′ proS primer for QRT-PCR |
| proS-R | AAT AGC TTC GTA AGC TTG ACG ATA AT | 3′ proS primer for QRT-PCR |
| proS-P | TCG TAG GTC ACA TCT AAT CTT CAT AGT TG | proS TaqMan probe for QRT-PCR |
Creation of ΔmalR isogenic mutant strain.
The ΔmalR isogenic mutant strain was created by nonpolar insertional mutagenesis from parental serotype M1 strain MGAS5005 using the PCR-mediated method described by Kuwayama et al. (23). Primers were designed to amplify the 5′ and 3′ ends of the malR gene region along with nucleotide sequences that were complementary to the 5′ or 3′ portion of the spectinomycin (spc) resistance cassette (Table 2). A third set of primers containing nucleotide sequences complementary to the 5′ and 3′ ends of the malR gene region was used to amplify the spc cassette from plasmid pSL60 (24). Fusion PCR was then used to link the 5′ and 3′ malR gene region PCR products to the spc cassette via the overlapping nucleotide sequence regions (11, 23). This resulted in a PCR product in which the spc cassette, flanked by the 5′ and 3′ malR gene regions, was inserted in place of nearly the entire malR open reading frame. The gene disruption PCR construct was used to transform competent GAS cells, which were then selected via spectinomycin resistance. The ΔmalR isogenic mutant strain was analyzed by Southern hybridization and DNA sequencing to confirm that the proper genetic construct was obtained (data not shown). Extensive efforts to complement the ΔmalR strain in trans were unsuccessful. Therefore, we addressed the possibility that the effects observed in the ΔmalR strain were due to unrelated spontaneous mutations acquired during construction of the mutant by creating a second, independent ΔmalR isogenic mutant strain. Southern hybridization and DNA sequencing were used to confirm that the proper genetic construct was obtained (data not shown).
Carbohydrate uptake assays.
Strain MGAS5005 and its ΔmalR isogenic mutant derivative were grown to the mid-exponential phase in CDM in the presence of the carbohydrate of interest added at a concentration of 1.0%. The bacteria were collected by centrifugation, washed, and suspended to an optical density at 600 nm (OD600) of 0.5 in 150 μl of carbohydrate-free CDM. [14C]maltose (600 mCi/mmol; 200 μCi/ml) and [14C]maltotriose (900 mCi/mmol; 100 μCi/ml) were purchased from American Radiolabeled Chemicals, St. Louis, MO. [14C]maltose was judged to be >98% pure by thin-layer chromatography, whereas [14C]maltotriose was found to be ∼90% pure (American Radiolabeled Chemicals, personal communication). Forty microliters of the 14C-labeled sugars was added to GAS cells at 37°C to obtain a final volume of 200 μl and a final concentration of radiolabeled carbohydrate of 40 μM. At the times indicated below, 40-μl samples were removed and filtered through a 0.45-μm nitrocellulose membrane (Millipore), which was then rinsed twice with carbohydrate-free CDM. The radioactivity retained on each filter was determined by liquid scintillation counting (Beckman model LS7500). Samples were taken every 30 s for the first 120 s, a period during which the carbohydrate transport rates were found to be linear. The uptake rates were adjusted to the amounts of protein in the cultures as determined by the Bradford assay (Bio-Rad). All experiments were performed in quadruplicate on two separate occasions.
Mouse colonization experiments.
All experiments with mice were performed according to a protocol approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. Mouse throat colonization studies were conducted with adult (18- to 20-g) female outbred CD-1 Swiss mice (Harlan Sprague-Dawley Inc.) (24, 34). The MGAS5005 and ΔmalR strains were grown in THY and harvested at an OD600 of approximately 0.5. The cells were washed once with sterile phosphate-buffered saline and suspended in phosphate-buffered saline at a concentration of 1 × 108 CFU/ml. Each nostril of each mouse was inoculated with 50 μl of the GAS suspension (total inoculum, 1 × 107 CFU), and there were 35 mice in each group. In previous experiments with MGAS5005 this dose resulted in colonization of approximately 65% of the inoculated animals at 72 h (24). The throat of each mouse was swabbed before inoculation and then daily thereafter. The throat swabs were streaked onto BSA and grown overnight to obtain beta-hemolytic colonies. These colonies were then tested for the presence of GAS carbohydrate antigen via latex agglutination (BD Biosciences). Blood collected by cardiac puncture from dead mice was cultured on BSA overnight.
Statistical analysis.
Growth and RNA transcript levels were compared using Student's two-sided t test. Radiolabeled carbohydrate uptake rates were compared using linear regression. The χ2 test was used to assess statistical differences in throat colonization rates between the mice infected with strain MGAS5005 and the mice infected its ΔmalR isogenic mutant derivative. Student's two-sided t test was used to test for differences in the number of CFU recovered from animals infected by the two strains. Statistical significance was assigned a two-sided P value of 0.05 using Bonferroni's adjustment for multiple comparisons when appropriate. Statistical calculations were performed using the NCSS software, version 2004.
RESULTS
GAS genome contains maltodextrin and pullulanase gene regions.
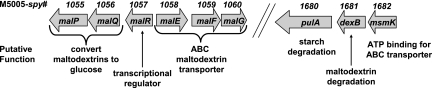
Maltodextrin is the term given to molecules composed of two to seven linked glucose monomers. Polysaccharide is the term for molecules containing more than seven glucose monomers. In gram-positive bacteria, genes encoding proteins putatively involved in the binding, transport, and utilization of maltodextrins and polysaccharides may be either contiguous or located in separate parts of the chromosome (22, 42). In all 12 GAS strains sequenced to date, including strain MGAS5005, these genes are found in two regions (2, 4, 5, 13, 16, 26, 36, 39). Putative maltodextrin binding, transport, and utilization genes are located in the gene region from M5005_Spy_1055 to M5005_Spy_1060 (Fig. 1). This region includes M5005_Spy_1058 (malE), a gene encoding a maltodextrin binding cell surface lipoprotein recently shown to be necessary for GAS to colonize the oropharynx (34). Genes encoding proteins putatively involved in the production of maltodextrin from polysaccharides, maltodextrin degradation, and ATP binding are located in a separate part of the chromosome, from M5005_Spy_1680 to M5005_Spy_1682 (Fig. 2). This region includes the gene encoding M5005_SPy_1680 (PulA), an LPXTG cell wall-anchored pullulanase thought to be involved in GAS virulence (20, 30, 31).
FIG. 1.
Schematic diagram of the maltodextrin and pullulanase gene regions in the chromosome of GAS strain MGAS5005. The M5005_spy number is the gene number in serotype M1 strain MGAS5005. The same gene arrangement is present in all 12 GAS strains sequenced to date (2, 4, 5, 13, 16, 26, 36, 39).
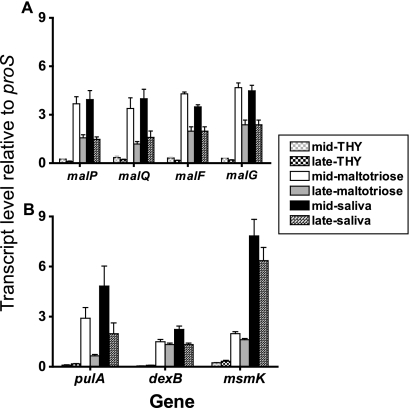
FIG. 2.
Transcript levels of maltodextrin (A) and pullulanase (B) region genes are increased during growth in a maltotriose-medium and human saliva. Strain MGAS5005 was grown to the mid-exponential or late-exponential phase in either standard laboratory medium (THY), CDM containing 1% maltotriose, or human saliva. TaqMan real-time QRT-PCR was performed using probes and primers listed in Table 2. The transcript levels of target genes were normalized to those of proS, a gene that is expressed constitutively throughout the GAS cell cycle and whose transcript levels are similar when the organism is grown in THY and when the organism is grown in saliva (35, 43). For each gene, the order of bars from left to right is as follows: mid- and late-exponential growth phases in THY, maltotriose-medium, and human saliva. The transcript levels are the means ± standard deviations of quadruplicate measurements obtained on two separate occasions.
In this paper, we refer to the region comprising the genes M5005_Spy_1055 to M5005_Spy_1060 as the maltodextrin gene region and the region comprising the genes M5005_Spy_1680 to M5005_Spy_1682 as the pullulanase gene region. The maltodextrin gene region contains three of the four genes that putatively encode an ATP binding cassette (ABC) transport system, a common mechanism used by bacteria to import low-molecular-weight substrates (18). The fourth gene, M5005_Spy_1682 (msmK), which encodes the putative ATP-binding portion of the maltodextrin ABC transport system, is located in the pullulanase gene region. The gene encoding the putative transcriptional regulator MalR (M5005_SPy_1057) is located in the maltodextrin gene region and is transcribed divergently from malE. The pullulanase gene region does not contain a putative transcriptional regulator. The lack of a gene encoding a transcriptional regulator in the pullulanase gene region suggests that expression of genes in this region is influenced by proteins encoded in other regions of the GAS genome.
Transcript levels of genes in the maltodextrin and pullulanase gene regions are increased in human saliva and a maltotriose-medium compared to a nutrient-rich medium.
Before the present study, we had demonstrated that malE transcript levels were significantly higher during growth in human saliva and in a maltodextrin-medium than during growth in a nutrient-rich medium (THY) (34). Now, we asked whether the same was true for putative polysaccharide utilization genes other than malE. As assessed by real-time QRT-PCR, the transcript levels of malP, malQ, malF, and malG during the mid-exponential phase of growth in human saliva and in a maltotriose-medium were increased an average of 75-fold compared to the levels in THY (P < 0.001 for both human saliva and in the maltotriose-medium) (Fig. 2A). Moreover, the transcript level for each of the maltodextrin region genes in a maltotriose-medium and in saliva was higher during the mid-exponential phase than during the late-exponential phase (Fig. 2A) (34). Similar to the findings for the maltodextrin region genes, the transcript levels of each of the three genes in the pullulanase gene region were significantly higher during growth in a maltotriose-medium and human saliva than during growth in THY (Fig. 2B). The pulA transcript levels followed a pattern nearly identical to the pattern for the maltodextrin utilization genes; they were higher during the mid-exponential growth phase than during the late exponential phase in a maltotriose-medium and human saliva (Fig. 2B). The transcript levels of dexB and msmK were less growth phase dependent. Taken together, these data suggest that gene transcript levels in the maltodextrin and pullulanase gene regions were increased during growth in a maltotriose medium and human saliva and that similar regulatory mechanisms influenced the two gene regions.
Putative maltodextrin and pullulanase transcriptional regulator MalR is highly conserved among diverse GAS strains.
As the marked elevation of the transcript levels for the maltodextrin and pullulanase region genes in a maltotriose-medium and human saliva suggested that the two gene regions share some aspects of transcriptional regulation, we next focused on MalR, the only putative transcriptional regulator encoded in the two gene regions. Some transcriptional regulatory proteins are highly variable, whereas others are well conserved among GAS strains (6). Moreover, single nucleotide changes or insertions have been shown to affect the function of GAS transcriptional regulators (4, 40). To determine whether MalR is conserved among genetically diverse GAS strains, we sequenced the malR gene from 35 strains representing 18 different M serotypes (Table 1). All strains tested had the malR gene. Nine nucleotide changes were identified, and the mean frequency of these changes was 2.46 nucleotide substitutions in the 1,017-bp open reading frame compared to the reference allele in serotype M1 strains. Eight of the nucleotide changes were synonymous substitutions resulting in less than one amino acid replacement per 339 amino acid sites (Table 1). There were no insertions, deletions, or nucleotide changes that resulted in truncated proteins. Thus, MalR is very highly conserved among genetically diverse GAS strains.
malR transcript levels are significantly higher during growth in human saliva and a maltotriose-medium than during growth in a nutrient-rich medium.
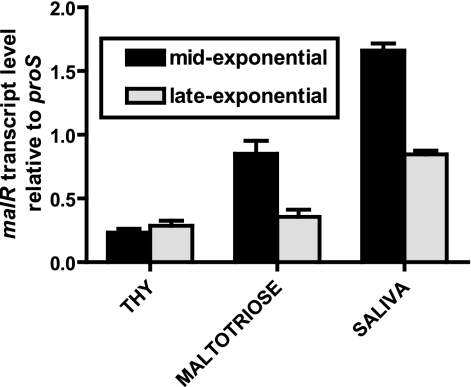
Data on how MalR functions in gram-positive organisms other than GAS are conflicting. For example, the homologue of MalR acts as a transcriptional repressor in Streptococcus pneumoniae, whereas in Staphylococcus xylosus and Enterococcus faecalis it acts as a gene activator (12, 19, 27). To begin to investigate the function of MalR in GAS, we determined the transcript levels of malR in various growth conditions. Similar to the levels of other genes in the maltodextrin gene region, malR transcript levels were significantly higher during the mid-exponential growth phase in a maltotriose-medium and in saliva than in THY (P < 0.01 for each comparison) (Fig. 3). Coupled with the increase in maltodextrin and pullulanase region gene transcript levels observed in a maltotriose-medium and human saliva, the pattern of malR transcript levels suggested that MalR may positively influence gene expression in GAS.
FIG. 3.
malR transcript levels are elevated during growth of GAS in a maltotriose-medium and human saliva. Strain MGAS5005 was grown to the mid-exponential phase and late-exponential phase in THY, a maltotriose-medium, and human saliva. TaqMan real-time QRT-PCR was performed using primers and probes listed in Table 2. The transcript levels of target genes were normalized to those of proS, a gene that is expressed constitutively throughout the GAS cell cycle and whose transcript levels are similar when the organism is grown in THY and when the organism is grown in saliva (35, 43). The transcript levels are the means ± standard deviations of quadruplicate measurements obtained on two separate occasions.
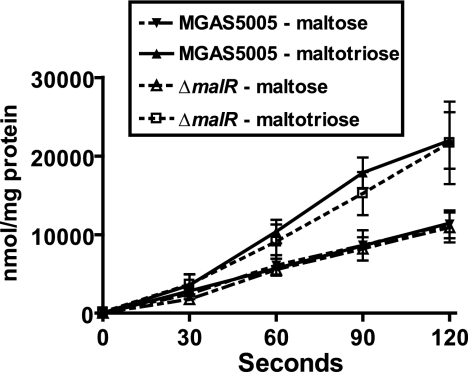
Uptake of maltodextrins is similar in parental and ΔmalR isogenic mutant strains.
To gain further insight into the role of MalR, we replaced the malR open reading frame in the serotype M1 strain MGAS5005 with a spectinomycin resistance cassette in a nonpolar fashion (24). Four genes in the maltodextrin and pullulanase gene regions (i.e., malE, malF, malG, and msmK) encode proteins that putatively form an ABC transport system for maltodextrins. To test the hypothesis that MalR activates maltodextrin transport, we studied the uptake of 14C-radiolabeled maltose and maltotriose by parental and ΔmalR GAS strains and found that it was similar for both strains (P = 0.84 for maltose and P = 0.57 for maltotriose) (Fig. 4). The finding that MalR was not needed for optimum transport of maltose or maltotriose suggests that MalR is not required for the activation of maltodextrin transport genes in GAS.
FIG. 4.
Uptake of maltodextrins in GAS is not affected by deletion of malR. Serotype M1 strain MGAS5005 and its isogenic ΔmalR derivative were grown in a maltose- or maltotriose-medium to the mid-exponential phase. Cells were harvested by centrifugation, washed with carbohydrate-free CDM, and suspended to an OD600 of 0.5. [14C]maltose or [14C]maltotriose was added to a final concentration of 40 μM. Samples were removed every 30 s for 120 s and passed through a 0.45-μm filter. The filters were washed twice with carbohydrate-free CDM, and the radioactivity retained was determined using a liquid scintillation counter. The data are the means ± standard deviations for four replicates done on two separate occasions.
ΔmalR mutant strain persists at a lower number of CFU than the parental strain in human saliva.
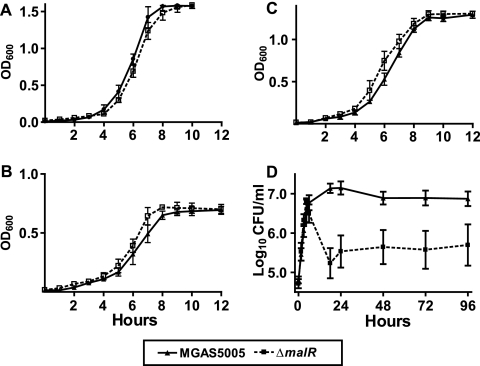
In light of the radioactive uptake data, we next tested the hypothesis that MalR is not necessary for the growth of GAS in media containing maltodextrin. Consistent with this hypothesis, there was no significant difference between the parental and ΔmalR strains in growth in THY, a maltose-medium, or a maltotriose-medium (Fig. 5A to C) (P = 0.791 for THY, P = 0.187 for maltose-medium, and P = 0.115 for maltotriose-medium).
FIG. 5.
Growth curves of parental and ΔmalR strains in various media. OD600 readings were taken at different times to measure growth in a nutrient-rich medium (THY) (A) and CDM with either 1% maltose (B) or 1% maltotriose (C). CFU were quantified and analyzed as a measure of growth in human saliva (D) as previously described (33). The data are the means ± standard deviations of duplicate measurements for five independent experiments.
Given the elevated malR transcript levels that we observed during growth in saliva, we hypothesized that MalR is important for optimal growth of GAS in human saliva. To test this hypothesis, we grew GAS in saliva and analyzed the resulting growth using CFU analysis. Numbers of CFU were used rather than density readings because the normal tendency of GAS to aggregate in saliva interferes with optical density readings (9, 33). In human saliva, strain MGAS5005 grew to a density of approximately 2.5 log10 CFU/ml from the baseline and persisted at a density of approximately 1 × 107 CFU/ml, whereas the ΔmalR isogenic mutant strain grew to a density of only approximately 1.9 log10 CFU/ml and persisted at a density of approximately 5.5 × 105 CFU/ml (Fig. 5D) (P = 0.03 for growth and P = 0.004 for persistence). Repeated Gram stain analysis of the saliva showed no difference between the aggregation of strain MGAS5005 and the aggregation of the ΔmalR strain, indicating that differences in cell clumping were unlikely to account for the growth disparity. Further analysis of the data revealed that the growth of the parental strain and the growth of the ΔmalR strain were initially identical, but the viability of the ΔmalR strain decreased rapidly during the shift from rapid growth to high-level persistence during the stationary phase (Fig. 5D). These data suggest that in human saliva, maintenance GAS from the rapid growth phase to the stationary phase requires MalR.
Despite extensive efforts, we were unable to genetically complement the ΔmalR strain. Therefore, to show that the changes observed in the ΔmalR strain were not due to spurious mutations that occurred during creation of the mutant strain, we created a second, independent ΔmalR strain (see Materials and Methods). The growth curves of the second ΔmalR strain in THY, maltose-medium, maltotriose-medium, and human saliva were identical to those of the original ΔmalR strain (data not shown).
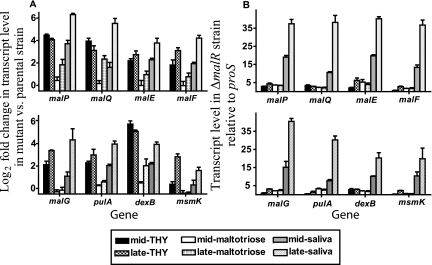
MalR negatively influences the transcription of maltodextrin and pullulanase region genes.
The radioactive uptake and growth data suggested that MalR does not activate maltodextrin utilization genes in GAS. To test the hypothesis that MalR negatively influences the transcription of genes in the maltodextrin and pullulanase gene regions, we assayed gene transcript levels in the MGAS5005 and ΔmalR strains. The transcript level of each of the eight genes assayed increased in the ΔmalR strain during growth in THY and saliva and to a lesser degree during growth in a maltotriose-medium compared to the transcript level in strain MGAS5005 (Fig. 6A). For all five genes in the maltodextrin gene region, the differences between the transcript levels in the mutant and parental strains were greater during the late-exponential phase of growth in human saliva than during the mid-exponential phase. This observation correlates with the finding that the significant difference in growth between the parental and ΔmalR strains in human saliva occurred during the transition from the late exponential phase to the stationary phase (Fig. 5D). The pattern of pulA transcript levels in the parental and ΔmalR strains was quite similar to the pattern observed for genes in the maltodextrin gene region, and the largest difference among the conditions tested occurred during the late-exponential phase in saliva (Fig. 6A). Similar to the results of the growth experiments, we found that the transcript levels of the maltodextrin and pullulanase region genes in the second ΔmalR strain were essentially identical to those in the original ΔmalR strain (data not shown).
FIG. 6.
MalR negatively influences the transcription of genes in the maltodextrin and pullulanase gene regions. Strain MGAS5005 and its isogenic ΔmalR derivative were grown to the mid-exponential or late-exponential phase in standard laboratory medium (THY), a maltotriose-medium, or human saliva. TaqMan real-time QRT-PCR was performed using probe and primers listed in Table 2. The transcript levels of target genes were normalized to those of proS, a gene that is expressed constitutively throughout the GAS cell cycle and whose transcript levels are similar during growth in THY and during growth in saliva (35, 43). (A) Gene transcript levels are expressed as the log2 fold difference between the ΔmalR mutant and the wild-type strain (ΔΔCT method); therefore, positive values indicate higher transcript levels in the ΔmalR strain. (B) Gene transcript levels in the ΔmalR strain relative to those of proS (ΔCT method). For each gene, the order of bars from left to right is as follows: mid- and late-exponential growth phases in THY, maltotriose-medium, and human saliva. The data are the means ± standard deviations of quadruplicate measurements obtained on two separate occasions.
We next sought to determine whether there was an increase in the maltodextrin and pullulanase region gene transcript levels during growth in a maltotriose-medium or human saliva in the ΔmalR strain. Unlike the results obtained for strain MGAS5005, for the ΔmalR strain there were no significant differences in the transcript levels for the maltodextrin and pullulanase region genes between growth in THY and growth in a maltotriose-medium (Fig. 6B). The lack of a difference between the two growth media was due to an increase in gene transcript levels in THY. However, an increase in maltodextrin and pullulanase region gene transcript levels was observed in the ΔmalR strain during growth in human saliva compared to growth in either THY or a maltotriose-medium (Fig. 6B). Taken together, these data suggest that MalR either directly or indirectly negatively influences the expression of all genes in the maltodextrin and pullulanase gene regions and that the influence of MalR is maximal during growth in human saliva.
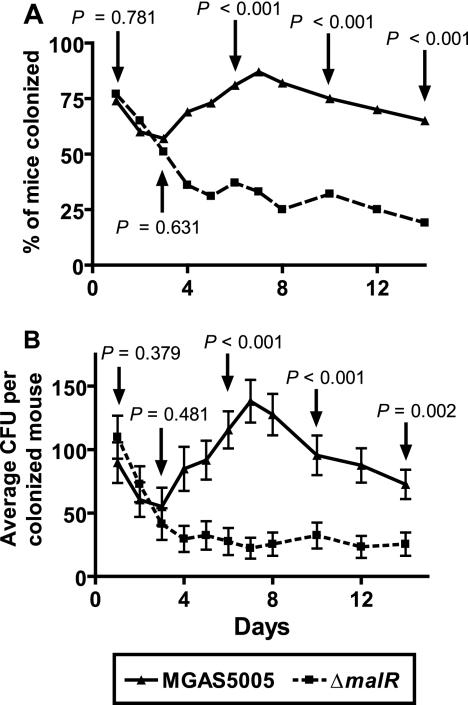
ΔmalR mutant strain persists at a significantly lower level than the parental strain in the mouse oropharynx.
In light of our finding that the ΔmalR strain was unable to persist at a high level in human saliva, we next tested the hypothesis that optimal GAS persistence in the oropharynx requires MalR. In the first 3 days after inoculation of adult outbred CD-1 mice with either the parental or ΔmalR GAS strain, there was no significant difference between the two groups of mice in terms of either the percent colonized or the average number of CFU per mouse recovered from the oropharynx (Fig. 7A). However, starting at day 4 and continuing through the remainder of the experiment, significantly fewer mice were colonized by the ΔmalR strain than by the parental strain. Moreover, starting at day 4 and continuing through the remainder of the experiment, the average numbers of GAS CFU were significantly greater in mice inoculated with the parental strain than in mice inoculated with the ΔmalR strain (Fig. 7B). Together, these data show that MalR is required for the optimal persistence of GAS in the oropharynx.
FIG. 7.
Failure of the ΔmalR isogenic mutant strain to persist in mouse orophyarnx. Adult outbred CD-1 mice (35 mice per group) were inoculated with ∼1.0 × 107 CFU of either MGAS5005 or its ΔmalR derivative. Mice oropharynges were swabbed daily, and the swabs were plated on BSA. The plates were incubated for 24 h, and beta-hemolytic colonies were counted and tested for GAS carbohydrate antigen using latex agglutination. (A) Percentages of mice with GAS isolated on different days. (B) Average numbers of CFU isolated on different days. The data are the means ± standard errors of the means. P values were determined by a χ2 test (A) or Student's t test (B).
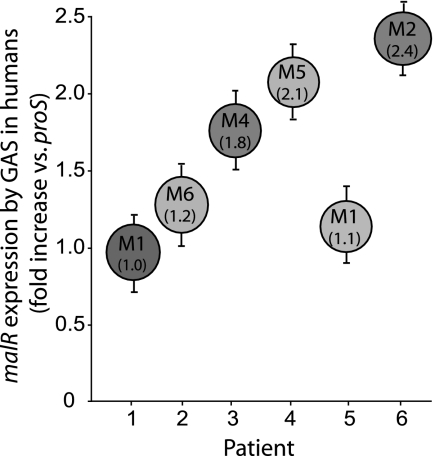
malR is expressed during human pharyngitis.
Inasmuch as malR contributes to the persistence of GAS in human saliva ex vivo and in the mouse oropharynx, we hypothesized that malR is transcribed and expressed in humans with GAS pharyngitis. We tested this hypothesis by using TaqMan real-time PCR to assay malR gene transcript levels in RNA isolated from throat swabs of six patients with GAS pharyngitis (43). Transcripts of malR were present in all six specimens at levels that were 1.5-fold higher than the levels of transcripts of the internal control gene proS (Fig. 8). These observations demonstrate that malR is actively transcribed during GAS pharyngitis in humans and further implicate MalR as a protein that participates in the GAS host-pathogen interaction.
FIG. 8.
Presence of malR transcripts in vivo during pharyngitis in humans. malR transcript levels for six patients with GAS pharyngitis were determined by TaqMan real-time PCR. The M serotypes of the infecting GAS strains are indicated in the circles, and the numbers in parentheses are the median fold increases in transcript levels relative to the levels of GAS proS gene transcripts (43). The error bars indicate the standard deviations of quadruplicate measurements obtained on two occasions.
DISCUSSION
Although GAS has long been known to be the leading cause of bacterial pharyngitis in humans, our understanding of the molecular mechanisms by which it colonizes and infects the human upper respiratory tract has remained relatively limited. To this end, we have been utilizing the interaction of GAS with human saliva as a model for investigation of previously unstudied portions of the GAS genome (33-35). The high transcript levels of putative polysaccharide utilization genes during GAS host-pathogen interactions suggest a role for polysaccharide utilization proteins in GAS pathogenesis (14, 15, 35, 44). Indeed, a recent investigation showed that MalE, a cell surface maltodextrin binding protein, was critical for initial colonization of the oropharynx by GAS (34).
The finding that interaction with human saliva markedly increased the transcript levels of malE (34) led us to investigate whether other genes encoding proteins putatively involved in polysaccharide utilization were similarly affected by growth in human saliva. Examination of the 12 available GAS genomes showed that genes encoding proteins putatively involved in polysaccharide utilization are located in two separate chromosomal regions (Fig. 1). All of the genes in these two locales, which we termed the maltodextrin and pullulanase gene regions, had marked increases in their transcript levels during growth in human saliva compared to the levels during growth in a standard laboratory medium (Fig. 2). These findings led us to focus on MalR, the only putative transcriptional regulator encoded in the maltodextrin and pullulanase gene regions.
The fact that the levels of malR transcripts were increased in saliva in a fashion similar to the fashion observed for the polysaccharide utilization genes studied suggested that MalR might function as a transcriptional activator in GAS (Fig. 3). However, as shown by the markedly elevated transcription of genes in the maltodextrin and pullulanase gene regions in the ΔmalR strain during growth in THY and saliva (Fig. 6), we surmised that MalR negatively influences gene expression. Whether MalR directly influences gene transcription, acts via a secondary transcriptional regulator, or affects gene expression through some other mechanism remains unclear. If MalR does function as a direct repressor of polysaccharide utilization genes, then one must ask why the transcript levels of MalR and polysaccharide genes both increase under the same conditions. Investigation of the mechanism by which MalR influences gene transcription is ongoing.
One critical finding of our transcriptional analyses was that the gene transcript levels of parental and ΔmalR strains differed more significantly during the late exponential growth phase in human saliva than during the rapid growth phase. This observation correlated well with our finding that the ΔmalR strain not only failed to make an effective transition from growth to high-level persistence in human saliva but also failed to persist in the mouse oropharynx. As experimental analysis of GAS pharyngitis in nonhuman primates has shown, malR transcript levels are highest during the persistence phase (44). These data have led us to conclude that MalR plays its most important role during the prolonged colonization phase of GAS in the oropharynx.
In an earlier investigation, the maltodextrin binding protein MalE was needed for optimal colonization of the oropharynx by strain MGAS5005 (34). If MalR represses expression of malE, our present finding that MalR contributes to the persistence of MGAS5005 in the oropharynx appears at first glance to contradict the MalE results. However, recent GAS infection experiments in nonhuman primates have demonstrated that gene expression by GAS varies significantly during the initial colonization, acute infection, and persistence phases of pharyngitis (44). Therefore, we propose that MalE and MalR differentially function in two distinct phases of GAS pathogenesis. The maltodextrin binding protein MalE appears to be most important during the initial colonization of the oropharynx by GAS, when rapid growth of the organism requires nutrient acquisition (34). Then, once the organism reaches maximal density, it must persist. In this situation, a protein that negatively influences gene expression, such as MalR, may become critical for fine-tuning the balance between energy utilization and protein production.
In addition to influencing the production of maltodextrin utilization genes, MalR also appears to affect the expression of genes in the distant pullulanase gene region. Our finding that MalR influences gene expression in two distinct chromosomal gene regions makes physiologic sense when the putative roles of the encoded proteins are considered. First, PulA is an LPXTG cell wall-anchored protein that degrades starch and other polysaccharides into maltodextrin and other transportable sugars (20, 31). Second, MsmK is the putative ATP-binding portion of the ABC transporter that mediates the uptake of maltodextrins. Third, the other three proteins that make up the ABC transporter are putatively encoded in the maltodextrin gene region. Finally, putative MalR binding sites are present in the maltodextrin and pullulanase regions (unpublished data). Thus, we hypothesize that MalR coordinates a system that is capable of breaking down starch into maltodextrins, incorporating maltodextrins into the cell, and converting maltodextrins into usable energy sources.
In conclusion, we used the results of multiple genome-wide expression microarray analyses to concentrate our investigations on the putative maltose/maltodextrin transcriptional regulator MalR. Our most important findings are that MalR influences the expression of at least eight GAS genes in two distinct chromosomal regions and that MalR is required for GAS to persist at high levels in human saliva and in the mouse oropharynx. These findings expand our insight into how GAS nutrient utilization contributes to pathogenesis. Comparative genomic analyses have shown that many mucosal human pathogens have conserved their mechanisms for the acquisition and selective utilization of nutrients. Therefore, further investigation of the links between nutrition and pathogenesis may yield broadly applicable insights into microbial host-pathogen interactions.
Acknowledgments
We thank Richard Hull for his advice on carbohydrate uptake.
This work was supported by American Heart Association grant 0565133Y (S.A.S.) and National Institutes of Health grant K08 RR17665-04 (S.A.S.).
Editor: D. L. Burns
Footnotes
Published ahead of print on 2 April 2007.
REFERENCES
- 1.Andes, D., A. Lepak, A. Pitula, K. Marchillo, and J. Clark. 2005. A simple approach for estimating gene expression in Candida albicans directly from a systemic infection site. J. Infect. Dis. 192:893-900. [DOI] [PubMed] [Google Scholar]
- 2.Banks, D. J., S. F. Porcella, K. D. Barbian, S. B. Beres, L. E. Philips, J. M. Voyich, F. R. DeLeo, J. M. Martin, G. A. Somerville, and J. M. Musser. 2004. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190:727-738. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou, R., M. A. Azcarate-Peril, T. Duong, S. B. Conners, R. M. Kelly, and T. R. Klaenhammer. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. USA 103:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beres, S. B., E. W. Richter, M. J. Nagiec, P. Sumby, S. F. Porcella, F. R. DeLeo, and J. M. Musser. 2006. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 103:7059-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessen, D. E., A. Manoharan, F. Luo, J. E. Wertz, and D. A. Robinson. 2005. Evolution of transcription regulatory genes is linked to niche specialization in the bacterial pathogen Streptococcus pyogenes. J. Bacteriol. 187:4163-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courtney, H. S., and D. L. Hasty. 1991. Aggregation of group A streptococci by human saliva and effect of saliva on streptococcal adherence to host cells. Infect. Immun. 59:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton, T. L., and J. R. Scott. 2004. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186:3928-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egeter, O., and R. Bruckner. 1995. Characterization of a genetic locus essential for maltose-maltotriose utilization in Staphylococcus xylosus. J. Bacteriol. 177:2408-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham, M. R., K. Virtaneva, S. F. Porcella, W. T. Barry, B. B. Gowen, C. R. Johnson, F. A. Wright, and J. M. Musser. 2005. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am. J. Pathol. 166:455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, M. R., K. Virtaneva, S. F. Porcella, D. J. Gardner, R. D. Long, D. M. Welty, W. T. Barry, C. A. Johnson, L. D. Parkins, F. A. Wright, and J. M. Musser. 2006. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am. J. Pathol. 169:927-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, N. M., S. Zhang, S. F. Porcella, M. J. Nagiec, K. D. Barbian, S. B. Beres, R. B. Lefebvre, and J. M. Musser. 2005. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192:760-770. [DOI] [PubMed] [Google Scholar]
- 17.Haralalka, S., S. Nandi, and R. K. Bhadra. 2003. Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J. Bacteriol. 185:4672-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293:381-399. [DOI] [PubMed] [Google Scholar]
- 19.Hufnagel, M., S. Koch, R. Creti, L. Baldassarri, and J. Huebner. 2004. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J. Infect. Dis. 189:420-430. [DOI] [PubMed] [Google Scholar]
- 20.Hytonen, J., S. Haataja, and J. Finne. 2003. Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, pullulanase. Infect. Immun. 71:784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 22.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 23.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukomski, S., N. P. Hoe, I. Abdi, J. Rurangirwa, P. Kordari, M. Liu, S. J. Dou, G. G. Adams, and J. M. Musser. 2000. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect. Immun. 68:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. [DOI] [PubMed]
- 26.Nakagawa, I., K. Kurokawa, A. Yamashita, M. Nakata, Y. Tomiyasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieto, C., M. Espinosa, and A. Puyet. 1997. The maltose/maltodextrin regulon of Streptococcus pneumoniae. Differential promoter regulation by the transcriptional repressor MalR. J. Biol. Chem. 272:30860-30865. [DOI] [PubMed] [Google Scholar]
- 28.Orihuela, C. J., J. N. Radin, J. E. Sublett, G. Gao, D. Kaushal, and E. I. Tuomanen. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 72:5582-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peter, G., and A. L. Smith. 1977. Group A streptococcal infections of the skin and pharynx (second of two parts). N. Engl. J. Med. 297:365-370. [DOI] [PubMed] [Google Scholar]
- 30.Reid, S. D., N. M. Green, J. K. Buss, B. Lei, and J. M. Musser. 2001. Multilocus analysis of extracellular putative virulence proteins made by group A Streptococcus: population genetics, human serologic response, and gene transcription. Proc. Natl. Acad. Sci. USA 98:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, S. D., N. M. Green, G. L. Sylva, J. M. Voyich, E. T. Stenseth, F. R. DeLeo, T. Palzkill, D. E. Low, H. R. Hill, and J. M. Musser. 2002. Postgenomic analysis of four novel antigens of group A Streptococcus: growth phase-dependent gene transcription and human serologic response. J. Bacteriol. 184:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roos, V., G. C. Ulett, M. A. Schembri, and P. Klemm. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelburne, S. A., III, C. Granville, M. Tokuyama, I. Sitkiewicz, P. Patel, and J. M. Musser. 2005. Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect. Immun. 73:4723-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shelburne, S. A., III, P. Sumby, I. Sitkiewicz, N. Okorafor, C. Granville, P. Patel, J. Voyich, R. Hull, F. R. Deleo, and J. M. Musser. 2006. Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. Infect. Immun. 74:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shelburne, S. A., III, P. Sumby, I. Sitkiewicz, C. N. Granville, F. R. DeLeo, and J. M. Musser. 2005. Central role of a two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc. Natl. Acad. Sci. USA 102:16037-16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiner, K., and H. Malke. 2001. relA-Independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sumby, P., S. F. Porcella, A. G. Madrigal, K. D. Barbian, K. Virtaneva, S. M. Ricklefs, D. E. Sturdevant, M. R. Graham, J. Vuopio-Varkila, N. P. Hoe, and J. M. Musser. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771-782. [DOI] [PubMed] [Google Scholar]
- 40.Sumby, P., A. R. Whitney, E. A. Graviss, F. R. DeLeo, and J. M. Musser. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tchawa Yimga, M., M. P. Leatham, J. H. Allen, D. C. Laux, T. Conway, and P. S. Cohen. 2006. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74:1130-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 43.Virtaneva, K., M. R. Graham, S. F. Porcella, N. P. Hoe, H. Su, E. A. Graviss, T. J. Gardner, J. E. Allison, W. J. Lemon, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2003. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect. Immun. 71:2199-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vise, P. D., K. Kodali, N. Hoe, A. Paszczynski, J. M. Musser, and G. W. Daughdrill. 2003. Stable isotope labeling of a group A Streptococcus virulence factor using a chemically defined growth medium. Protein Expr. Purif. 32:232-238. [DOI] [PubMed] [Google Scholar]
- 46.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]