Abstract
The natural history, microevolution, and patterns of interindividual transmission and global dissemination of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans were studied by population genetic analysis. The JP2 clone is strongly associated with aggressive periodontitis in adolescents of African descent and differs from other clones of the species by several genetic peculiarities, including a 530-bp deletion in the promoter region of the leukotoxin gene operon, which results in increased leukotoxic activity. Multilocus sequence analysis of 82 A. actinomycetemcomitans strains, 66 of which were JP2 clone strains collected over a period of more than 20 years, confirmed that there is a clonal population structure with evolutionary lineages corresponding to serotypes. Although genetically highly conserved, as shown by alignment of sequences of eight housekeeping genes, strains belonging to the JP2 clone had a number of point mutations, particularly in the pseudogenes hbpA and tbpA. Characteristic mutations allowed isolates from individuals from the Mediterranean area and from West Africa, including the Cape Verde Islands, to be distinguished. The patterns of mutations indicate that the JP2 clone initially emerged as a distinct genotype in the Mediterranean part of Africa approximately 2,400 years ago and subsequently spread to West Africa, from which it was transferred to the American continents during the transatlantic slave trade. The sustained exclusive colonization of individuals of African descent despite geographical separation for centuries suggests that the JP2 clone has a distinct host tropism. The colonization of family members by JP2 clone strains with unique point mutations provides strong evidence that there is intrafamilial transmission and suggests that dissemination of the JP2 clone is restricted to close contacts.
Aggressive periodontitis is a major clinical problem that eventually results in loss of the tissues supporting teeth at an early age. It is an infectious disease, but the complexity of the microflora that constitutes the biofilm in the affected periodontal pockets makes it difficult to determine the exact etiology (44). Early studies demonstrated that there is an association between the disease and the presence of bacteria belonging to the species Actinobacillus actinomycetemcomitans (51), which recently was reclassified in the new genus Aggregatibacter together with its close relatives Aggregatibacter (Haemophilus) aphrophilus and Aggregatibacter (Haemophilus) segnis (42). The primary habitat of A. actinomycetemcomitans is dental plaques, and this organism may also be isolated from part of the healthy population (32, 51). It is a recognized opportunistic pathogen that occasionally causes endocarditis and abscesses at various body sites and internal organs (43, 45).
A. actinomycetemcomitans isolates from healthy individuals, from patients with periodontitis, and from patients with systemic infections exhibit significant genetic diversity, in agreement with the status of this organism as an opportunistic pathogen. However, the majority of isolates obtained from aggressive periodontitis in adolescents of African descent living in different parts of the world are genetically homogeneous and belong to a single clone termed the JP2 clone (19, 21, 22, 25). Studies of adolescents in Morocco have demonstrated that the JP2 clone is endemic in that population and that it is strongly associated with disease, in contrast to other clones of A. actinomycetemcomitans (24, 25). The concept of the high pathogenic potential of the JP2 clone was also supported by the results of a longitudinal study of African-American families with aggressive periodontitis (9). The association between the JP2 clone and periodontitis in adolescents has been demonstrated for several study populations living in geographically widespread areas, however, supporting the hypothesis that the JP2 clone is associated with patients of African descent (10, 11, 18-22, 31, 35). Isolation of the JP2 clone from occasional periodontitis patients having other ethnic origins was mentioned recently, but detailed information was not provided (18). The possible spread of the JP2 clone to other ethnic groups requires further study and should take into account the inherent complexity of defining genetic background in some human populations (3, 6). The primary association of the JP2 clone with patients of various African descents and its putative unusual virulence may contribute to observed differences in the prevalence of aggressive periodontitis in different countries (2).
The JP2 clone is named after the first recognized isolate belonging to the clone, which was obtained from an 8-year-old African-American child with prepubertal periodontitis (56). It was first noted because of its enhanced leukotoxic activity caused by a specific 530-bp deletion in the promoter region of the leukotoxin gene operon (8). This clone belongs to serotype b and is further characterized by a distinct profile of alleles encoding intracellular metabolic enzymes, as revealed by multilocus enzyme electrophoresis, by a distinct MspI DNA fingerprint, and by clear beta-hemolytic zones on blood agar caused by the enhanced leukotoxic activity (5, 21, 22). In addition, as a result of a mutational event affecting the gene encoding the hemoglobin-binding protein (hbpA), members of the JP2 clone are unable to use human hemoglobin as an iron source (26).
According to Maynard Smith (37), a bacterial clone is defined as “a set of genetically similar cells, recently derived from a common ancestor, without chromosomal recombination.” This definition takes into account the minor evolutionary diversification that inevitably occurs over time as a result of accumulation of mutations. Unusually, in A. actinomycetemcomitans genomic reorganizations facilitated by intragenomic homologous recombination between the multicopy rRNA operons and IS150-like elements contribute to this genetic diversification process (15), which, even among members of the JP2 clone, results in differences in ribotyping patterns and in pulsed-field gel electrophoresis (PFGE) patterns of restriction fragments after cleavage of genomic DNA with rare cutting endonucleases (e.g., XhoI) (15, 21, 22).
In the study reported here we used the markers of genetic diversification in the JP2 clone of A. actinomycetemcomitans to obtain information about its natural history and patterns of interindividual transmission and global dissemination.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A total of 82 A. actinomycetemcomitans strains, including 66 strains with a 530-bp deletion in the promoter region of the leukotoxin (ltx) gene operon (JP2 strains) and 16 strains without this deletion (non-JP2 strains), were included in this study. The strains without the 530-bp deletion included four serotype a strains, five serotype b strains, two serotype c strains, two strains serotype d, two serotype e strains, and one nonserotypeable strain, representing the major evolutionary lineages observed by Poulsen and coworkers (48). Among the non-JP2 serotype b strains were HK1605 (= UP14), which belongs to restriction fragment length polymorphism (RFLP) group II described by DiRienzo and coworkers (12, 13), and strain HK975 (= Y4) of Tsai et al. (55). The JP2 strains with the 530-bp deletion were isolated from 66 subjects living in geographically widespread areas over a period of more than 20 years. Strains were obtained from individuals from Morocco, Algeria, Israel, Turkey, Portugal, and the Cape Verde Islands, some of whom had migrated to northern European countries (Denmark, Sweden, Switzerland, and The Netherlands), and from individuals who are now living in Brazil and the United States but are of African and Hispanic ethnicity. It is generally accepted that Hispanics have a mixture of native American, African, and European ancestry (6). Five of the strains assigned to the JP2 clone were obtained from laboratories in The Netherlands (two strains) and the United States (three strains) and were isolated from periodontitis patients having unspecified ethnic backgrounds. Of the 66 JP2 strains, 17 were obtained from individuals belonging to seven families.
Leukotoxin promoter subtyping by PCR.
The two promoter region types with and without the 530-bp deletion upstream of the ltx gene operon were differentiated by PCR as previously described using primers ltx3 and ltx4 (46).
MspI DNA fingerprinting.
Whole-cell DNA was prepared as described previously (47). Approximately 10 μg was digested with MspI, electrophoresed in a 1% agarose gel overnight at 1.5 V cm−1 in Tris-acetate-EDTA buffer, and visualized by staining with ethidium bromide (50).
XhoI DNA fingerprinting.
XhoI DNA fingerprinting was performed as previously described (15). Briefly, bacteria were grown on chocolate agar plates and washed off the plates in buffer. Most strains had the adherent rough phenotype, and to resolve aggregation mediated by proteins, each bacterial suspension was treated with proteinase K. After the agarose plugs were prepared, the proteinase K was inactivated by incubation with phenylmethylsulfonyl fluoride. After washing, the plugs were treated with lysozyme. The plugs were digested with proteinase K, which was followed by treatment with phenylmethylsulfonyl fluoride. DNA in the plugs was digested with XhoI in the buffer supplied with the enzyme by adding the restriction enzyme three times at 20-h intervals. XhoI was added three times because we found that it was difficult to obtain complete digestion of the genomic DNA in the plugs. The resulting DNA fragments were resolved by PFGE.
Sequence analysis of genes encoding housekeeping enzymes and pseudogenes encoding hemoglobin-binding and transferrin-binding proteins.
Fragments of the following genes encoding six housekeeping enzymes were selected for multilocus sequence typing analysis: pgi encoding glucose-6-phosphate isomerase, recA encoding the RecA protein, adk encoding adenylate kinase, frdB encoding fumarate reductase, atpG encoding the gamma subunit of ATP synthase F1, and mdh encoding malate dehydrogenase. These housekeeping genes were selected from the seven genes included in the Haemophilus influenzae multilocus sequence typing scheme (http://haemophilus.mlst.net). One copy of each of the six housekeeping genes was found in the A. actinomycetemcomitans HK1651 genome (http://www.genome.ou.edu/act.html), from which the primers were designed (Table 1). The seventh H. influenzae gene (fucK) was not found in A. actinomycetemcomitans. The six genes were located in widespread areas of the 2.1-Mb genome of HK1651 (Table 1) and were not close to any virulence genes that might be expected to be under selection pressure. In addition, two fragments of the hemoglobin-binding protein gene hbpA (which is a pseudogene in JP2 strains) and one fragment of the pseudogene for transferring-binding protein (tbpA) were sequenced (Table 1). The PCR and DNA sequencing were performed as previously described (15). The regions sequenced are shown in Table 1. The selected fragments from serotype a, c, d, and e strains were sequenced in both directions. Fragments from strains belonging to the serotype b cluster of strains were sequenced in only one direction except if polymorphic sites were revealed. Fragments with polymorphic sites were sequenced in both directions for confirmation.
TABLE 1.
Primers used in multilocus sequence analysis and lengths and positions of the fragments sequenced
| Gene | PCR and sequencing primers (5′-3′)
|
Length of fragment sequenced (bp) | Position of sequenced fragment in HK1651 genomea | |
|---|---|---|---|---|
| Forward primer | Reverse primer | |||
| adk | TGACATAAAGAGGACAACATGA | TTGTTAAGGGATTAAGCCAAGATT | 559 | 1269235-1269793 |
| atpG | AGATGGCAGGTGCAAAAGAGATAA | TGTCTTTCGCTTAGGTGATCGTTT | 500 | 998161-998660 |
| frdB | TGCTTGATGCCTTAGGTTACATTA | TCGGTTTAAGCATGGAAATCAGAT | 510 | 551859-552368 |
| mdh | GTTCCGAATTGTCACTTTATGATA | GGCTTTCGCGTTTACTACTTCGGTG | 440 | 1213887-1214326 |
| pgi | CAAGCGAAAGAAGCCATGTTCAAC | CGCGATGGATAAACCGATGGCTGA | 527 | 1098608-1099134 |
| recA | CTCGACGCGGCGTTAGGTCAAATT | ATTACCGAACATCACGCCGATT | 495 | 1045025-1045519 |
| hbpA-1 | CATTGATTTGGTATACAAAGGC | TTGTACCAAATGGGCGGATTGATC | 439 | 526858-527296 |
| hbpA-2 | TATTTTGACGCAATTGCTGTTC | TAGGGCACTTATCATTTCCATC | 329 | 525072-525400 |
| tbpA | CAACACAAATACTGCTTAAGATGC | GAAATTCTATTACGATCCATTCCG | 344 | 1500560-1500903 |
For alignment of the sequences we used the program ClustalX (54). A cluster analysis of the 82 isolates, based on concatenated sequences consisting of nine sequenced DNA fragments in the order adk atpG frdB mdh pgi recA hbpA-1 hbpA-2 tbpA, was conducted by using the Minimum Evolution algorithm in MEGA, version 3.0 (33).
Nucleotide sequence accession numbers.
The DNA sequences determined in this study have been deposited in the GenBank database (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov) under the following accession numbers: adk alleles, EF142117 to EF142198; atpG alleles, EF142199 to EF142280; frdB alleles, EF142281 to EF142362; hbpA-1 alleles, EF142363 to EF142443; hbpA-2 alleles, EF142444 to EF142524; mdh alleles, EF142525 to EF142606; pgi alleles, EF142607 to EF142688; recA alleles, EF142689 to EF142770; and tbpA alleles, EF142771 to EF142852.
RESULTS
Genomic variation as determined by MspI fingerprinting of whole-cell DNA.
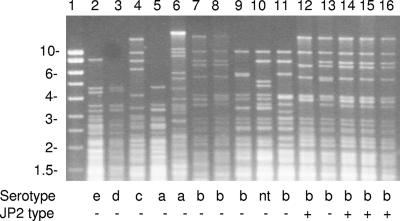
A total of 16 different RFLP patterns were identified for the 82 isolates after cleavage of whole-cell DNA with the restriction enzyme MspI and analysis by conventional 1% agarose gel electrophoresis. Most of the 66 JP2 strains had the same DNA fingerprint; the only exception was strain HK1986, whose fingerprint deviated slightly due to loss of two bands (Fig. 1, lane 12). Different MspI fingerprints were obtained for most of the 16 non-JP2 strains without the 530-bp deletion which belonged to different serotypes; the exception was two strains belonging to serotype d which had the same unique pattern (Fig. 1, lane 3). Among the non-JP2 serotype b strains, strain HK911 had a DNA fingerprint identical to the characteristic DNA cleavage pattern for JP2 strains (Fig. 1, lane 8), strains HK912 and HK1605 (= UP14) had very similar but distinct fingerprints (Fig. 1, lanes 7 and 13), and strains HK908, HK913, and HK975 also had patterns similar to the pattern for JP2 clone strains (Fig. 1, lanes 9, 10, and 11). Thus, the JP2 clone is closely related to other non-JP2 serotype b strains. In the study of DiRienzo and coworkers (12, 13), strain HK1605 (= UP14) was an exception as it belonged to RFLP group II, exhibiting the strongest correlation with disease and conversion to disease, but it did not have the 530-bp deletion in the promoter region of the leukotoxin gene operon that is characteristic of the JP2 clone (9).
FIG. 1.
Genotyping of A. actinomycetemcomitans strains by restriction of whole-cell DNA with MspI and resolution by 1% agarose gel electrophoresis. The positions of molecular weight markers (in kilobases) are indicated on the left. Since MspI is a frequently cutting enzyme, most of the resulting DNA fragments are smaller than 2 kb. These small fragments form a smear in the front of the gel, and only a few larger fragments produce bands in the gel. Thus, the bands of DNA fragments larger than 2 kb represent only a small fraction of the genome (approximately 5%). Lane 1, molecular weight marker; lane 2, HK929; lane 3, HK928; lane 4, HK978; lane 5, HK780; lane 6, HK974; lane 7, HK912; lane 8, HK911; lane 9, HK913; lane 10, HK908; lane 11, HK975 (= Y4); lane 12, HK1986; lane 13, HK1605 (= UP14); lane 14, HK1630; lane 15, HK921 (= JP2); lane 16, HK1651.
Genomic variation as revealed by XhoI DNA fingerprinting.
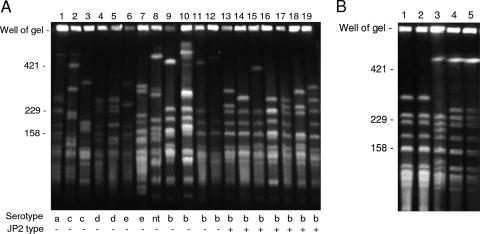
We have previously shown that PFGE using the rare cutting restriction enzyme XhoI can reveal genomic rearrangements in strains belonging to the JP2 clone (15). By using this method 34 different XhoI DNA fingerprints were obtained for the 82 A. actinomycetemcomitans strains studied. Representative examples of XhoI DNA fingerprints are shown in Fig. 2A. Fifteen different XhoI DNA fingerprints were obtained for the 16 non-JP2 strains, meaning that most of the non-JP2 strains had unique XhoI DNA fingerprints; the only exception was two serotype b strains, HK911 and HK1605 (= UP14), which had the same pattern (Fig. 2A, lanes 9 and 11). For the 66 JP2 clone strains 19 different patterns were identified. One XhoI DNA fingerprint was dominant (obtained for 33 of the 66 strains) and was obtained only for JP2 clone strains (Fig. 2A, lane 13, and Fig. 2B, lanes 1 and 2). Five different XhoI DNA fingerprints for the remaining 33 JP2 strains were produced by six, five, four, three, and two strains, and the remaining 13 strains each produced a unique pattern. The second and third most dominant XhoI DNA fingerprints among JP2 clone strains were very similar to XhoI DNA fingerprints obtained for serotype b non-JP2 strains that were produced by one (HK913) and two (HK911 and HK1605 [= UP14]) strains (Fig. 2A, lanes 9, 10, 11, 14, and 15). Of 17 isolates from members of seven families, 12 produced the most frequent XhoI DNA fingerprint, including all isolates from families 1, 2, and 4, HK1507, HK1512, and HK1514 from family 3, HK1520 from family 5, and HK1613 from family 7 (Fig. 2A, lane 13, and Fig. 2B, lanes 1 and 2). The XhoI DNA fingerprints of isolates from families 3, 5, 6, and 7 revealed that there were two different patterns in each of the families.
FIG. 2.
XhoI DNA fingerprinting of A. actinomycetemcomitans strains. Bacteria in agarose plugs were digested with XhoI, and the resulting DNA fragments were resolved by PFGE. The positions of molecular weight markers (in kilobases) are indicated on the left. (A) Representative XhoI DNA fingerprints of A. actinomycetemcomitans strains without and with the 530-bp deletion in the leukotoxin gene operon (non-JP2 and JP2 strains, respectively). The serotype and leukotoxin promoter type of each A. actinomycetemcomitans strain are indicated below the gel. Lane 1, HK892; lane 2, HK907; lane 3, HK978; lane 4, HK928; lane 5, HK1002; lane 6, HK929; lane 7, HK961; lane 8, HK908; lane 9, HK911; lane 10, HK913; lane 11, HK1605; lane 12, HK975; lane 13, HK1507; lane14, HK1985; lane 15, HK1651; lane 16, HK 1607; lane 17, HK1548; lane 18, HK1615; lane 19, HK1659. The fingerprints in lanes 9 and 11 were considered identical, whereas the fingerprints in the remaining lanes were considered distinct. (B) XhoI DNA fingerprints of JP2 clone strains from related and unrelated individuals. Lane 1, HK1536 (family 4); lane 2, HK1631 (family 4); lane 3, HK2006; lane 4, HK1548 (family 6); lane 5, HK1517 (family 3). Lanes 1 and 2 show the prevailing PFGE pattern of the JP2 clone from two related individuals. Lane 3 shows an example of a unique PFGE pattern. Lanes 4 and 5 show the same PFGE pattern for two unrelated individuals, one from an African-American family and one from a family living in Sweden and originating from the Cape Verde Islands. The upper band in lanes 3, 4, and 5 represents undigested DNA.
Overall population structure of A. actinomycetemcomitans and microevolution in the serotype b cluster of strains as revealed by multilocus sequence analysis.
The multilocus sequence analysis was based on fragments of genes encoding six housekeeping enzymes, two fragments of the hbpA gene, which is a pseudogene in the JP2 strains, and a fragment of the tbpA pseudogene. Concatenated sequences of these nine gene fragments, which altogether consisted of 4,143 nucleotides, were used to determine the overall genetic similarity among strains. The PCR using the primer sets for hbpA did not result in amplification of a product for strain HK975 (= Y4). Strain HK929 had an insertion sequence in the hbpA-2 fragment, which left only 187 bp (positions 525072 to 525258 in the HK1651 genome) of this fragment in the concatenated sequence of this strain.
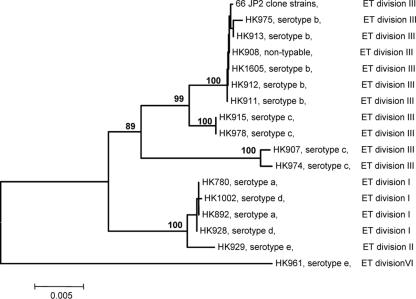
Cluster analysis demonstrated that there was a close relationship between clusters and serotypes (Fig. 3). The serotype b strains, including JP2 clone strains, constituted a separate cluster in the dendrogram, and the close genetic relatedness among serotype b JP2 and non-JP2 strains is in agreement with their very similar MspI fingerprints.
FIG. 3.
Phylogenetic relationships among 82 A. actinomycetemcomitans strains based on cluster analysis using the Minimum Evolution algorithm for concatenated sequences of fragments of six housekeeping enzyme genes and three DNA fragments from genes encoding hemoglobin-binding and transferring-binding proteins. A total of 66 strains with the 530-bp deletion in the leukotoxin gene operon (JP2 clone strains) obtained from geographically widespread areas and 16 strains without the deletion (non-JP2 strains) were studied. The scale bar indicates genetic distance. The numbers at the nodes indicate the percentages of bootstrap replicates that support the corresponding clusters. Electrophoretic type (ET) divisions, as determined by multilocus enzyme electrophoresis in a previous population genetic study of A. actinomycetemcomitans, are indicated on the right (48).
The 66 JP2 strains were genetically very homogeneous, and only 11 sequence types (STs) with minor differences were identified (Table 2). For the sequenced DNA fragments of the six housekeeping enzymes a total of five point mutations were found in JP2 clone strains in the sequenced fragments of adk, atpG, frdB, and recA, whereas no variation was found in the fragments of mdh and pgi. Thus, the frequency of polymorphic sites in the six functional housekeeping gene fragments varied from 0 (mdg and pgi) to 1:250 nucleotides (atpG). Three of the five mutations were transitions, two were transversions, and all were silent mutations. For the JP2 clone strains the frequency of mutations detected in the three fragments of the two nonfunctional pseudogenes was higher (1:165 to 1:220) than the frequency in the sequenced fragments of the housekeeping enzyme genes, in agreement with the lack of functional constraints on sequences of pseudogenes.
TABLE 2.
Polymorphic sites for 72 serotype b strains
| ST | No. of isolates | A. actinomycetemcomitans type | Nucleotides in gene fragments sequenced
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| adk position 1269464a |
atpG
|
frdB
|
recA
|
hbpA-1
|
hbpA-2
|
tbpA
|
|||||||||||||
| Position 998226 | Position 998430 | Position 551989 | Position 552223 | Position 1045096 | Position 1045144 | Position 526897 | Position 527177 | Position 525077 | Position 525152 | Position 525187 | Position 525285 | Position 1500566 | Position 1500848b | Position 1500872 | Position 1500879 | ||||
| 1 | 30 | JP2 | T | C | G | A | G | G | C | G | C | T | C | C | G | C | A | A | A |
| 2 | 2 | JP2 | T | C | G | A | G | G | C | G | C | T | C | C | G | C | A | G | A |
| 3 | 1 | JP2 | T | C | G | A | G | G | C | G | C | T | C | C | G | A | A | A | A |
| 4 | 3 | JP2 | T | C | G | A | A | G | C | G | C | T | C | C | G | C | A | A | A |
| 5 | 1 | JP2 | T | C | G | A | G | G | C | G | C | T | G | C | G | C | A | A | A |
| 6 | 1 | JP2 | T | C | G | A | G | G | C | G | T | T | C | C | A | C | A | A | A |
| 7 | 1 | JP2 | T | C | C | A | G | G | C | T | C | T | C | C | A | C | A | A | A |
| 8 | 20 | JP2 | T | C | G | A | G | G | C | G | C | T | C | C | A | C | A | A | A |
| 9 | 3 | JP2 | T | T | G | A | G | G | C | G | C | T | C | C | A | C | A | A | A |
| 10 | 2 | JP2 | C | C | G | A | G | G | C | G | C | T | C | C | A | C | A | A | A |
| 11 | 2 | JP2 | T | C | G | A | G | G | A | G | C | T | C | C | A | C | A | A | A |
| 12 | 1 | Non-JP2 | T | C | G | A | G | G | C | G | C | G | C | C | G | C | A | A | A |
| 13 | 1 | Non-JP2 | T | C | G | A | G | T | C | G | C | G | C | C | G | C | — | A | A |
| 14 | 1 | Non-JP2 | T | C | G | A | G | G | C | G | C | G | C | A | G | C | A | A | A |
| 15 | 1 | Non-JP2 | T | C | G | G | G | G | C | G | C | G | C | C | G | C | — | A | G |
| 16 | 2 | Non-JP2 | T | C | G | A | G | G | C | G | C | G | C | C | G | C | — | A | A |
Position in the HK1651 genome (http://www.genome.ou.edu/act.html).
—, deletion of a single nucleotide at this position.
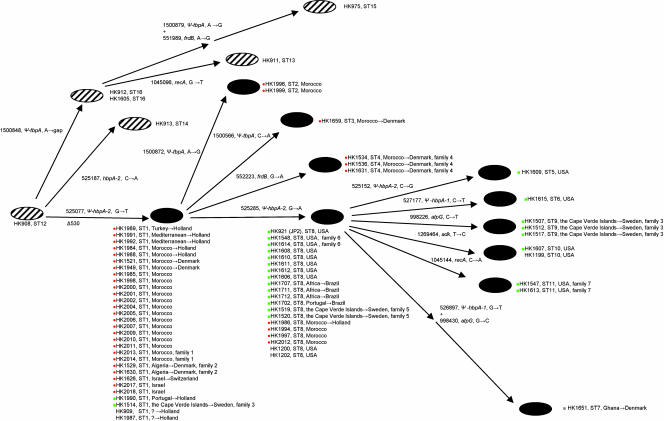
The genetic diversity among serotype b non-JP2 strains has previously been shown to be much higher than the genetic diversity among JP2 strains. Thus, for 26 serotype b non-JP2 strains 18 distinct electrophoretic types were revealed by multilocus enzyme electrophoresis (48), whereas for 37 of 38 JP2 clone strains the electrophoretic types were identical (22). Based on this difference in diversity and because all JP2 clone strains have a unique deleterious mutation in the hbpA gene not found in other strains of the species, it is conceivable that the JP2 clone is relatively new and that a serotype b non-JP2 strain was the progenitor of the JP2 clone. Assuming that the sequence variation observed in the serotype b cluster (Table 2) was caused by single site mutations and that each mutation occurred only once, it is possible to deduce the evolutionary scenario for the 72 strains in the serotype b cluster, as shown in Fig. 4. The cluster of serotype b strains consisted of two evolutionary lineages in which all six non-JP2 strains clustered separately primarily due to one polymorphic site (position 525077 in the HK1651 genome) in the hbpA gene. In addition, a specific mutation in the nonfunctional hbpA pseudogene in the JP2 strains (position 525285 in the HK1651 genome) divided the group of JP2 strains further into two evolutionary lineages (Table 2 and Fig. 4). The vast majority of isolates in one cluster of JP2 clone strains (ST1 to ST4) were obtained from individuals who lived in the Mediterranean area (32 of 34 individuals with known origins [94%]), whereas in the other cluster (ST5 to ST11) 23 of 27 isolates (85%) were obtained from individuals who lived in West Africa or were West African descendants living in the Cape Verde Islands, in Brazil, or in the United States (Fig. 4). In addition to this mutation, which resulted in separation of JP2 clone strains into two major clusters, 15 of the JP2 clone strains had a single mutation, and one strain, HK1651, had two mutations in the 4,143 nucleotides analyzed.
FIG. 4.
Model for microevolution in the serotype b cluster consisting of 66 JP2 clone and 6 non-JP2 A. actinomycetemcomitans strains based on genetic variation revealed by multilocus sequence analysis and based on the assumptions described in the Results. The cluster of non-JP2 strains is indicated by cross-hatched ellipses, and the JP2 clone strains are indicated by solid ellipses. Each arrow in the model indicates a single mutation in the 4,143 nucleotides analyzed, and each mutation is indicated by the position in the HK1651 genome, the designation of the gene involved, and the nucleotide substitution. The 530-bp deletion in the ltx promoter is indicated by Δ530. Bacterial isolates from related family members in seven families are indicated, as are the countries of origin of the individuals from whom the JP2 clone isolates originated. If the individual was living in a country other than the country of origin, this is indicated after the small arrow. Isolates from individuals who originated in the Mediterranean area are indicated by a red dot, and isolates from individuals with a West African origin, including African-Americans from the United States, are indicated by a green square. No information about their origins was available for five JP2 clone-positive individuals, three living in the United States and two living in The Netherlands. Most of the JP2 strains from the United States were obtained from African-Americans; the only exceptions were three strains isolated from individuals of unknown descent (HK1199, HK1200, and HK1202). ψ, pseudogene.
Comparison of JP2 clone strains isolated from family members as revealed by multilocus sequence analysis.
Of the 66 JP2 clone strains included in this study, 17 originated from seven families, and two to four individuals represented each family (Fig. 4). Two pairs of related individuals, a sister and a brother from Algeria living in Denmark (family 2) and a mother and her daughter from Morocco (family 1), were all colonized by the dominant ST1 strains (Fig. 4 and Table 2). Two individuals from each of two families were colonized by ST8 isolates (families 5 and 6). Each of the remaining three families was colonized by a unique type (ST4, ST9, and ST11) of the JP2 clone (Fig. 4 and Table 2). Strains isolated from three related siblings (family 4) who were living in Denmark and originated from Morocco had a single unique point mutation in the sequenced fragment of frdB (ST4). Isolates from three siblings who originated from the Cape Verde Islands and were living in Sweden (family 3) carried a unique type of the JP2 clone with a point mutation in the atpG gene (ST9), whereas the fourth sibling in this family carried A. actinomycetemcomitans with the prevailing ST, ST1. Finally, two African-American family members (family 7) were colonized by a strain with a unique point mutation in the sequenced recA gene fragment (ST11).
DISCUSSION
Identification of bacterial clones clearly associated with disease often leads to a better understanding of decisive virulence properties, routes of dissemination, and potential specific host adaptations (39, 41, 52). In this study we examined the natural history, microevolution, and patterns of global dissemination and interindividual transmission of the JP2 clone of A. actinomycetemcomitans, which is strongly associated with aggressive periodontitis in adolescents of African descent (9, 24, 25). Our previous studies demonstrated that the JP2 clone differs from other clones of the species by several genetic peculiarities, including a 530-bp deletion in the promoter region of the leukotoxin gene operon, resulting in increased leukotoxic activity (8, 31).
Our study was based on 66 JP2 clone strains collected over a period of more than 20 years from individuals living on five continents plus 16 non-JP2 strains representing the different evolutionary lineages of the A. actinomycetemcomitans population (48). The JP2 clone strains obtained from the continents and countries clearly do not provide a complete picture of the global distribution of the clone. The collection was affected to a significant degree by the availability of isolates and by the strains that we have been able to collect during our long-term interest in this species. In spite of the geographic biases of the strain collection, the study revealed interesting new information about the JP2 clone.
The population genetic analysis performed in this study was based on sequences at eight loci. Multilocus sequence analysis of basically selectively neutral sequences provided a quantitative measure of the similarity between strains and data that were suitable for inferring phylogeny. Although it is conceivable that examination of additional loci would reveal additional polymorphic sites, a sample consisting of seven loci is considered a reasonable sample of the genome for estimating the overall population structure of bacteria (36). As A. actinomycetemcomitans has a clonal population structure, there is no reason to assume that inclusion of additional loci would affect the structure observed.
According to the sequence polymorphism observed, which is quite compatible with the current definition of bacterial clones (37), the 66 JP2 clone strains were divided into two major clusters, which provide important epidemiological information (Fig. 4). A mutation in the hbpA pseudogene divided the JP2 clone into strains that originated from two groups of human hosts that belong to distinct ethnic groups (i.e., Arabs and Africans) and differ by culture, religion, and history (Fig. 4). Thus, the characteristic mutation distinguished isolates obtained from individuals from the northern Mediterranean part of Africa from isolates obtained from individuals from West Africa, including the Cape Verde Islands. Isolates obtained from patients in Brazil and the United States were indistinguishable from isolates obtained from patients in West Africa, supporting the assumption that the JP2 clone was disseminated to the American continents by the transatlantic slave trade.
The presence of a characteristic nucleotide (G in hbpA at position 525285 in the HK1651 genome) in serotype b non-JP2 strains and JP2 clone strains with ST1 to ST4 (Fig. 4) strongly suggests that the JP2 clone originated in the northern Mediterranean part of Africa, where it is still endemic and associated with an unusually high prevalence of aggressive periodontitis according to our studies of Moroccan adolescents (25). A strain with a point mutation that changed G to A in the hbpA pseudogene (position 525285 in the HK1651 genome) characteristic of the other cluster of JP2 clone strains (ST5 to ST11) subsequently spread to the ethnically different population of West Africa and, during the transatlantic slave trade, to North and South America.
Identification of strains without the 530-bp deletion that are identical to the JP2 strain in the six housekeeping genes indicates that this is the allelic composition of the immediate ancestor of the JP2 clone. This provides an opportunity to estimate the approximate age of the JP2 clone. In the 56 independent isolates of the JP2 clone, a total of five synonymous mutations, with a maximum of one mutation per isolate, were found in the combined 3,031 nucleotides of six housekeeping genes (Table 2). It is generally assumed that sequence diversity at synonymous sites accumulates by mutation at a relatively constant rate (molecular clock hypothesis). Assuming that the mutation rate in A. actinomycetemcomitans is 1.4 × 10−10, as demonstrated for Escherichia coli, that 23.7% of the nucleotides in the sequence are at risk for synonymous mutations (14, 34), and that the generation time in biofilms in vivo is 24 h, it can be calculated that the JP2 clone emerged as a distinct genotype some 2,400 years ago. This is fully compatible with the necessary assumption, based on history and our findings described here, that the characteristic leukotoxin promoter deletion and the divergence into at least two separate lineages now colonizing Arabs and Africans took place after that time and prior to the transatlantic slave trade.
We have previously hypothesized that the JP2 clone developed a distinct host tropism for individuals with genetic origins in northern and western parts of Africa (22). Studies of several other pathogens have revealed geographically structured populations with distinct types or lineages causing infection in distinct populations of human hosts, in some cases also discernible after human migration (1, 16, 17, 29, 39, 40, 58). Such patterns may be due to geographic and/or social separation rather than to specific tropisms resulting from coevolution of specific lineages of bacteria with distinct lineages of hosts. Several findings support the conclusion that specific host tropism is the major factor behind the restricted epidemiology of the JP2 clone. First, there is no direct evidence that this clone has been disseminated to Caucasian and Asian populations despite its widespread geographical dissemination for centuries with hosts of African descent (10, 11, 18-20, 28, 35, 38, 48, 53). Second, the two lineages of JP2 clone isolates identified in this study have remained separated according to host ethnicity within a restricted geographic area of Africa. Third, although social separation of races was the rule in most countries for centuries, there was a long tradition of African-American foster mothers and maids in Caucasian families in the United States. If the clone were not restricted by host tropism, it is likely that this habit would have resulted in transfer of the JP2 clone to Caucasian children as the majority of the oral microflora is acquired early in life from the mother or other close contacts.
Only in a few cases has the molecular mechanism of specific tropisms of microorganisms been identified (7, 49). However, it would obviously be interesting to study the molecular mechanisms of the host tropism of the JP2 clone in order to be able to evaluate the possibility that this pathogen, and the high prevalence of aggressive periodontitis, eventually may spread to other human populations as a consequence of changing demographic patterns.
Our identification of identical STs of the JP2 clone strains in several members of each of seven families, and in particular in three families each of which has a unique ST (Fig. 4), provides strong evidence that there is transmission of the JP2 clone of A. actinomycetemcomitans within families and supports the results of previous studies (4, 57).
PFGE is a tool that is frequently used in studies of bacterial transmission. We recently demonstrated that this technique reveals considerable polymorphism, even among members of the JP2 clone, due to intragenomic inversions resulting from homologous recombination between multicopy sequences in the genome (15). We therefore tested the applicability of this technique for studies of the local epidemiology of the JP2 clone. A high number of XhoI fingerprints were obtained for both JP2 and non-JP2 strains of A. actinomycetemcomitans. However, due to the possibility of homologous intragenomic recombination and the potentially reversible nature of the events, interpretation of evolutionary issues based on results obtained by PFGE should be performed with caution. Two of three families (families 3 and 7) harbored JP2 clone strains with identical unique point mutations in the housekeeping genes sequenced, yet with different XhoI fingerprints as determined by the PFGE analysis. This finding suggests that the intragenomic inversions leading to changing patterns take place at a very high rate. As a result, the method is useful only for very short-term epidemiological studies.
The clonal population structure of A. actinomycetemcomitans (20, 27, 30, 48) confirmed by the data obtained in this study makes it possible to trace distinct properties that determine virulence. In line with other bacterial species in the oral flora, it is plausible to consider A. actinomycetemcomitans an opportunistic pathogen as phylogenetically diverse and unique strains were recovered from Caucasian individuals with periodontal or various systemic diseases (20, 30, 43, 48). However, the JP2 clone occupies a special position due to its strong association with disease and its global dissemination with particular ethnic groups. It has been argued by Kaplan and coworkers (30) that the JP2 clone simply may be more prevalent in some populations of Arabs and Africans and that the increased prevalence of aggressive periodontitis in these ethnic groups may be due to increased susceptibility intrinsic to the host rather than to a particular pathogenic potential of the JP2 clone. The mechanisms of the increased susceptibility to aggressive periodontitis in hosts of Arabic and African descent needs to be explored further. However, substantial evidence supports the conclusion that the highly toxic JP2 clone constitutes a particular virulent subpopulation of A. actinomycetemcomitans (9, 23, 24, 31). Combined with the successful global dissemination of members of the JP2 clone, which implies that horizontal transmission is an important mechanism of its spread in addition to the well-established vertical transmission, these findings support the conclusion that the JP2 clone is a clone that has traits similar to those of traditional pathogens.
In conclusion, this study suggests that the JP2 clone of A. actinomycetemcomitans originated in the northern Mediterranean part of Africa and spread to West Africa, from which it was transmitted to other continents initially during the transatlantic slave trade in the 16th to 18th centuries. Distinct lineages of the JP2 clone are still associated with Arabic and African populations and most likely developed tropism for these hosts. With its highly conserved genome, as reflected by the results of multilocus sequence analysis, the JP2 clone may be a valuable marker for tracking the population migration due to sustained signatures at the nucleotide level. Results obtained in this study can provide a better understanding of the global epidemiology of aggressive periodontitis and may help workers anticipate future global trends in the clinical picture of the disease.
Acknowledgments
We thank Lise Hald, Bente Hansen, and Tove Findahl for excellent technical assistance.
This study was supported by grants 22-01-0265 ct/mp and 22-02-0306 ch/mp from the Danish Medical Research Council and by grants from the Danish Dental Association.
Editor: A. Camilli
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z.-J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L.-J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Albandar, J. M., and E. M. B. Tinoco. 2002. Global epidemiology of periodontal diseases in children and young persons. Periodontol. 2000 29:153-176. [DOI] [PubMed] [Google Scholar]
- 3.Alves-Silva, J., M. da Silva Santos, P. E. Guimaraes, A. C. Ferreira, H. J. Bandelt, S. D. Pena, and V. F. Prado. 2000. The ancestry of Brazilian mtDNA lineages. Am. J. Hum. Genet. 67:444-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asikainen, S., and C. Chen. 1999. Oral ecology and person-to-person transmission of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontol. 2000 20:65-81. [DOI] [PubMed] [Google Scholar]
- 5.Balashova, N. V., J. A. Crosby, L. Al Ghofaily, and S. C. Kachlany. 2006. Leukotoxin confers beta-hemolytic activity to Actinobacillus actinomycetemcomitans. Infect. Immun. 74:2015-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedoya, G., P. Montoya, J. Garcia, I. Soto, S. Bourgeois, L. Carvajal, D. Labuda, V. Alvarez, J. Ospina, P. W. Hedrick, and A. Ruiz-Linares. 2006. Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate. Proc. Natl. Acad. Sci. USA 103:7234-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borén, T., P. Falk, K. A. Roth, G. Larson, and S. Normark. 1993. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262:1892-1895. [DOI] [PubMed] [Google Scholar]
- 8.Brogan, J. M., E. T. Lally, K. Poulsen, M. Kilian, and D. R. Demuth. 1994. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect. Immun. 62:501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bueno, L. C., M. P. Mayer, and J. M. DiRienzo. 1998. Relationship between conversion of localized juvenile periodontitis-susceptible children from health to disease and Actinobacillus actinomycetemcomitans leukotoxin promoter structure. J. Periodontol. 69:998-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras, A., T. Rusitanonta, C. Chen, W. G. Wagner, B. S. Michalowicz, and J. Slots. 2000. Frequency of 530-bp deletion in Actinobacillus actinomycetemcomitans leukotoxin promoter region. Oral Microbiol. Immunol. 15:338-340. [DOI] [PubMed] [Google Scholar]
- 11.Cortelli, J. R., S. C. Cortelli, S. Jordan, V. I. Haraszthy, and J. J. Zambon. 2005. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J. Clin. Periodontol. 32:860-866. [DOI] [PubMed] [Google Scholar]
- 12.DiRienzo, J. M., and T. L. McKay. 1994. Identification and characterization of genetic cluster groups of Actinobacillus actinomycetemcomitans isolated from human oral cavity. J. Clin. Microbiol. 32:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiRienzo, J. M., J. Slots, M. Sixou, M. A. Sol, R. Harmon, and T. L. McKay. 1994. Specific genetic variants of Actinobacillus actinomycetemcomitans correlate with disease and health in a regional population of families with localized juvenile periodontitis. Infect. Immun. 62:3058-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elena, S. F., T. S. Whittam, C. L. Winkworth, M. A. Riley, and R. E. Lenski. 2005. Genomic divergence of Escherichia coli strains: evidence for horizontal transfer and variation in mutation rates. Int. Microbiol. 8:271-278. [PubMed] [Google Scholar]
- 15.Eriksen, K., D. Haubek, and K. Poulsen. 2005. Intragenomic recombination in the highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans. Microbiology 151:3371-3379. [DOI] [PubMed] [Google Scholar]
- 16.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Megraud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582-1585. [DOI] [PubMed] [Google Scholar]
- 17.Gagneux, S., K. DeRiemer, T. Van, M. Kato-Meada, B. C. de Jong, S. Narayanan, M. Nicol, S. Niemann, K. Kremer, M. C. Gutierrez, M. Hilty, P. C. Hopewell, and P. M. Small. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103:2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthmiller, J. M., E. T. Lally, and J. Korostoff. 2001. Beyond the specific plaque hypothesis: are highly leukotoxic strains of Actinobacillus actinomycetemcomitans a paradigm for periodontal pathogenesis? Crit. Rev. Oral Biol. Med. 12:116-124. [DOI] [PubMed] [Google Scholar]
- 19.Haraszthy, V. I., G. Hariharan, E. M. B. Tinoco, J. R. Cortelli, E. T. Lally, E. Davis, and J. J. Zambon. 2000. Evidence for the role of highly leukotoxic Actinobacillus actinomycetemcomitans in the pathogenesis of localized juvenile and other forms of early-onset periodontitis. J. Periodontol. 71:912-922. [DOI] [PubMed] [Google Scholar]
- 20.Haubek, D., K. Poulsen, S. Asikainen, and M. Kilian. 1995. Evidence for absence in Northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans. J. Clin. Microbiol. 33:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haubek, D., K. Poulsen, J. Westergaard, G. Dahlen, and M. Kilian. 1996. Highly toxic clone of Actinobacillus actinomycetemcomitans in geographically widespread cases of juvenile periodontitis in adolescents of African origin. J. Clin. Microbiol. 34:1576-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haubek, D., J. M. DiRienzo, E. M. Tinoco, J. Westergaard, N. J. Lopez, C. P. Chung, K. Poulsen, and M. Kilian. 1997. Racial tropism of a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J. Clin. Microbiol. 35:3037-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haubek, D., O.-K. Ennibi, N. Benzarti, and S. Poulsen. 2002. Attachment loss in Moroccan early-onset periodontitis patients in relation to infection with the JP2-type of Actinobacillus actinomycetemcomitans. J. Clin. Periodontol. 29:657-660. [DOI] [PubMed] [Google Scholar]
- 24.Haubek, D., O.-K. Ennibi, K. Poulsen, N. Benzarti, and V. Baelum. 2004. The highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans and progression of periodontal attachment loss. J. Dent. Res. 83:767-770. [DOI] [PubMed] [Google Scholar]
- 25.Haubek, D., O.-K. Ennibi, K. Poulsen, S. Poulsen, N. Benzarti, and M. Kilian. 2001. Early-onset periodontitis in Morocco is associated with the highly leukotoxic clone of Actinobacillus actinomycetemcomitans. J. Dent. Res. 80:1580-1583. [DOI] [PubMed] [Google Scholar]
- 26.Hayashida, H., K. Poulsen, and M. Kilian. 2002. Differences in iron acquisition from human haemoglobin among strains of Actinobacillus actinomycetemcomitans. Microbiology 148:3993-4001. [DOI] [PubMed] [Google Scholar]
- 27.Hayashida, H., K. Poulsen, O. Takagi, and M. Kilian. 2000. Phylogenetic associations of ISAa1 and IS150-like insertion sequences in Actinobacillus actinomycetemcomitans. Microbiology 146:1977-1985. [DOI] [PubMed] [Google Scholar]
- 28.He, T., T. Nishihara, D. R. Demuth, and I. Ishikawa. 1999. A novel insertion sequence increases the expression of leukotoxicity in Actinobacillus actinomycetemcomitans clinical isolates. J. Periodontol. 70:1261-1268. [DOI] [PubMed] [Google Scholar]
- 29.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan, J. B., H. C. Schreiner, D. Furgang, and D. H. Fine. 2002. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J. Clin. Microbiol. 40:1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilian, M., E. V. G. Frandsen, D. Haubek, and K. Poulsen. 2006. The etiology of periodontal disease revisited by population genetic analysis. Periodontol. 2000 41:1-22. [DOI] [PubMed] [Google Scholar]
- 32.Kilian, M., and C. R. Schiott. 1975. Haemophili and related bacteria in the human oral cavity. Arch. Oral Biol. 20:791-796. [DOI] [PubMed] [Google Scholar]
- 33.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefs Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 34.Lenski, R. E., C. L. Winkworth, and M. A. Riley. 2003. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J. Mol. Evol. 56:498-508. [DOI] [PubMed] [Google Scholar]
- 35.Macheleidt, A., H. P. Muller, T. Eger, M. Putzker, A. Fuhrmann, and L. Zoller. 1999. Absence of an especially toxic clone among isolates of Actinobacillus actinomycetemcomitans recovered from army recruits. Clin. Oral Investig. 3:161-167. [DOI] [PubMed] [Google Scholar]
- 36.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maynard Smith, J. 1995. Do bacteria have population genetics, p. 1-12. In S. Baumberg, J. P. W. Young, E. M. H. Wellington, and J. R. Saunders (ed.), Population genetics of bacteria. Cambridge University Press, Cambridge, United Kingdom.
- 38.Mombelli, A., R. Gmür, N. P. Lang, E. Corbet, and J. Frey. 1999. Actinobacillus actinomycetemcomitans in Chinese adults. J. Clin. Periodontol. 26:505-510. [DOI] [PubMed] [Google Scholar]
- 39.Monot, M., N. Honore, T. Garnier, R. Araoz, J. Y. Coppee, C. Lacroix, S. Sow, J. S. Spencer, R. W. Truman, D. L. Williams, R. Gelber, M. Virmond, B. Flageul, S. N. Cho, B. Ji, A. Paniz-Mondolfi, J. Convit, S. Young, P. E. Fine, V. Rasolofo, P. J. Brennan, and S. T. Cole. 2005. On the origin of leprosy. Science 308:1040-1042. [DOI] [PubMed] [Google Scholar]
- 40.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, G. Hammond, E. A. Hoiby, K. E. Jonsdottir, M. Kabeer, I. Kallings, W. N. Khan, M. Kilian, K. Knowles, H. J. Koornhof, B. Law, K. I. Li, J. Montgomery, P. E. Pattison, J.-C. Piffaretti, A. K. Takala, M. L. Thong, R. A. Wall, J. I. Ward, and R. K. Selander. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 41.Nallapareddy, S. R., H. Wenxiang, G. M. Weinstock, and B. E. Murray. 2005. Molecular characterization of a widespread, pathogenic and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J. Bacteriol. 187:5709-5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norskov-Lauritsen, N., and M. Kilian. 2006. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int. J. Syst. Evol. Microbiol. 56:2135-2146. [DOI] [PubMed] [Google Scholar]
- 43.Paju, S., P. Carlson, H. Jousimies-Somer, and S. Asikainen. 2000. Heterogeneity of Actinobacillus actinomycetemcomitans strains in various human infections and relationships between serotype, genotype, and antimicrobial susceptibility. J. Clin. Microbiol. 38:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paturel, L., J. P. Casalta, G. Habib, and D. Raoult. 2004. Actinobacillus actinomycetemcomitans endocarditis. Clin. Microbiol. Infect. 10:98-118. [DOI] [PubMed] [Google Scholar]
- 46.Poulsen, K., O.-K. Ennibi, and D. Haubek. 2003. Improved PCR for detection of the highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans in subgingival plaque samples. J. Clin. Microbiol. 41:4829-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poulsen, K., J. P. Hjorth, and M. Kilian. 1988. Limited diversity of the immunoglobulin A1 protease gene (iga) among Haemophilus influenzae serotype b strains. Infect. Immun. 56:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poulsen, K., E. Theilade, E. T. Lally, D. R. Demuth, and M. Kilian. 1994. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiology 140:2049-2060. [DOI] [PubMed] [Google Scholar]
- 49.Rios, M., and C. Bianco. 2000. The role of blood group antigens in infectious diseases. Semin. Hematol. 37:177-185. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 51.Slots, J., H. S. Reynolds, and R. J. Genco. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect. Immun. 29:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spratt, B. G. 2004. Exploring the concept of clonality in bacteria. Methods Mol. Biol. 266:323-352. [DOI] [PubMed] [Google Scholar]
- 53.Tan, K. S., C. H. Woo, G. Ong, and K. P. Song. 2001. Prevalence of Actinobacillus actinomycetemcomitans in an ethnic adult Chinese population. Clin. Periodontol. 28:886-890. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai, C.-C., W. P. Mc Arthur, P. C. Baehni, B. F. Hammond, and N. S. Taichman. 1979. Extraction and partial characterization of a leukotoxin from a plaque-derived gram-negative microorganism. Infect. Immun. 25:427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai, C.-C., B. J. Shenker, J. M. DiRienzo, D. Malamud, and N. S. Taichman. 1984. Extraction and isolation of a leukotoxin from Actinobacillus actinomycetemcomitans with polymyxin B. Infect. Immun. 43:700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Winkelhoff, A. J., and K. Boutaga. 2005. Transmission of periodontal bacteria and models of infection. J. Clin. Periodontol. 32:16-27. [DOI] [PubMed] [Google Scholar]
- 58.Wirth, T., X. Wang, B. Linz, R. Novick, J. K. Lum, M. Blaser, G. Morelli, D. Falush, and M. Achtman. 2004. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc. Natl. Acad. Sci. USA 101:4746-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]