Abstract
Previous studies have demonstrated that animals exposed to Streptococcus pneumoniae while recovering from influenza A virus infection exhibit exacerbated disease symptoms. However, many of the current animal models exploring dual viral and bacterial synergistic exacerbations of respiratory disease have utilized mouse-adapted influenza virus and strains of Streptococcus pneumoniae that in themselves are highly lethal to mice. Here we describe a mouse model of bacterial superinfection in which a mild, self-limiting influenza virus infection is followed by mild, self-limiting superinfection with S. pneumoniae serotype 3. S. pneumoniae superinfection results in rapid dissemination of the bacterium from the respiratory tract and systemic spread to all major organs of the mice, resulting in fatal septicemia. This phenomenon in mice was observed in superinfected animals undergoing an active viral infection as well as in mice that had completely cleared the virus 7 to 8 days prior to superinfection. Neutrophils were the predominant cellular inflammatory infiltrate in the lungs of superinfected mice compared to singly infected animals. Among other cytokines and chemokines, the neutrophil activator granulocyte colony-stimulating factor (G-CSF) was found to be significantly overexpressed in the spleens, lungs, and brains of superinfected animals. High G-CSF protein levels were observed in sera and lung lavage fluid from superinfected animals, suggesting that G-CSF is a major contributor to synergistic exacerbation of disease leading to fatal septicemia.
Influenza A virus infections are usually limited to the upper respiratory tract and cause mild symptoms, such as sore throat, sneezing, fever, headache, muscle fatigue, and inactivity (35). Influenza virus infection is not often fatal in normal healthy adults, and mortality is usually due to secondary bacterial superinfections (1, 12, 17, 24, 27). Interestingly, 44 to 57% of patients hospitalized with influenza test positive for bacterial pneumonia (30). Clinical data have shown that the majority of all fatal secondary bacterial infections are associated with bacterial septicemia (34). Streptococcus pneumoniae is the causative agent in the majority of patients suffering from bacterial and viral coinfections (23) and is the leading cause of community-acquired pneumonia. S. pneumoniae colonizes the nasopharynx of 20 to 50% of healthy children and 8 to 30% of healthy adults (23). In addition to causing pneumonia, Streptococcus pneumoniae infection can result in otitis media, sinusitis, meningitis, and bacteremia (36). Worldwide, S. pneumoniae is the leading cause of respiratory illness among children less than 5 years of age, causing approximately 1 million deaths per year (38). In addition, S. pneumoniae accounts for over 100,000 infections yearly in the United States, with approximately 40,000 of these cases resulting in death (6). Serotype 3 pneumococcus is the most common serotype colonizing adult throats (16). It is also the serotype most often associated with invasive disease and is associated with a greater risk of death than other serotypes (16). Importantly, the current seven-valent pneumococcal conjugate vaccine does not provide protection against serotype 3 pneumococcus (16).
Influenza A virus is a highly pleomorphic virus that produces both spherical and filamentous virions (3, 4, 8). Primary human isolates of influenza virus are predominantly filamentous, suggesting that the filamentous phenotype may play a role in disease progression (3, 4, 8). The filamentous phenotype of influenza A viruses is not often used for research purposes, since it is readily lost during serial passage of the virus in egg or tissue culture (8). Filamentous influenza A virus particles can reach lengths up to 20 μm in length, which is much larger than the 80- to 120-nm spherical particles (33) typically observed in most laboratory-adapted strains of influenza virus.
Previous research demonstrated that mice infected with influenza virus prior to being infected with S. pneumoniae had increased colonization of the bacterium in the respiratory epithelium (31). Unmasking of the pneumococcal receptor by virus neuraminidase (24, 25) and a depression in neutrophil function by influenza virus, which allows the bacterium to evade the immune system (1), have been proposed as contributing factors that may account for increased bacterial colonization. However, the majority of previous studies have utilized mouse-adapted influenza A/PR8/34 virus to predispose mice. This strain of influenza virus itself induces significant respiratory disease and is highly lethal to BALB/c mice. This does not mimic the typical mild course of influenza disease progression observed in humans. In order to more accurately monitor the natural course of influenza in humans, we chose to examine dual synergistic exacerbation of respiratory disease in vivo between the filamentous influenza A/Udorn/72 virus strain and S. pneumoniae serotype 3. This strain of influenza virus as noted is highly filamentous and results in a mild influenza infection that is confined to the respiratory tract and is rapidly resolved within 6 to 7 days. Here we report that filamentous influenza virus infection predisposes mice to fatal septicemia following superinfection with Streptococcus pneumoniae serotype 3. Further, our data suggest that high levels of granulocyte colony-stimulating factor (G-CSF) play a major role in synergistic exacerbation of disease.
MATERIALS AND METHODS
Cell culture and influenza virus propagation.
Madin-Darby canine kidney (MDCK) cells were grown and maintained in Dulbecco's modified Eagle's medium (DMEM; Mediatech) supplemented with 10% fetal bovine serum (Atlanta Biologicals). Stocks of influenza A/Udorn/72 (H3N2) virus, a human clinical isolate that has been laboratory-adapted for growth in cell culture, were prepared in MDCK cells that were grown to 80% confluence. Briefly, cells were infected with influenza A/Udorn/72 (H3N2) at a low multiplicity of infection (0.002) for 1 h. Following virus adsorption, DMEM supplemented with 1.5 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma) was added to the cells, and virus was allowed to replicate for 48 h at 37°C. Virus was recovered from supernatants, and cellular debris was removed from the viral suspension by centrifugation at 580 × g for 10 min. The virus was then further purified and concentrated by ultracentrifugation of the supernatant at 55,000 × g for 60 min through a 14% Opti-prep (Axis-Shield) cushion. The virus pellet was then resuspended in phosphate-buffered saline (PBS) supplemented with 1% bovine serum albumin, and the titer was determined using a tissue culture infectious dose (TCID) assay, which determined the dose required to cause cytopathic effect in 50% of the cells (32). The stocks were diluted to 5 × 103 50% TCID (TCID50)/50 μl in sterile PBS. After removing aliquots for bacterium samples, the tissue homogenates (or bronchoalveolar lavage fluids [BALF]) were centrifuged at 580 × g for 15 min to remove cellular debris. The supernatants were collected, 100-μl aliquots were serially diluted, and titers were determined via a tissue culture infectious dose assay essentially as described by Reed and Munch (32). Briefly, MDCK monolayers (48-well tissue culture plates) were incubated with 100-μl aliquots per dilution (n = 8/dilution) of serially diluted tissue, BALF, or nasal wash samples. The virus was allowed to adsorb for 1 hour at 37°C under 5% CO2 in the incubator. Following removal of the virus suspension, cells were incubated in DMEM supplemented with 1.5 μg/ml TPCK-treated trypsin for 48 to 72 h at 37°C under 5% CO2. The titers are expressed as the reciprocal of the highest dilution that results in at least 50% of the wells displaying cytopathic effect (32).
Bacterium propagation and quantitation of bacterial loads.
ATCC 6303 serotype 3 S. pneumoniae was grown in brain heart infusion broth to mid-logarithmic phase as determined by optical density measurements at a wavelength of 600 nm. The culture was diluted to 5 × 103 CFU/50 μl in sterile PBS. The titer was confirmed by plating in triplicate on trypticase soy agar plates supplemented with 5% sheep's blood (BBL Inc.). Aliquots of tissue homogenate or BALF (100 μl) were collected from animals, serially diluted, and plated onto trypticase soy agar plates supplemented with 5% sheep erythrocytes (BBL Inc.). The plates were incubated for 16 to 18 h at 37°C in 5% CO2. The colonies of bacteria were manually counted, and titers were expressed as the log10 CFU/ml of BALF or CFU/gram of tissue. Plating was repeated in triplicate to maintain accuracy.
Animal infections.
Female BALB/c mice (6 weeks in age) were obtained from the National Institutes of Health (NCI, Charles River Laboratories) and were housed in sterile microisolators in a biosafety level 2 facility under the care of the Wayne State University Department of Laboratory Animal Resources. Acclimated mice were inoculated intranasally with influenza A/Udorn/72 at 5 × 103 TCID50/50 μl or PBS (sham infected) while under light isoflurane anesthetic. Virus- and sham-infected mice were euthanized with sodium pentobarbital (intraperitoneal administration) at days 3, 5, 8, 10, 14, and 17 to determine viral loads and inflammatory states in the bronchoalveolar lavage fluid.
Udorn- and sham-infected mice were also superinfected at 3, 5, 7, and 14 days post-influenza virus infection with S. pneumoniae serotype 3. Here, S. pneumoniae (5 × 103 CFU) was administered intranasally to lightly anesthetized mice in a volume of 50 μl. PBS was used as a negative control for sham superinfections. Mice were monitored twice daily for overt disease symptoms (ruffled fur, inactivity, and weight loss) and were euthanized by intraperitoneal administration of a lethal dose of sodium pentobarbital. Lungs, spleens, brains, kidneys, and livers were excised from at least seven mice per group and homogenized in sterile DMEM (Mediatech) to determine bacterial and viral loads. RNA was also isolated from individual organs using the Bio-Rad Aurum total RNA mini kit. Blood was collected from the animals via heart puncture and was analyzed for bacterial loads and serum cytokine levels. Nasal washes were also performed to determine viral and bacterial loads. Half of each organ from at least three mice in each group was formalin fixed and processed for histopathology by AML Laboratories in Baltimore, MD. Bronchoalveolar lavages were performed on at least three mice per group.
BALF.
Bronchoalveolar lavage fluid was collected from sham, singly infected, and superinfected groups (n = 3) on day 3 postsuperinfection. Following isolation of the trachea, a 20-gauge oral gavage needle was inserted into the trachea and 3 1-ml volumes of sterile PBS were instilled, removed, and then combined. The cells were removed from suspension by centrifugation at 3,000 rpm and were resuspended in sterile PBS for counting and differential analysis. The cells were spun onto glass slides at 50 × g and were then fixed and stained using the Hema-3 differential staining kit. Ten separate fields of view were counted microscopically to determine differential cell numbers that were based on morphological assessment of cell type.
Cytokine/chemokine quantitative real-time PCR.
RNA was isolated from 350 μl of tissue homogenate using the Bio-Rad total RNA mini kit. The RNA was then quantified using the Ribogreen RNA quantitation kit (Molecular Probes). One μg of RNA was used to make cDNA via the Bio-Rad cDNA synthesis kit. The cDNA was then analyzed by real-time PCR using cytokine/chemokine-specific primers (7, 27, 29), SYBR Green, and a Bio-Rad iCycler. All primer sequences are available upon request. For normalization, the mRNA for mouse mitochondrial ribosomal protein L19 was quantified in parallel for all samples. The change in critical threshold (CT) between the cytokine primer set and the L19 primer set was determined for all samples, and the change over mock (PBS) infection was determined using the 2−ΔΔCt method as described elsewhere (21).
Cytokine/chemokine protein levels.
Protein levels of selected cytokines and chemokines (keratinocyte-derived chemokine [KC], interleukin-10 [IL-10], IL-6, tumor necrosis factor alpha [TNF-α], and IL-1β) from the bronchoalveolar lavage fluid or sera were analyzed by a five-plex antibody assay performed by the Chemicon Corporation. G-CSF and KC protein levels were measured from the sera and BALF using cytokine-specific enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer's protocol (Biosource).
Statistical analysis.
To determine whether differences in survival, mRNA, and protein levels were significant from each other, analysis of variance, Bonferroni's multiple comparison test, and Student's t test (Prism; GraphPad) were performed, and a P value of <0.05 was considered significant. Significance values are listed in the figure legends.
RESULTS
Filamentous influenza A/Udorn/72 and S. pneumoniae serotype 3 both produced a mild, nonlethal infection in BALB/c mice when administered individually.
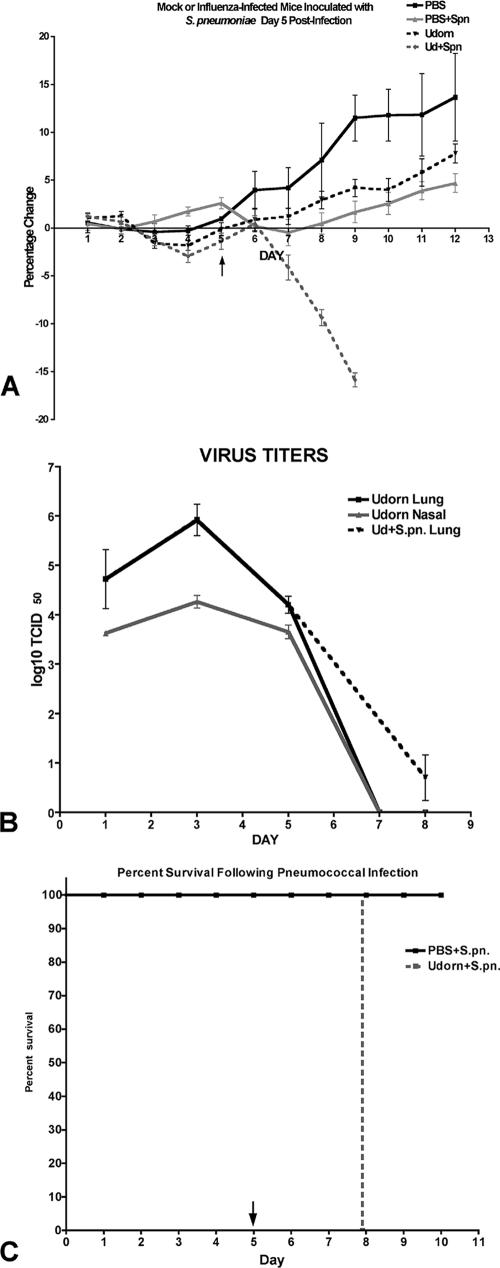
Since influenza virus in the clinical setting generally results in a mild, self-limiting upper respiratory tract infection with little or marginal involvement of the lower airways, we selected a strain of influenza virus for these studies that replicates to detectable levels in the respiratory tract but results in a mild, self-limiting viral infection in BALB/c mice. Further, to mimic the filamentous phenotype typically observed from clinical isolates, we chose a filamentous strain of influenza virus, influenza A/Udorn/72. As shown in Fig. 1A, juvenile BALB/c mice that were infected intranasally with 5 × 103 TCID50 of influenza A/Udorn/72 did not experience overt clinical symptoms associated with serious influenza disease, such as drastic weight loss or ruffled fur. Minimal transient weight loss was observed 1 to 3 days immediately after infection, but the mice rapidly regained weight by 4 days postinfection. Virus replication was detectable in the lung and nasal tract at 24 h postinfection, with peak levels observed at day 3 (1 × 106 TCID50/g tissue), which was followed by gradual clearance of the virus from days 4 to 6, with no detectable virus loads present by day 7 postinfection (Fig. 1B). Thus, this dose of virus results in a mild, self-limiting viral infection that is restricted to the upper and lower respiratory tract. At no time was virus isolated from the brain, spleen, or kidneys in singly infected animals.
FIG. 1.
Percent weight change (A), nasal tract and lung viral titers (B), and percent survival (C) in influenza virus-infected (Udorn) or mock-infected (PBS) mice that received type 3 pneumococcus on day 5 post-virus infection. Mice were weighed daily, and survival was recorded as a function of time postinfection. Viral loads were determined from nasal washes (nasal tract) and lung tissue homogenates harvested at different times post-virus infection and are expressed as TCID50 per gram tissue or ml of nasal wash. Singly infected mice did not succumb to infection.
Similarly, a single low-dose infection with serotype 3 S. pneumoniae (5 × 103 CFU) resulted in a mild, self-limiting infection in BALB/c mice when administered intranasally. Transient bacteremia (2 × 103 CFU/ml) was observed in 30% of infected animals (n = 10), but systemic bacterial loads declined rapidly and by day 7 the bacteria had been completely cleared from the body (data not shown).
Type 3 pneumococcus administered during active influenza A/Udorn/72 virus infection resulted in fatal septicemia.
To examine dual synergistic exacerbation of disease, animals that had been initially infected with a mild, self-limiting dose of influenza (5 × 103 TCID50) were then superinfected with a mild, self-limiting dose of S. pneumoniae (5 × 103 CFU) at different times post-virus infection.
Superinfection at either day 3 (data not shown) or day 5 post-virus infection resulted in bacteremia in 100% of the animals, and the mice succumbed to exacerbated disease within 3 days (Fig. 1C and Table 1). These superinfections represent time points when viral loads are peaking (day 3) or on the decline (day 5). Clinical signs of exacerbated disease included severe rapid weight loss detected within 24 h of superinfection (Fig. 1A) which was followed by labored breathing, ruffled fur, and inactivity. In contrast to the transient and cleared bacteremia observed in singly infected animals, superinfected animals exhibited significantly higher bacterial loads in all target organs, including the nasal tract (Table 1) and blood (3 × 106 to 4 × 106 CFU/ml). Interestingly, a delay in viral clearance from the lungs was also observed in the superinfected mice. Influenza A/Udorn is generally completely cleared from the mouse lung by day 6 to 7 postinfection (Fig. 1B), but when S. pneumoniae was administered during active viral infection, there were still detectable viral loads in two of seven mice analyzed at day 8 (Fig. 1B). Thus, an active filamentous influenza virus infection is synergistically exacerbated by superinfecting S. pneumoniae, and this leads to lethal disease.
TABLE 1.
Bacterial loads recovered from organs of mock- or influenza virus-infected mice superinfected with pneumococci on day 5 or day 14 post-virus infectiona
| Time and type of superinfection | Log10 CFU/g tissue (SEM)
|
|||||
|---|---|---|---|---|---|---|
| Lung | Nasal | Spleen | Brain | Kidney | Liver | |
| Day 5 | ||||||
| PBS + S. pneumoniae | 1.286 (0.609) | 3.330 (0.158) | 1.047 (0.680) | 0.736 (0.476) | 0.843 (0.544) | 1.327 (0.631) |
| UD + S. pneumoniae | 6.993 (0.120)*** | 4.312 (0.129)* | 4.321 (0.276)* | 5.807 (0.350)** | 5.137 (0.344)** | 4.427 (0.414)* |
| Day 14 | ||||||
| PBS + S. pneumoniae | 1.398 (0.555) | 3.641 (0.202) | 0.644 (0.430) | 0.606 (0.397) | 0.735 (0.504) | 0.821 (0.538) |
| UD + S. pneumoniae | 6.545 (0.417)** | 4.382 (0.138)* | 4.298 (0.616)* | 4.706 (0.227)*** | 4.366 (0.644)* | 3.955 (0.660)* |
Mice received PBS (sham) or influenza A/Udorn (UD) on day zero and were superinfected at day 5 or day 14 post-virus infection with Streptococcus pneumoniae. Bacterial loads were determined from animals upon reaching certain end points (e.g., >20% weight loss).
P < 0.01;
P < 0.001;
P < 0.0001 (n = 7).
The increased susceptibility to fatal septicemia is maintained well after virus clearance.
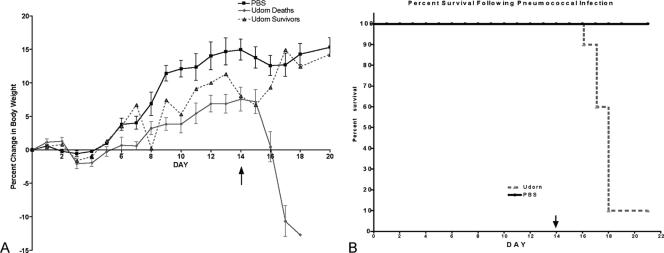
To determine if synergistic exacerbation of disease is only dependent on active viral replication, mice were inoculated with S. pneumoniae (ATCC 6303) on day 14 post-influenza virus infection. This time point was chosen since there are no detectable levels of influenza A/Udorn/72 virus beyond 7 days post-viral infection (Fig. 1B). This was confirmed in three independent studies. Therefore, superinfection on day 14 allowed for ample recovery time from the virus infection. Superinfection at day 14 post-virus infection (7 to 8 days post-viral clearance) resulted in 90% fatality by 4 days postsuperinfection (Fig. 2B). Superinfected animals exhibited identical symptoms observed in mice that were superinfected during an active influenza virus infection, i.e., weight loss, lethargy, and labored breathing (Fig. 2A). Similarly, these mice also had significantly higher bacterial loads in all of the major target organs compared to animals that received pneumococcus alone (Table 1). These results indicate that a “predisposed state” is generated by prior exposure to nonlethal doses of influenza virus and that a synergistic exacerbation of disease is not dependent on active viral replication.
FIG. 2.
Percent weight change (A) and percent survival (B) in influenza virus-infected (Udorn) or mock-infected (PBS) mice that received type 3 pneumococcus on day 14 post-virus infection. Mice were weighed daily, and survival was recorded as a function of time postsuperinfection. Singly infected mice did not succumb to infection.
Neutrophils dominate the lung inflammatory cell infiltrate in influenza virus-“predisposed” mice.
To further characterize the synergistic exacerbative response, we analyzed inflammatory cell infiltrates obtained by bronchoalveolar lavage. Here, a subset of the mice in each group (singly, superinfected, or sham-infected mice) was sacrificed at a time point judged to be immediate to pending death to determine the inflammatory response existing within the lungs.
Animals that were superinfected on day 5 and sacrificed 3 days later (day 8 post-influenza virus infection) revealed distinct lung cellular infiltrate profiles. BALF from sham-infected and singly infected mice consisted largely of alveolar macrophages, which represented 99.9% ± 0.1% (sham infected), 62.1% ± 1.72% (influenza virus infected), and 79.2% ± 1.12% (S. pneumoniae infected) of the total lung immune cell population (Table 2). In contrast, polymorphonuclear neutrophils were the predominant cell infiltrate in BALF from superinfected animals, comprising greater than 50% of the total lung immune cell population (Table 2). Significant accumulation of neutrophils was not observed in the influenza virus or S. pneumoniae singly infected animals (Table 2). However single infections did result in varied degrees of lymphocyte infiltration: 16% in S. pneumoniae and 34% of total cellular infiltrate in influenza virus singly infected animals (Table 2).
TABLE 2.
Total cell counts from bronchoalveolar lavages and cell types in inflammatory infiltrates from superinfected micea
| Treatment group | Total cell count (log10)
|
% Macrophages
|
% Neutrophils
|
% Lymphocytes
|
||||
|---|---|---|---|---|---|---|---|---|
| Day 5 | Day 14 | Day 5 | Day 14 | Day 5 | Day 14 | Day 5 | Day 14 | |
| Sham | 5.81 (0.28) | 5.45 (0.12) | 99.9 (0.01) | 99.7 (0.03) | 0.1 (0.10) | 0.3 (0.09) | ND | ND |
| S. pneumoniae | 6.17 (0.48) | 6.33 (0.55) | 79.2 (1.12) | 82.8 (0.87) | 4 (0.51) | 6 (1.20) | 16.8 (2.21) | 11.2 (3.24) |
| Udorn | 6.27 (0.11) | 6.22 (0.32) | 62.1 (1.72) | 85.6 (3.32) | 3.6 (0.73) | 1.4 (0.12) | 34.3 (1.32) | 13.0 (2.31) |
| Udorn + S. pneumoniae | 7.01 (0.06) | 7.01 (0.67) | 34.6 (2.02) | 29.7 (3.22) | 54.3 (5.41) | 65.3 (1.31) | 11.1 (2.22) | 5.0 (2.12) |
SEM values are in parentheses. ND, this cell type was not detected in any of the fields of view. Mice received PBS (sham) or influenza A/Udorn (UD) on day zero and were superinfected at day 5 or day 14 post-viral infection with Streptococcus pneumoniae. Bronchioalveolar lavage fluid was collected post mortem from at least three mice upon reaching end points (e.g., >20% weight loss). As controls, mice exposed to single (S. pneumoniae or Udorn) or sham infections were included.
Similar to mice that were superinfected on day 5 post-influenza virus infection, superinfection on day 14 also resulted in a significant influx of neutrophils into the lungs (Table 2). On day 17 postinfection (day 3 postsuperinfection), BALF from sham-infected and singly infected mice consisted mostly of alveolar macrophages. In contrast, polymorphonuclear neutrophils comprised approximately 65% of the lung inflammatory cell infiltrates in superinfected mice (Table 2), suggesting that the granulocyte is an important cell type in the synergistic exacerbation of disease. Interestingly, even at day 17 post-influenza virus infection, 13% of the cellular infiltrate was compromised of lymphocytes. This suggests that the influenza virus-induced lymphocytic infiltration may be responsible for the establishment of the “predisposed state” leading to exacerbative responses following superinfection.
Histopathological analysis confirmed neutrophil accumulation in tissues.
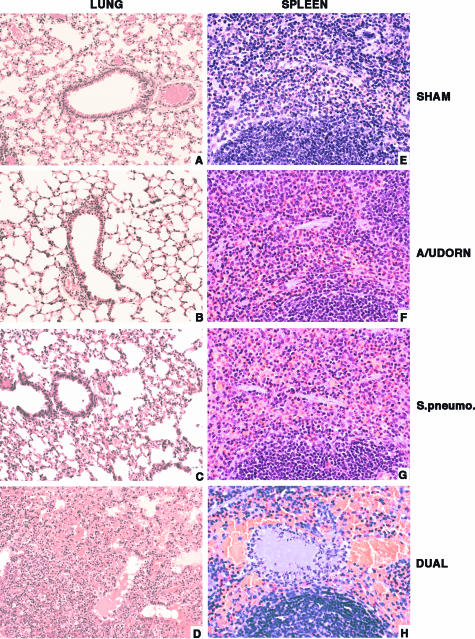
Microscopic examination of the hematoxylin and eosin (H&E)-stained lung sections from the superinfected mice showed edema in four out of seven tissue samples, and acute bronchopneumonia characterized by intra-alveolar hemorrhage and neutrophilic infiltrate was confirmed in seven out of seven animals (Fig. 3D). Lung sections from the singly infected mice group were similar to the sham-infected control lungs (Fig. 3A to C) and revealed no pathological changes. All splenic sections from the superinfected mouse group (Fig. 3H) showed significant congestion and neutrophilic infiltrate with focal fibrin thrombi compared to the control and singly infected mice group, which were unremarkable (Fig. 3E to G). These results suggest that the animals died from a fatal septicemia that was synergistically exacerbated in virus-predisposed animals.
FIG. 3.
Histopathology of lungs and spleens recovered from sham-infected, singly infected, or superinfected mice. The lung sections from sham, influenza A/Udorn, and S. pneumoniae (A to C) groups are unremarkable, whereas lung sections from a superinfected mouse (D) show edema and hemorrhage with intra-alveolar neutrophilic infiltrates. The spleen section from a superinfected mouse (H) has a fibrin thrombus in the red pulp associated with congestion and neutrophilic infiltration. The spleen sections from sham-infected, influenza A/Udorn singly infected, and S. pneumoniae singly infected animals (E to G) are unremarkable. Magnification, ×200 (A to D) and ×400 (E to H).
Cytokine and chemokine gene expression levels were selectively upregulated in superinfected mice.
Since the results described above suggested that a synergistic recruitment of neutrophils into the lungs was a result of the superinfections, we next determined the mRNA expression levels of several key neutrophil and lymphocyte chemoattractants as well as proinflammatory cytokines within the lungs of singly infected, superinfected, or sham-infected mice. In addition, due to the systemic spread of bacteria throughout the body of superinfected mice, the cytokine/chemokine mRNA expression profiles of brain and splenic tissues from superinfected animals were included in the analyses.
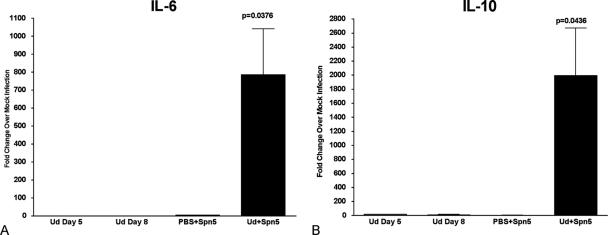
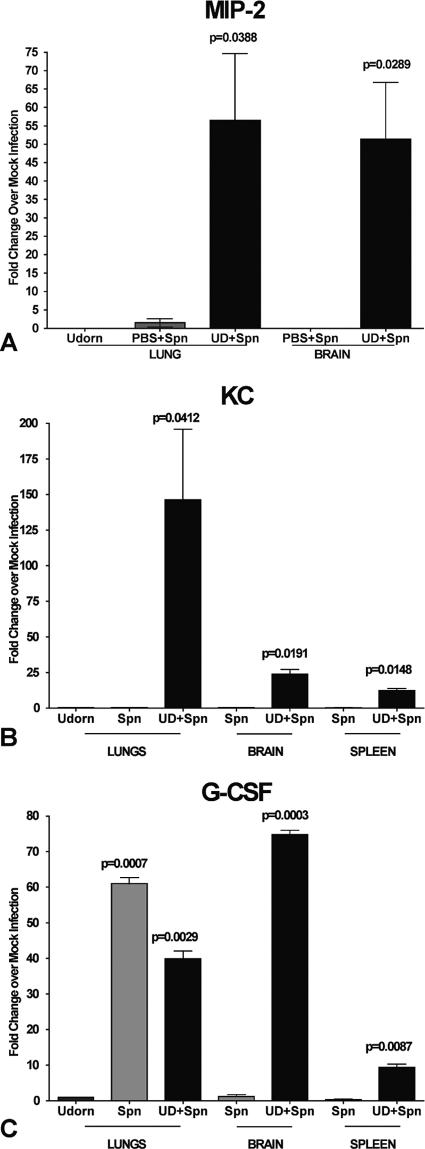
In contrast to singly infected or sham-infected animals, superinfection at day 5 post-viral infection resulted in significant upregulation of proinflammatory cytokines TNF-α, IL-1β (data not shown), and IL-6 (Fig. 4A), which were synergistically upregulated in the lungs 110-fold, 8-fold, and 500-fold, respectively. Gene expression of the anti-inflammatory cytokine IL-10 was also significantly upregulated in the lungs from superinfected animals compared to sham- or singly infected mice (Fig. 4B). Importantly, the chemoattractants macrophage inflammatory protein 1β (MIP-1β), gamma interferon-inducible 10-kDa protein (IP-10) (data not shown), MIP-2, and KC (Fig. 5A and B) were also significantly upregulated in superinfected animals; the latter two chemokines are likely responsible for inducing the synergistic infiltration of neutrophils, as was noted earlier. The mRNA levels of IL-2, IL-4, IL-12, IL-13, and gamma interferon were also determined in various tissues from the superinfected animals (data not shown), but they did not differ significantly over levels observed for singly or sham-infected animals.
FIG. 4.
Cytokine mRNA expression levels in mice superinfected at day 5 post-virus infection. RNA was isolated from the lung tissue of infected animals sacrificed just prior to mortality (day 8 post-virus infection), and mRNA levels of IL-6 (A) and IL-10 (B) were quantified using real-time PCR. Values are expressed as fold change over mock infection after normalization to L19 mRNA.
FIG. 5.
Gene expression levels of neutrophil chemoattractants MIP-2 and KC and the neutrophil activator G-CSF in day 5 superinfected mice. RNA was isolated from the lung, brain, and spleen tissue of infected animals at day 8 post-virus infection (day 3 postsuperinfection), and mRNA expression levels of MIP-2 (A), KC (B), and G-CSF (C) were quantified using real-time PCR. Values are expressed as fold change over mock infection after normalization to L19 mRNA.
In the brains of day 5 superinfected animals, notable differences in mRNA levels were only observed for inducible nitric oxide synthase, IL-1β (data not shown), MIP-2, and KC, which were all synergistically upregulated compared to singly infected animals (Fig. 5A and B). In the spleens, IL-6, IL-10 (data not shown), and KC (Fig. 5B) were the predominately upregulated mRNA species in superinfected animals; MIP-2 expression was not detected in the spleens. Finally, of particular note is the significant upregulation of G-CSF mRNA levels in lungs, brains, and spleens of superinfected animals; G-CSF is a potent activator of neutrophils, and this is the first report of its upregulation in response to superinfection (Fig. 5C).
The chemokine/cytokine mRNA expression profiles observed in the lungs, spleens, and brains of mice that were superinfected on day 14 post-influenza virus infection were very similar to the expression profiles observed in mice superinfected during an active influenza virus infection. Notably, in day 14 superinfected animals, there were significant increases in gene expression levels of the proinflammatory cytokine IL-6 (>1,000-fold increase), the anti-inflammatory cytokine IL-10 (700-fold increase), the chemoattractant MIP-2 (15-fold), and the neutrophil activator G-CSF (25-fold increase) observed in lung tissue. In contrast, neither IL-1β, TNF-α, nor KC (data not shown) mRNA levels in the lungs were significantly increased in mice superinfected at day 14 post-virus infection. The spleens and brains of mice superinfected on day 14 also exhibited increases in MIP-2, KC, and G-CSF mRNA levels similar to those in mice superinfected at day 5 post-virus infection (data not shown), although the levels of their expression were reduced by a factor of 3.
Cytokine and chemokine protein levels were synergistically upregulated in superinfected mice.
To confirm that the observed upregulation in gene expression in superinfected animals correlates with an increase in cytokine/chemokine protein production and secretion, both the BALF and sera from infected animals following superinfection at day 5 and day 14 were analyzed by a five-plex antibody array and/or a KC- and G-CSF-specific ELISA.
Dramatic increases in IL-10, IL-6, TNF-α, and KC production were observed in the BALF and sera of superinfected animals, regardless of whether virus was present (Table 3) or had been cleared 7 days previously (Table 3). These increases correlate well with the observed increases in mRNA expression levels. Surprisingly, we did not observe a dramatic increase in IL-1β protein levels in the sera, but there was a significant increase of IL-1β in the BALF of superinfected animals (Table 3). Interestingly, G-CSF protein levels increased to greater than 2,400 pg/ml in the BALF and 4,600 pg/ml in the sera of animals superinfected at day 5 (Table 3) compared to levels of less than 2 pg/ml determined in the sera of sham-infected and singly infected animals. In animals superinfected at day 14, G-CSF was measured in excess of 1,600 and 950 pg/ml in the BALF and sera, respectively, which is well above the levels detected in any of the singly or sham-infected animals.
TABLE 3.
Cytokine and chemokine protein levels in mice superinfected at day 5 or day 14 post-influenza A/Udorn virus infectiona
| Time and type of treatment and sample assayed | Cytokine or chemokine level (pg/ml) (SEM; n = 3)
|
|||||
|---|---|---|---|---|---|---|
| KC | G-CSF | IL-1β | IL-10 | IL-6 | TNF-α | |
| Day 5 post-viral infection | ||||||
| BALF | ||||||
| Sham | 12.0 | 3.3 (2.0) | 26.0 | ND | ND | ND |
| Udorn | 18.5 (5.5) | 30.7 (26.2) | 31.0 (10.0) | ND | ND | ND |
| PBS + S. pneumoniae | 10.0 | 3.2 (3.2) | ND | ND | ND | ND |
| UD + S. pneumoniae | 4,998.0 (722.5) | 2,417.7 (539.2) | 276.5 (45.5) | 41.0 (7.0) | 3,748.0 (287.5) | 389.0 (29.5) |
| Serum | ||||||
| Sham | 20.0 | ND | ND | 19 | ND | ND |
| Udorn | 62.5 (15.5) | ND | ND | 22.5 (2.5) | ND | ND |
| PBS + S. pneumoniae | 108.0 (20.5) | ND | ND | ND | ND | ND |
| UD + S. pneumoniae | 3,055.0 (285.0) | 4,632.0 (1,149.50) | 31.0 | 86.0 (12.5) | 1,094.0 (531.0) | 78.0 (2.0) |
| Day 14 post-viral infection | ||||||
| BALF | ||||||
| Sham | 12.0 | 3.3 (2.0) | ND | ND | ND | ND |
| Udorn | 30.0 (5.0) | 35.6 (30.1) | ND | 14.0 (2.0) | ND | 9.0 |
| PBS + S. pneumoniae | ND | 3.2 (3.2) | ND | ND | ND | ND |
| UD+Spn | 2,628.0 (1,687.0) | 1,632.2 (723.6) | 28.5 (2.5) | 49.5 (17.5) | 3,560.0 (715.0) | 233.0 (79.5) |
| Serum | ||||||
| Sham | 20.0 | ND | ND | 19.0 | ND | ND |
| Udorn | 60.0 (15.0) | ND | ND | ND | ND | ND |
| PBS+Spn | 80.0 (5.5) | ND | ND | ND | ND | ND |
| UD+Spn | 2,670.0 (225.0) | 951.0 (240.3) | 29.0 (4.5) | 179.0 (21.5) | 2,295.0 (245.0) | 98.0 (4.5) |
ND, below the level of detection (<7 pg/ml for the Chemicon five-plex antibody array of BALF samples, <14 pg/ml for the Chemicon five-plex antibody array of sera, and <2 pg/ml for the G-CSF ELISA).
Together, these results demonstrate that the predisposed state induced by a prior influenza virus infection can be maintained for at least 7 to 8 days following viral clearance and results in upregulated secretion of the anti-inflammatory cytokine IL-10 together with several proinflammatory cytokines and, more specifically, the neutrophil activator G-CSF.
DISCUSSION
Our goal was to investigate the role of synergistic bacterial superinfections in exacerbation of influenza virus-associated mortality using mild, self-limiting viral and bacterial infections. Singly infected animals did not die, nor did they exhibit overt signs of disease. Both the mild virus and bacterial infections and the transient bacteremia found in about 30% of S. pneumococcus-infected mice were cleared within 7 days. In contrast, superinfected mice rapidly became sick and succumbed to infection within 2 to 4 days postsuperinfection. In all influenza virus-infected mice, type 3 pneumococcus superinfection resulted in the prolonged systemic dissemination of bacteria. The high bacterial loads observed in all major organs likely overwhelm the host immune system such that it is no longer able to control the infection, and this leads to septic shock. The in vivo studies described here using mild, self-limiting viral and bacterial infections are highly reminiscent of synergistic disease observed in the clinical setting and should provide for an informative, more clinically relevant model to evaluate the effects of antivirals, antibiotics, and vaccine approaches on controlling synergistic respiratory disease.
In order to more closely mimic clinical establishment of synergistic disease, we used a filamentous non-mouse-adapted strain of influenza virus. Original human isolates of influenza virus are highly filamentous in morphology; even the avian H5N1 strain of influenza virus exhibits filamentous morphology. In contrast, most laboratory-adapted strains, including the commonly used influenza A/PR/8/34 strain of influenza virus, produce strictly spherical virions. Previous studies examining virus-bacterium synergistic interactions in vivo have utilized mouse-adapted, spherical strains of influenza virus that are themselves highly lethal to mice (20, 24, 30, 31, 35). Several mechanisms have been proposed that may contribute to synergistic exacerbation of respiratory disease. Early studies demonstrated that Staphylococcus aureus adheres more efficiently to influenza virus-infected MDCK cells (11, 12). More recently, Streptococcus pneumoniae has also been shown to bind more efficiently to influenza virus-infected epithelial cells in vitro (25) and to mouse respiratory epithelium in vivo (31). This increased binding is believed to be a result of destruction of ciliated cells and complete desquamation of the respiratory epithelium. In our studies, we also found that S. pneumoniae colonizes the respiratory tracts of mice more efficiently in animals with an ongoing influenza virus infection. However, the observation that animals superinfected after virus clearance remained highly susceptible to synergistic fatal septicemia suggests that direct viral and bacterial interactions are not required for enhanced colonization or exacerbative disease.
A recent study has suggested that increased levels of the anti-inflammatory cytokine IL-10 may be responsible for increased severity of pneumococcal infection in mice recovering from influenza virus infection (35). However, administration of an anti-IL-10 monoclonal antibody that was able to marginally reduce bacterial loads in superinfected animals did not protect animals and only delayed mortality (35). In our study, IL-10 mRNA levels in the lungs and spleens of superinfected mice were upregulated by four- to fivefold in comparison to singly infected mice. However, dramatic increases in mRNA levels for a variety of proinflammatory molecules in superinfected animals, including IL-6, TNF-α, IL-1β, inducible nitric oxide synthase (not shown), and notably G-CSF, were also observed. These cytokines all result in local inflammation and tissue damage when produced in excess amounts. Thus, our results suggest that multiple inflammatory mediators are synergistically exacerbated rather than IL-10 alone, and these all contribute to septic shock and death.
Depression of the immune system has been implicated in the reduced bacterial clearance from influenza virus-predisposed mice. In vitro studies have shown that influenza virus is able to cause apoptosis in neutrophils (9) and inhibit phagocytic and chemotaxic function of macrophages (19, 26). Other studies have observed a decrease in chemotactic activity of neutrophils at the time of secondary infection in an in vitro model (2). In our synergy model, however, we did not observe overt signs of neutropenia. In contrast, superinfection resulted in a massive influx of neutrophils into the lungs. This coincided with upregulated levels of the neutrophil chemoattractants MIP-2 and KC, mouse homologues of human IL-8, in the lungs of superinfected animals. Both have been implicated to play significant roles in acute lung injury (ALI) of patients (18, 22), a disease characterized by accumulation of inflammatory infiltrates such as activated neutrophils in the lung tissue and BALF that leads to tissue damage and respiratory distress. Immune system deregulation predisposes patients to ALI. Our studies suggest that the massive influx of neutrophils observed in our superinfected mice is contributing to the pathogenesis of the synergistic infection. Upon activation, neutrophils produce proinflammatory cytokines, which cause fever and local inflammation as well as aid in recruitment of additional inflammatory cells, including T and B lymphocytes (14). Activated neutrophils also produce antimicrobial agents, such as hydrogen peroxide, H2O2, superoxide anion, O2−, and nitrous oxide, NO (14). Thus, the enhanced bacterial colonization of the lungs following influenza virus infection may lead to the sustained production of toxic compounds by neutrophils that together with the bacteria increases tissue destruction. In addition, protein levels of G-CSF, which regulates the maturation, differentiation, and proliferation of neutrophils, were highly elevated in both serum and BALF from superinfected animals.
Recombinant human G-CSF (rhG-CSF) has been utilized in conjunction with antibiotics to prevent neutropenia-associated infections in cancer patients receiving chemotherapy. However, recent clinical studies have examined a possible correlation between the administration of G-CSF and acute lung injury and acute respiratory distress syndrome (37). Filgrastim (Roche), a commonly used rhG-CSF cancer therapeutic, when administered subcutaneously leads to serum levels of 48 to 56 ng/ml (10, 15). Due to the short half-life of rhG-CSF, patients receive daily administration of filgrastim until neutropenia subsides (10, 15). In neutropenic rats, lipopolysaccharide administered following treatment with recombinant G-CSF resulted in exacerbated lung injury compared to control rats that received no G-CSF therapy (5). This suggests that high G-CSF levels in the serum may be sufficient to predispose animals and/or patients to heightened bacterial infections. Interestingly, in our study G-CSF levels in excess of 4,600 pg/ml in the serum and over 2,400 pg/ml in the BALF following bacterial superinfection were determined, levels within the range at which rats and humans are predisposed to exacerbated lung disease by rG-CSF treatment. Together, these studies and the data presented here suggest that the release of G-CSF by the inflamed endothelium and alveolar monocytes and macrophages likely contributes significantly to the synergistic pathogenesis leading towards fatal septicemia.
The lung is not the only organ subject to neutrophil-induced tissue damage. Dysfunction of the blood-brain barrier in traumatic brain injury patients is associated with an increase of IL-8 production by brain microvascular endothelial cells and astrocytes (28). The production of IL-8 in the brain is induced by the synergistic activity of TNF-α and IL-1β (28). IL-1β was consistently upregulated at the mRNA level in the brains of superinfected mice. In a rat model of bacterial meningitis, administration of neutralizing antibodies targeting MIP-2 significantly decreased the influx of neutrophils into the meninges, further illustrating the contributing role of neutrophils in disease progression (13). KC, MIP-2, and G-CSF mRNA levels were all upregulated in the brains of the superinfected mice. Both KC and G-CSF mRNA expression were upregulated in the spleens from superinfected animals, suggesting that neutrophils may also be infiltrating the spleens and causing damage as well. KC and G-CSF were also elevated in the serum, which further implies the presence of circulating activated neutrophils.
Clearly evident from our study and the reports by others is that influenza virus infection establishes an environment in the respiratory tract, “the predisposed state,” that allows for an exacerbative response to subsequent bacterial infections. Singly infected animals did not display any adverse upregulation of proinflammatory cytokines that could be directly attributed to the synergistic effect. However, virus infection did result in an influx of predominantly lymphocytes and monocytes into the bronchoalveolar spaces, which were maintained well after the virus had been cleared. This suggests that these inflamed, or activated, lymphocytes/monocytes are predisposed to secrete massive levels of chemokines and proinflammatory cytokines once a superinfection results. Future studies will focus on identifying and characterizing these cells and how they respond to different stimuli.
In summary, we have established a murine model of fatal septicemia that closely mimics clinical exacerbation of influenza virus-associated respiratory disease by superinfection with serotype 3 pneumococci. The use of mild, nonlethal doses of each organism is sufficient to establish a synergistic and highly virulent relationship between the two pathogens. Further, our results suggest that bacteremia and the accompanying systemic tissue damage is of paramount importance in virus-bacterium synergistic pathogenesis. Whereas the neutrophil is likely to be the major host cellular infiltrate responsible for the exacerbation of disease in predisposed animals, it is the virus-induced lymphocytes infiltrating the lungs that are likely predisposed for massive upregulation of neutrophil chemoattractants and activators, such as MIP-2, KC, and G-CSF. G-CSF may be acting to prolong neutrophil survival, allowing for sustained secretion of tissue-damaging molecules in the lungs, spleens, and brains of superinfected animals. The present study further illustrates the complexity of factors that synergistically contribute to exacerbative influenza virus-associated illness. As we learn more about the nature of the influenza virus-induced predisposed state, we can then begin to design new therapeutic treatment strategies to reduce this synergistic exacerbation of disease. Future studies will focus on evaluating whether other bacterial pathogens frequently associated with influenza virus infections, i.e., Staphylococcus aureus and Haemophilus influenzae, also exacerbate disease in a similar fashion.
Acknowledgments
We thank Jocelyn Ang and Roy S. Sundick for many helpful discussions.
This study was supported in part by Public Health Service grants (AI47783 and AI065591) from the National Institute of Allergy and Infectious Diseases.
Editor: A. Camilli
Footnotes
Published ahead of print on 2 April 2007.
REFERENCES
- 1.Abramson, J. S., and E. L. Mills. 1988. Depression of neutrophil function induced by viruses and its role in secondary microbial infections. Rev. Infect. Dis. 10:326-341. [DOI] [PubMed] [Google Scholar]
- 2.Abramson, J. S., J. C. Lewis, D. S. Lyles, K. A. Heller, E. L. Mills, and D. A. Bass. 1982. Inhibition of neutrophil lysosome-phagosome fusion associated with influenza infection in vitro: role in depressed bactericidal activity. J. Clin. Investig. 69:1393-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ada, G. L., and B. T. Perry. 1958. Properties of the nucleic acid of the Ryan strain of filamentous influenza virus. J. Gen. Microbiol. 19:40-54. [DOI] [PubMed] [Google Scholar]
- 4.Ada, G. L., B. T. Perry, and A. Abbot. 1958. Biological and physical properties of the Ryan strain of filamentous influenza virus. J. Gen. Microbiol. 19:23-39. [DOI] [PubMed] [Google Scholar]
- 5.Azoulay, E., H. Attalah, K. Yang, S. Herigault, H. Jouault, C. Brun-Buisson, L. Brochard, A. Harf, B. Schlemmer, and C. Delclaux. 2003. Exacerbation with granulocyte colony-stimulating factor of prior acute lung injury during neutropenia recovery in rats. Crit. Care Med. 31:157-165. [DOI] [PubMed] [Google Scholar]
- 6.Briles, D. E., R. C. Tart, E. Swiatlo, J. P. Dillard, P. Smith, K. A. Benton, B. A. Ralph, A. Brooks-Walter, M. J. Crain, S. K. Hollingshead, and L. S. McDaniel. 1998. Pneumococcal diversity: consideration for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA). Clin. Microbiol. Rev. 11:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carollo, M., C. M. Hogaboam, S. L. Kunkel, S. Delaney, M. I. Christie, and M. Peretti. 2001. Analysis of the temporal expression of chemokines and chemokine receptors during experimental granulomatous inflammation: role and expression of MIP-1α and MCP-1. Br. J. Pharmacol. 134:1166-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choppin, P. W., J. S. Murphy, and I. Tamm. 1960. Studies of two kinds of virus particles which comprise influenza A virus strains. J. Exp. Med. 112:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colamussi, M. L., M. R. White, E. Crouch, and K. L. Hartshorn. 1999. Influenza A virus accelerates neutrophil apoptosis and markedly potentiates apoptotic effects of bacteria. Blood 93:2395-2403. [PubMed] [Google Scholar]
- 10.Dale, D. C., M. A. Bonilla, M. W. Davis, A. M. Nakanishi, W. P. Hammond, J. Kurtzberg, W. Wang, A. Jakubowski, E. Winton, P. Lalezari, et al. 1993. A randomized controlled phase III trial of recombinant human granulocyte colony-stimulating factor (filgrastim) for treatment of severe chronic neutropenia. Blood 81:2496-2502. [PMC free article] [PubMed] [Google Scholar]
- 11.Davison, V. E., and B. A. Sanford. 1981. Adherence of Staphylococcus aureus to influenza A virus-infected Madin-Darby canine kidney cell cultures. Infect. Immun. 32:118-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison, V. E., and B. A. Sanford. 1982. Factors influencing adherence of Staphylococcus aureus to influenza A virus-infected cell cultures. Infect. Immun. 37:946-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diab, A., H. Abdalla, H. L. Li, F. D. Shi, J. Zhu, B. Hojberg, L. Lindquist, B. Wretlind, M. Bakhiet, and H. Link. 1999. Neutralization of macrophage inflammatory protein 2 (MIP-2) and MIP-1α attenuates neutrophil recruitment in the central nervous system during experimental bacterial meningitis. Infect. Immun. 67:2590-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis, T. N., and B. L. Beaman. 2004. Interferon-γ activation of polymorphonuclear neutrophil function. Immunology 112:2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heil, G., D. Hoelzer, M. A. Sanz, K. Lechner, J. A. Liu Yin, G. Papa, L. Noens, J. Szer, A. Ganser, C. O'Brien, J. Matcham, and A. Barge. 1997. A randomized, double-blind placebo-controlled, phase III study of filgrastim in remission induction and consolidation therapy for adults with de novo acute myeloid leukemia. Blood 90:4710-4718. [PubMed] [Google Scholar]
- 16.Hitt, E. 2004. S. pneumoniae serotype 3 not covered by 7-valent vaccine. Medscape Med. News 2004:470964. [Google Scholar]
- 17.Jones, W. T., and J. H. Menna. 1982. Influenza type A virus-mediated adherence of type 1a group B Streptococci to mouse tracheal tissue in vivo. Infect. Immun. 38:791-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinoshita, M., H. Mochizuki, and S. Ono. 1999. Pulmonary neutrophil accumulation following human endotoxemia. Chest 116:1709-1715. [DOI] [PubMed] [Google Scholar]
- 19.Kleinerman, E. S., C. A. Daniels, R. P. Polisson, and R. Synderman. 1976. Effect of virus infection of the inflammatory response: depression of macrophage accumulation in influenza-infected mice. Am. J. Pathol. 85:373-382. [PMC free article] [PubMed] [Google Scholar]
- 20.LeVine, A. M., V. Koeningsknecht, and J. M. Stark. 2001. Decreased pulmonary clearance of S. pnuemoniae following influenza A infection in mice. J. Virol. Methods 94:173-186. [DOI] [PubMed] [Google Scholar]
- 21.Livak, K. J., and T. D. Schittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 22.Lomas-Neira, J. L., C.-S. Chung, D. E. Wesche, M. Perl, and A. Ayala. 2005. In vivo gene silencing (with siRNA) of pulmonary expression of MIP-2 versus KC results in divergent effects on hemorrhage-induced, neutrophil-mediated septic acute lung injury. J. Leukoc. Biol. 77:846-853. [DOI] [PubMed] [Google Scholar]
- 23.McCullers, J. A. 2006. Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. 19:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCullers, J. A., and J. E. Rehg. 2002. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J. Infect. Dis. 186:341-350. [DOI] [PubMed] [Google Scholar]
- 25.McCullers, J. A., and K. C. Bartmess. 2003. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 187:1000-1009. [DOI] [PubMed] [Google Scholar]
- 26.Nickerson, C. L., and G. J. Jakab. 1990. Pulmonary antibacterial defenses during mild and severe influenza virus infection. Infect. Immun. 58:2809-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto, S., S. Kawabata, I. Nakagawa, Y. Okuno, T. Goto, K. Sano, and S. Hamada. 2003. Influenza A virus-infected hosts boost an invasive type of Streptococcus pyogenes infection in mice. J. Virol. 77:4104-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto, V. I., U. E. Heinzel-Pleines, S. M. Gloor, O. Trentz, T. Kossmann, and M. C. Morganti-Kossmann. 2000. sICAN-1 and TNF-alpha induce MIP-2 with distinct kinetics in astrocytes and brain microvascular endothelial cells. J. Neurosci. Res. 60:733-742. [DOI] [PubMed] [Google Scholar]
- 29.Overbergh, L., A. Giuletti, D. Valckx, R. Decallonne, R. Bouillon, and C. Mathieu. 2003. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J. Biomol. Tech. 14:33-43. [PMC free article] [PubMed] [Google Scholar]
- 30.Peltola, V. T., K. G. Murti, and J. A. McCullers. 2005. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J. Infect. Dis. 192:249-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plotkowski, M.-C. 1986. Adherence of type I Streptococcus pneumoniae to tracheal epithelium of mice infected with influenza A/PR8 virus. Am. Rev. Respir. Dis. 134:1040-1044. [DOI] [PubMed] [Google Scholar]
- 32.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 33.Roberts, P. C., R. A. Lamb, and R. W. Compans. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127-137. [DOI] [PubMed] [Google Scholar]
- 34.Stevens, K. M. 1976. Cardiac stroke volume as a determinant of influenzal fatality. Med. Intell. 295:1363-1366. [DOI] [PubMed] [Google Scholar]
- 35.van der Sluijs, K., L. J. van Elden, M. Nijhuis, R. Schuurman, J. M. Pater, S. Florquin, M. Goldman, H. M. Jansen, R. Lutter, and T. van der Poll. 2004. IL-10 is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. J. Immunol. 172:7603-7609. [DOI] [PubMed] [Google Scholar]
- 36.Watt, J. P., K. L. O'Brien, S. Katz, M. A. Bronsdon, J. Elliott, J. Dallas, M. J. Perilla, R. Reid, L. Murrow, R. Facklam, M. Santosham, and C. G. Whitney. 2004. Nasopharyngeal versus oropharyngeal sampling for detection of pneumococcal carriage in adults. J. Clin. Microbiol. 42:4974-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiedermann, F. J. 2005. Acute lung injury during G-CSF-induced neutropenia recovery: effect of G-CSF on pro- and anti-inflammatory cytokines. Bone Marrow Transpl. 36:731. [DOI] [PubMed] [Google Scholar]
- 38.Wu, H.-Y., A. Virolainen, B. Mathews, J. King, M. W. Russell, and D. E. Briles. 1997. Establishment of a Streptococcus pneumoniae nasopharyngeal colonization model in adult mice. Microb. Pathog. 23:127-137. [DOI] [PubMed] [Google Scholar]