Abstract
Two-partner secretion (TPS) systems are a family of proteins being rapidly identified and characterized in a growing number of gram-negative bacteria. TPS systems mediate the secretion of proteins, many involved in virulence traits such as hemolysis, adherence to epithelial cells, inhibition of bacterial growth, and immunomodulation of the host. A TPS system typically consists of a transporter located in the bacterial outer membrane (OM) which is responsible for the recognition and secretion of at least one large exoprotein. Two of the better-characterized TPS systems specify the Bordetella pertussis FHA and Haemophilus influenzae HMW1/HMW2 proteins. We identified three gene products of Moraxella catarrhalis strain O35E that resemble TPS proteins and designated them MhaC (transporter), MhaB1 (exoprotein), and MhaB2 (exoprotein). Western blot analysis using anti-MhaC, or antibodies reacting to both MhaB1 and MhaB2 (MhaB-reactive), revealed that these antigens are expressed in the OM of 63% of isolates tested. Mutations in the mhaC gene specifying the putative transporter of the M. catarrhalis wild-type strains O35E, O12E, and McGHS1 resulted in the absence of MhaB1/MhaB2 in the OM of mutants. These results are therefore consistent with the Mha proteins functioning as a TPS system. Furthermore, we discovered that these mhaC mutants exhibit markedly decreased binding to human epithelial cells relevant to pathogenesis by M. catarrhalis (Chang, HEp2, A549, and/or 16HBE14o−). Expression of O12E MhaC and MhaB1 in a nonadherent strain of Escherichia coli was found to increase the adherence of recombinant bacteria to HEp2 monolayers by sevenfold, thereby demonstrating that this M. catarrhalis TPS system directly mediates binding to human epithelial cells. The construction of isogenic mutants in the mhaB1 and mhaB2 genes of strain O35E also suggests that the MhaB proteins play distinct roles in M. catarrhalis adherence.
Moraxella catarrhalis is a gram-negative, unencapsulated diplococcus that is a major pathogen of the middle ear in children (10, 16, 28, 39) and of the respiratory tract in adults, particularly those with chronic obstructive pulmonary disease (34). There is currently no vaccine for M. catarrhalis, and pathogenesis by this bacterium is not well understood. One key event leading to infection by most human bacterial pathogens is adherence to mucosal surfaces (6, 44). M. catarrhalis expresses multiple adhesins, allowing the organism to bind to human epithelial cell lines of various anatomical origins that are relevant to its pathogenesis. UspA1, which mediates attachment to conjunctival cells (29), is expressed by most isolates tested to date (9, 32) and binds to the extracellular matrix protein laminin (48). The closely related UspA2H protein also facilitates binding to conjunctival cells, but is expressed by a smaller proportion of M. catarrhalis isolates (29). The autotransporter Hag (also designated MID) is one of the better-characterized adherence factors expressed by the bacterium. Virtually all isolates tested to date contain a hag gene (33), and its expression is important for adherence to A549 human type II pneumocytes (7, 13, 22), primary cultures of human middle ear epithelial cells (7), and human immunoglobulin D (14, 38).
In addition to these major adherence factors, several outer membrane (OM) proteins have been shown to contribute to the binding of M. catarrhalis to epithelial cells. McaP (49), which also exhibits phospholipase B activity, mediates attachment to several human cell lines, including A549, NCIH292, and HEp2, while another previously characterized OM protein, OMPCD (23, 35, 36), facilitates binding to A549 cells (21). Preincubation of M. catarrhalis cells with a monoclonal antibody against lipooligosaccharides was reported to significantly reduce adherence to Chang human conjunctival cells, implying a role for lipooligosaccharides in adherence (24).
This array of adherence factors likely allows the organism to efficiently colonize several different areas of the human respiratory tract and thus represent attractive vaccine candidates. Antibodies to these surface molecules would presumably opsonize M. catarrhalis and block its binding to human mucosal surfaces, thereby interfering with colonization and subsequent development of infection. Adhesins have proven to be effective vaccine antigens. For instance, all vaccines currently licensed for use in the United States against Bordetella pertussis, the causative agent of whooping cough, target the filamentous hemagglutinin adhesin (FHA) (3). In this study, we report the identification and characterization of two filamentous hemagglutinin-like adhesins expressed in the OM of M. catarrhalis strain O35E.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture of cell lines, and growth conditions.
Strains and plasmids are described in Table 1. M. catarrhalis was cultured in Todd-Hewitt (TH) broth (BD Diagnostic Systems) or on TH agar plates at 37°C in the presence of 7.5% CO2. Where appropriate, M. catarrhalis mutants were grown in medium supplemented with kanamycin (20 μg/ml), zeocin (5 μg/ml), streptomycin (75 μg/ml), or spectinomycin (15 μg/ml). For the preparation of electrocompetent cells as well as OM vesicles, M. catarrhalis was grown in broth with shaking. For all other experiments, M. catarrhalis was cultured on agar plates. Escherichia coli was grown in Luria-Bertani (LB) broth (Fisher Bioreagents) or on solidified LB medium. Recombinant E. coli bacteria were cultured in medium containing 15 μg/ml chloramphenicol as well as additional antibiotics (kanamycin, 50 μg/ml; zeocin, 50 μg/ml; spectinomycin, 100 μg/ml) where indicated. For adherence assays, the preparation of plasmid DNA and the extraction of Sarkosyl-insoluble OM proteins, recombinant E. coli bacteria were grown in broth supplemented with CopyControl Induction Solution (Epicenter) as reported by Holm and colleagues (21). Human epithelial cell lines including Chang (human conjuctival epithelium; ATCC CCL20.2), HEp2 (human laryngeal epithelium; ATCC CCL-23), 16HBE14o− (polarized bronchial epithelium), NCIH292 (human lung mucoepidermoid; ATCC CRL-1848), and A549 (human type II alveolar lung epithelium; ATCC CCL85) were cultured as reported by Timpe and colleagues (49).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| M. catarrhalis | ||
| O35E | WT isolate | 2 |
| O35E.SM100 | Streptomycin-resistant derivative of O35E | This study |
| O35E.B1 | Isogenic mhaB1 mutant, Spcr | This study |
| O35E.B2 | Isogenic mhaB2 mutant, Zeor | This study |
| O35E.B1B2 | Double isogenic mutant, Spcr Zeor | This study |
| O35E.C | Isogenic mhaC mutant, Kanr | This study |
| O35E.CR1 | Repaired isogenic mhaC mutant, Strr Kans | This study |
| O12E | WT isolate | 29 |
| O12E.C | Isogenic mhaC mutant, Kanr | This study |
| O12E.CR1 | Repaired isogenic mhaC mutant, Strr Kans | This study |
| McGHS1 | WT isolate | 7 |
| McGHS1.C | Isogenic mhaC mutant, Kanr | This study |
| McGHS1.CR1 | Repaired isogenic mhaC mutant, Strr Kans | This study |
| O46E | WT isolate | 29 |
| P44 | WT isolate | 29 |
| TTA24 | WT isolate | 9 |
| TTA37 | WT isolate | 29 |
| V1171 | WT isolate | 9 |
| 11P29B1 | WT isolate (Buffalo, NY) | Tim Murphy |
| 32P11B1 | WT isolate (Buffalo, NY) | Tim Murphy |
| 13P18B1 | WT isolate (Buffalo, NY) | Tim Murphy |
| 7P94B1 | WT isolate (Buffalo, NY) | Tim Murphy |
| McGH | WT isolate (Toledo, OH) | George Hageage |
| Mc34F | WT isolate (Toledo, OH) | George Hageage |
| McGHS1 | WT isolate | 7 |
| McGHS2 | WT isolate (Toledo, OH) | George Hageage |
| 7169 | WT isolate | A. Campagnari |
| E. coli | ||
| EPI300 | Cloning strain | Epicenter |
| TUNER | Expression strain | Novagen |
| Plasmids | ||
| pETcoco-1 | Protein expression vector, Cmr | Novagen |
| pRBHis.MhaB.72.399 | pETcoco-1 expressing O12E MhaB1 aa 72 to 399 joined to 6 N-terminal histidine residues, Cmr | This study |
| pRB.His.MhaC.61.690 | pETcoco-1 expressing O12E MhaC aa 61 to 690 joined to 6 N-terminal histidine residues, Cmr | This study |
| pCC1 | Cloning vector, Cmr | Epicenter |
| pCC1.3 | pCC1 in which the control insert provided by the manufacturer was cloned; adherence-negative control | 21 |
| pELO12EC | pCC1 containing a 2.2-kb fragment corresponding to O12E mhaC, Cmr | This study |
| pELO12EC-K | Modified from pELO12EC by adding a kanamycin cassette near the middle of the O12E mhaC ORF, Cmr Kanr | This study |
| pRBO12EB1 | pCC1 containing a 6-kb insert containing the entire O12E mhaB1, Cmr | This study |
| pRBO12EB1NT | pCC1 containing part of O12E mhaC, the intergenic region and the first 3 codons of mhaB1; used to construct pRBO12EB1KO, Cmr | This study |
| pRBO12EB1KO | pCC1 containing the C-terminal fragment of mhaB1 from pRBO12EB1 ligated into pRBO12EB1NT, Cmr | This study |
| pRBO12EB1KO-S | Modified from pRBO12EB1KO by insertion of a spectinomycin cassette; used for making mhaB1 isogenic mutants, Cmr Spcr | This study |
| pRBO12EB2NT | pCC1 containing DNA upstream of O12E mhaB2 and the first 3 codons of mhaB2; used to construct pRBO12EB2KO, Cmr | This study |
| pRBO12EB2CT | pCC1 containing 3′ end of O12E mhaB2, Cmr | This study |
| pRBO12EB2KO | pCC1 containing the mhaB2 C-terminal fragment from pRBO12EB2CT ligated into pRBO12EB2NT, Cmr | This study |
| pRBO12EB2KO-Z | Modified from pRBO12EB2KO by inserting a zeocin cassette; used for construction of mhaB2 isogenic mutants, Cmr Zeor | This study |
| pSVCdwn | pCC1 containing a 2.1-kb insert corresponding to intergenic DNA between O12E mhaC and mhaB2, Cmr | This study |
| pSVCdwn-K | pSVCdwn containing a Kanr gene near the middle of the insert; used to construct O12E.dwn, Cmr Kanr | This study |
| pRB.O12E.B1C | pCC1 containing an ∼30-kb DNA fragment from strain O12E.dwn, expresses MhaB1 and MhaC, adherence positive, Cmr Kanr | This study |
| pUC4K | Source of the Kanr cassette | GE HealthCare Life Sciences |
| pSPECR | Source of the Spcr cassette | 56 |
| pEM7/ZEO | Source of the Zeor cassette | Invitrogen |
PCR and recombinant DNA techniques.
Standard molecular biology procedures were performed as described elsewhere (42). M. catarrhalis genomic DNA was isolated using the Invitrogen Easy DNA kit under the conditions recommended by the manufacturer. Plasmid DNA was obtained with the QIAprep Spin Miniprep Kit (QIAGEN) according to the manufacturer's specifications. Unless otherwise stated, all PCR experiments were performed using Platinum Pfx DNA polymerase (Invitrogen). PCR products of 2.1-kb containing mhaC open reading frames (ORFs) were amplified from strains O35E, O12E, McGHS1, Mc34F, O46E, TTA37, and V1171 with the oligonucleotide primers P1 (5′-AGC GAT TCA TGG TGG ACT GTC CTT-3′) and P2 (5′-ACT TTG GCA GAA AGA AAG GCG GTG-3′). The Failsafe PCR System (Epicenter) was used to amplify DNA fragments of 6 kb specifying mhaB1 from strains O35E, O12E, and McGHS1 with the oligonucleotides P3 (5′-AAC GCA TGG GTT TGT CCT GTC CTT-3′) and P4 (5′-TTG GGC GTT GAT AGG AAT GCG TTG-3′). The Failsafe PCR system was also used to generate amplicons of 7 kb specifying the mhaB2 genes with primers P5 (5′-TTA GGC TCG CAT TGC ACA-3′) and P6 (5′-ACG GTA TCG GTG TGG GTT AGC TTT-3′) for strain O35E and P5 and P8 (5′-TCT TTG GTG ATG TGT TCG GCA GGA-3′) for strain O12E. These PCR products were used as templates in sequencing reactions. In addition, the PCR fragments specifying O12E mhaC and O12E mhaB1 were cloned using the CopyControl PCR cloning kit (Epicenter), yielding the plasmids pELO12EC and pRBO12EB1, respectively. The 0.9-kb PCR product corresponding to the rpsL gene of M. catarrhalis strain O35E.SM100 was amplified with primers PsmF (5′-CGC GGA TCC GCG ACT CAA GTG AAA ATA CGC A-3′) and PsmR (5′-CCG GAA TTC CGG ACA CGA CGT CTT GGC ATA A-3′).
Construction of isogenic mutations in the mhaC, mhaB1, and mhaB2 genes.
The Kanr cassette from pUC4K (GE HealthCare Life Sciences) was introduced into a unique NsiI site near the middle of the O12E mhaC ORF of pELO12EC, yielding the plasmid pELO12EC-K. This construct was used to electroporate strains O12E, McGHS1, and O35E using a method previously described by Holm et al. (22). These electroporated cells were next plated on medium containing kanamycin, and colonies were screened by PCR with primers P1 and P2 (see above) to identify the mhaC isogenic mutants O12E.C, McGHS1.C, and O35E.C (data not shown). Lack of expression of MhaC in these mutants was verified by Western blotting (see Fig. 2 and Fig. 5).
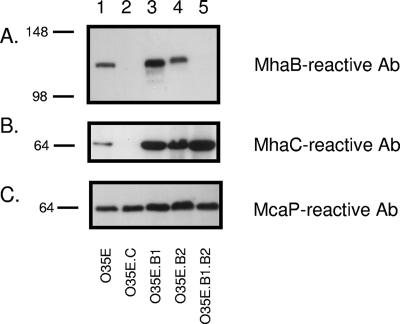
FIG. 2.
Western blot analysis of selected OM proteins expressed by M. catarrhalis strain O35E and isogenic mha mutants. OM vesicles were obtained from the WT isolate O35E (lane 1) and from its mhaC (lane 2), mhaB1 (lane 3), mhaB2 (lane 4), and mhaB1 mhaB2 (lane 5) mutant strains and analyzed by immunoblotting with the indicated antibodies (Ab). Molecular mass markers are shown to the left in kilodaltons.
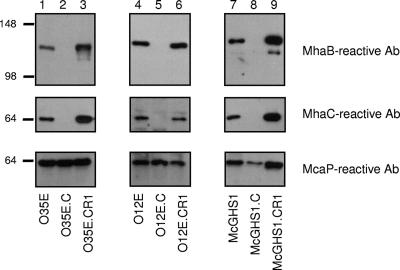
FIG. 5.
Western blot analysis of selected OM proteins expressed by M. catarrhalis O35E, O12E, and McGHS1 and strains derived from these isolates. OM preparations from the WT, mhaC mutants, and repaired mhaC strains were electrophoresed, transferred to PVDF membranes, and analyzed by immunoblotting with the indicated antibodies (Ab). Molecular mass markers are shown to the left in kilodaltons.
To construct a mutation in mhaB1, we first amplified an ∼1.4-kb PCR fragment from strain O12E with primers P9 (5′-TGA CTG CGG TCC ATC CTA ATG AGT-3′) and P10 (5′-GCC GTC AGT TAA TTA AGC GAT TCA TGG TGG ACT GTC CTT G-3′; PacI site underlined). This amplicon corresponded to 1.3 kb at the 5′ end of mhaC, the intergenic region between mhaC and mhaB1, and the first 3 codons of mhaB1 at the end of which a PacI site was engineered. This PCR product was cloned into pCC1 using the CopyControl PCR cloning kit (Epicenter) to yield the plasmid pRBO12EB1NT. The plasmid pRBO12EB1, which contains the entire O12E mhaB1 (see above), was restricted with PacI and EcoRI to generate a 1.5-kb fragment that corresponds to the 3′ end of mhaB1. This DNA fragment was excised from an agarose gel, purified with the High-Pure PCR product purification kit (Roche) according to the manufacturer's guidelines, and ligated into the plasmid pRBO12EB1NT which had been previously digested with PacI and EcoRI. The resulting plasmid, pRBO12EB1KO, was linearized with PacI and ligated with the spectinomycin resistance cassette from pSPECR (56) to yield pRBO12EB1KO-S. This construct, corresponding to the mhaB1 gene in which 4.4 kb of the 5.2-kb ORF had been replaced with the 1.2-kb spectinomycin-resistant cassette, was introduced into strain O35E by electroporation, and spectinomycin-resistant transformants were screened by PCR with primers P3 (see above) and P11 (5′-TTG GGC GTT GAT AGG AAT GCC TTG-3′). This primer pair yielded a PCR product of ∼6-kb in the WT strain O35E and a smaller amplicon of 2.8-kb in the isogenic mutant O35E.B1, thereby confirming proper allelic exchange (data not shown).
To engineer a mutation in the mhaB2 gene, a 1-kb amplicon corresponding to the DNA located upstream of O12E mhaB2 and including the first 3 codons of the ORF was first amplified using oligonucleotides P12 (5′-ACA GGC AAC CCA AGG ATA CCA TCT-3′) and P10 (the end of which contains the first 3 codons of mhaB2 preceded by a PacI site, see above) and cloned into the vector pCC1; the resulting plasmid was designated pRBO12EB2NT. Concurrently, a PCR product of 1 kb specifying the 3′ end of O12E mhaB2 was amplified with P13 (5′-TGA ATC GCT TAA TTA ATG TCG CCA GTG AGA TCT TTA CCG A-3′; PacI site underlined) and P14 (5′-TTA TGC ACC CAC ACC CCA TA-3′) and cloned in pCC1 to yield pRBO12EB2CT. The latter was restricted with PacI and EcoRI, and a digestion product of 1 kb was purified from agarose slices and ligated with the plasmid pRBO12EB2NT, digested with PacI and EcoRI, to generate the construct pRBO12EB2KO. This plasmid was finally linearized with PacI to introduce the 0.4-kb zeocin-resistant cassette from pEM7/ZEO (Invitrogen), and the resulting construct was named pRBO12EB2KO-Z. This construct, specifying the O12E mhaB2 gene in which 4 kb of the 5,385-nucleotide (nt) ORF has been replaced with the 0.4-kb zeocin cassette, was used to electroporate strain O35E, yielding the zeocin-resistant strain O35E.B2. Allelic exchange was confirmed by PCR (data not shown).
The double-mutant strain O35E.B1B2 was obtained by electroporating the mhaB1 mutant, O35E.B1, with pRBO12EB2KO-Z and selecting for zeocin-resistant, spectinomycin-resistant colonies. Allelic exchange was confirmed by PCR (data not shown). Lack of expression of MhaB1 and MhaB2 was also verified by Western blotting (see Fig. 2A). It should be noted that all cloned inserts described in this section were sequenced to ensure fidelity.
Repair of isogenic mutations in the mhaC gene of strains O35E.C, O12E.C, and McGHS1.C by the use of congression.
A streptomycin-resistant mutant of strain O35E, designated O35E.SM100, was obtained as previously reported by Attia and colleagues (4). The rpsL gene of O35E.SM100, which specifies resistance to streptomycin, was amplified with primers PsmF and PsmR, purified, and mixed with PCR products corresponding to the wild-type (WT) mhaC genes of strain O35E, O12E, or McGHS1. These amplicons were then mixed with a few colonies of the appropriate recipient strain (i.e., O35E.C, O12E.C, or McHGS1.C), placed in the middle of a TH agar plate, and incubated at 37°C for 3 h (natural transformation). Bacteria were aseptically suspended in broth and spread onto agar plates supplemented with streptomycin. Resistant colonies were next tested for the loss of Kanr, which is the marker that was used to disrupt the mhaC gene in the recipient strains O35E.C, O12E.C, and McGHS1.C (see above). This overall strategy yielded strains O35E.CR1, O12E.CR1, and McGHS1.CR1 in which the WT mhaC gene has been reintroduced in the chromosome. PCR was used to verify allelic exchange (data not shown), and the repaired strains were shown to express MhaC in their OM (see Fig. 5).
Construction of a plasmid expressing O12E MhaB1 and MhaC.
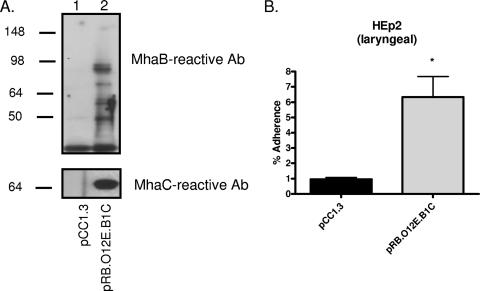
A 2.1-kb amplicon corresponding to the intergenic region between O12E mhaC and mhaB2 was generated with primers P17 (5′-CAC CGC CTT TCT GCC AAA GT-3′) and P18 (5′-TTG GCA AAT GCA GAA CCG CTA CAG-3′) and cloned into pCC1 to yield the plasmid pSVCdwn. The plasmid was then mutagenized with the EZ::TN <KAN2> system (Epicenter) to obtain a construct containing a kanamycin-selectable marker near the middle of this region. The resulting plasmid, pSVCdwn-K, was electroporated in strain O12E, and kanamycin-resistant colonies were screened by PCR with primers P17 and P18 to identify the isogenic mutant strain O12E.dwn. Chromosomal DNA was isolated from this strain and used to create a plasmid-based library of large DNA fragments using the CopyControl Fosmid Library production kit (Epicenter). This library was introduced in the nonadherent E. coli cloning strain EPI300, and recombinant clones were selected for resistance to both chloramphenicol (vector marker) and kanamycin (chromosomal DNA from O12E.dwn downstream of the mhaC gene). This strategy identified the plasmid pRB.O12E.B1C, which specifies expression of O12E MhaC and MhaB1 (see Fig. 7A) and contains a kanamycin resistance marker in the intergenic region between mhaC and mhaB2. This construct was also used as a sequencing template.
FIG. 7.
Western blot analysis and adherence assays of recombinant E. coli bacteria expressing O12E MhaC and MhaB1. (A) Sarkosyl-insoluble OM proteins were extracted from E. coli strains carrying the plasmids pCC1.3 (lane 1) and pRB.O12E.B1C (lane 2) and were then analyzed by immunoblotting with murine sera against MhaB and MhaC; molecular mass markers are shown in kilodaltons. (B) Duplicate adherence assays were performed on at least four separate occasions. The adherence is expressed as the percentage (±standard error) of inoculum bound to epithelial cells following a 3-h incubation. The Mann-Whitney test was used to determine whether the increase in adherence was statistically significant (*, P < 0.05). Ab, antibody.
Adherence assays.
Quantitative adherence assays were performed with modifications to the procedure previously described by Holm et al. (22). Briefly, 105 human cells were seeded into each well of a 24-well tissue culture plate and incubated overnight before use. The day of the assay, bacteria were aseptically suspended to an optical density of 230 Klett units in 5 ml of phosphate-buffered saline supplemented with 0.15% (wt/vol) gelatin. Portions of these bacterial suspensions (25 μl; 107 CFU) were used to inoculate duplicate wells of the 24-well tissue culture plate containing monolayers of human epithelial cells. The tissue culture plate was centrifuged for 5 min at 165 × g to facilitate contact between bacteria and human cells and incubated at 37°C for 5 min (assays with M. catarrhalis), or 3 h (assays with recombinant E. coli). Following this incubation, nonadherent bacteria were removed by washing each well four times with 0.5 ml of phosphate-buffered saline supplemented with 0.15% (wt/vol) gelatin, and the human cells were lysed using 500 μl/well of a solution containing the detergent Saponin (Acros) in addition to EDTA (Sigma). The well contents were serially diluted, spread onto agar plates, and incubated overnight at 37°C. The following day, CFU were counted to determine the number of viable bacteria attached to human cells. The level of adherence is expressed as the percentage (±standard error) of bacteria attached to epithelial cells relative to the inoculum used to infect monolayers. Statistical analyses were performed with the GraphPad Prism software using the Mann-Whitney test; P values of <0.05 were reported as statistically significant. Adherence assays were repeated on at least four separate occasions for each strain.
Production of polyclonal antibodies.
To obtain polypeptides for antibody production, a 0.7-kb PCR product encompassing amino acids (aa) 72 to 399 of O12E MhaB1 was amplified with primers P19 (5′-CCC AAG CTT GTT ATT TCT GAC AGT CAA GCA-3′; HindIII site underlined) and P20 (5′-CCT TAA TTA ACC AAT ACC TTG CAA GTT GGC AGT-3′; PacI site underlined), restricted, and cloned into the HindIII and PacI sites of the pETcoco-1 vector (Novagen). This yielded the plasmid pRBHis.MhaB.72.399 which encodes the in-frame, N-terminally histidine-tagged protein designated His-MhaB. A similar strategy was used to construct the plasmid pRB.His.MhaC.61.690 using the oligonucleotides P21 (5′-CCC AAG CTT CTT TCT GCA ATA ACG GAT GAT-3′; HindIII site underlined) and P22 (5′-CCT TAA TTA ACC AAA TTC AGG CGT CTT AAT TCC-3′; PacI site underlined). This construct specifies aa 61 to 690 of O12E MhaC joined to six N-terminal histidines and is referred to as His-MhaC. Both plasmid inserts were sequenced to verify fidelity.
The aforementioned recombinant proteins were individually overexpressed in the E. coli strain TUNER, extracted from inclusion bodies with the BugBuster HT protein extraction reagent and rLysozyme, and purified under denaturing conditions using the His-Bind resin system (Novagen). The protein refolding kit from Athena Enzyme Systems Group was used to determine the buffer composition suitable for refolding purified His-MhaC and His-MhaB proteins, and urea was removed from these preparations by dialyzing overnight at 4°C while gradually decreasing its concentration (4 M→2 M→1 M→no urea) over a period of 4 days. BALB/c mice were immunized with His-MhaB or His-MhaC as described elsewhere (30). Of note, the serum raised against His-MhaB reacts both with MhaB1 and MhaB2 because of the sequence identity in the N-terminal region of these proteins. Antibodies were demonstrated to specifically recognize MhaC or MhaB1/MhaB2 using whole-cell or OM preparations of the WT strain O35E, the mhaC mutant O35E.C, and the mhaB1 mhaB2 mutant O35E.B1B2 (data not shown). The MhaB antibodies cross-reacted with only one E. coli protein of ∼20 kDa (see Fig. 7, lane 1). The MhaC antibodies cross-reacted with a single E. coli protein of ∼20 kDa (data not shown). When testing M. catarrhalis preparations, the MhaB antibodies showed a low level of cross-reactivity with one protein of ∼60 kDa (data not shown), and the MhaC antibodies reacted only with the 65-kDa MhaC protein (Fig. 2B and 5B). The Western blotting results in Fig. 2, 4, and 5 only show the gel areas pertaining to the antigens under study (i.e., MhaC, MhaB1, and MhaB2).
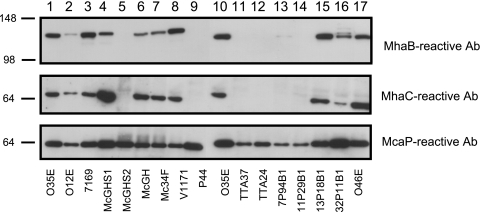
FIG. 4.
Western blot analysis of selected OM proteins expressed by M. catarrhalis isolates of various origins. OM vesicles were resolved on 7.5% polyacrylamide gels, transferred to PVDF membranes, and probed with murine sera against MhaC and MhaB. McaP antibodies (Ab) were also included in these experiments as loading indicators. Molecular mass markers are shown to the left in kilodaltons. This figure is a composite of separate Western blots, and therefore, O35E (lanes 1 and 10) was included as a reference strain.
OM preparations and immunoblotting.
OM proteins were purified from M. catarrhalis strains by the EDTA procedure described by Murphy and Loeb (37). Sarkosyl-insoluble OM proteins were extracted from recombinant E. coli bacteria following a protocol previously outlined by Carlone and colleagues (8). These preparations were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 7.5% polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes, and immunoblots were performed as reported elsewhere (7) with polyclonal sera against His-MhaC and His-MhaB at dilutions of 1:100,000. The previously described antibodies against McaP (7) were used at a dilution of 1:10,000. Protein bands were visualized by chemiluminescence.
Nucleotide sequence analysis.
PCR products and plasmids were sequenced at the University of Michigan Sequencing Core. Chromatograms were assembled using the Chromatool software (Biotools). Nucleotide sequences were deposited in GenBank (see “Nucleotide sequence accession numbers” below). Sequence analysis was performed using Vector NTI 10.1 (Invitrogen).
Nucleotide sequence accession numbers.
The nucleotide sequences of the O12E mhaC (EF362391), O35E mhaC (EF362390), McGHS1 mhaC (EF362392), Mc34F mhaC (EF362393), O46E mhaC (EF362394), TTA37 mhaC (EF362395), V1171 mhaC (EF362396), O12E mhaB1 (EF362386), O35E mhaB1 (EF362385), McGHS1 mhaB1 (EF362387), O12E mhaB2 (EF362389), and O35E mhaB2 (EF362388) ORFs were deposited in GenBank under the accession numbers indicated.
RESULTS
Identification of ORFs in M. catarrhalis strain O35E that resemble the Bordetella pertussis fhaC and fhaB gene products.
Analysis of the patented genomic sequence of M. catarrhalis strain ATCC 43617 using tblastn (NCBI) identified an ORF (nt 58937 to 61048 of GenBank AX067458) with similarity (expect value, 7e−18) to the fhaC gene product of B. pertussis Tomaha1 (NP880575, GenBank). FhaC is a 64.4-kDa protein possessing 19 β-strands which form a porin-like structure in the OM of B. pertussis that mediates the secretion and proper surface display of the FHA (17, 18, 25, 57). Primers were designed to amplify the fhaC-like gene of M. catarrhalis strain O35E, and sequence analysis predicted a 78.5-kDa protein of 705 aa that is 99% identical to the gene product of strain ATCC 43617. Using SignalP 3.0, a putative signal sequence cleavage site was detected between residues 50 and 51 (KA▾QI), indicating that the M. catarrhalis gene product is likely secreted. Further analysis with PSIPRED revealed that the ORF potentially contains 18 to 22 β-strands, suggesting a porin conformation. Database searches using Pfam also demonstrated that the N terminus of the O35E gene product possesses a POTRA2 domain (PF08479), which is present in a number of porin-like proteins responsible for the transport of polypeptides across the OM of gram-negative bacteria. Together, these observations suggest structural and functional similarities between the B. pertussis fhaC gene product and the M. catarrhalis O35E ORF. Thus, the ORF was named mhaC for Moraxella catarrhalis FhaC-like protein.
Sequence analysis upstream of O35E mhaC revealed the presence of a diverging ORF beginning 50 nt from the mhaC ATG start codon (Fig. 1). This ORF was found to be 5,181 nt in length and predicted to specify a protein of 184 kDa that resembles FhaB (expect value, 2e−28), the precursor of the adhesin FHA from B. pertussis BP536 (GenBank accession no. AAA22974) (40). A signal sequence cleavage site was detected between aa 69 and 70 (FA▾NV), indicating that this M. catarrhalis gene product is likely secreted, and database searches revealed the presence of a carbohydrate-dependent hemagglutination activity domain (pfam05860.4) between aa 74 to 197. Hemagglutination activity domains are found in a number of proteins including the adhesins FhaB (B. pertussis) (31) and HMW1/HMW2 (Haemophilus influenzae) (11, 46), the Serratia marcescens hemolysin ShlA (43), and the large secreted proteins LspA1 and LspA2 of Haemophilus ducreyi (53-55). These proteins belong to the TpsA family of molecules, which are exoproteins secreted in a two-partner secretion (TPS) manner (27). Upon closer examination, we noted that residues 143 to 146 (NPFL) and 183 to 187 (NPSGI) of the O35E ORF correspond to two amino acid motifs present in the hemagglutination domain of most TpsA molecules and have been shown to be important for the proper secretion as well as activation of these exoproteins. Analysis of the O35E ORF also revealed another domain between aa 1221 to 1385 that is frequently associated with TpsA molecules (pfam04830.6). The similarities to B. pertussis FhaB and other TPS exoproteins prompted us to designate this ORF MhaB1 (Moraxella catarrhalis FhaB-like protein 1).
FIG. 1.
Schematic representation of the M. catarrhalis mhaB1 mhaC mhaB2 locus. The open bars represent the regions of the mhaB1 and mhaB2 gene products that are virtually identical, whereas the patterned arrowheads illustrate the regions of divergence.
DNA sequence analysis of the flanking regions revealed a third ORF 2.1 kb downstream of, and in the same orientation as, mhaC (Fig. 1). This 5,664-nt ORF was predicted to encode a protein of 201 kDa that is 69.5% identical to O35E MhaB1. The first 1,200 aa of MhaB1 and this downstream ORF are perfectly conserved, whereas the remainder of the proteins share only 16% identity. Because of the striking similarity, this gene was designated mhaB2. Based on these sequence analyses revealing similarities to B. pertussis FHA and FhaC as well as to other related TPS systems, we hypothesized that MhaB1, MhaB2, and MhaC function as a TPS system that contributes to the binding of M. catarrhalis strain O35E to human epithelial cells.
MhaB1, MhaB2, and MhaC are expressed in the OM of the M. catarrhalis strain O35E and are involved in attachment to epithelial cells in vitro.
To test the hypothesis that these gene products are involved in adherence, we disrupted the ORFs specifying these proteins and measured the binding of the resulting isogenic mutant strains to epithelial cell lines that are relevant to pathogenesis by M. catarrhalis. To verify that the mutants lacked expression of the intended gene products, OM preparations were obtained from WT and isogenic mha mutants and tested by Western blotting with antibodies against MhaC or antibodies binding to both MhaB1 and MhaB2 (MhaB reactive). The latter antibodies were obtained by immunizing mice with the purified recombinant protein His-MhaB which corresponds to aa 72 to 399 of MhaB1 fused to six N-terminal histidine residues (see Material and Methods). This portion of MhaB1 is identical to residues 72 to 399 of MhaB2 (data not shown), and therefore, antibodies raised against this polypeptide were expected to react with both MhaB1 and MhaB2. Blots were also probed with antibodies to the M. catarrhalis adhesin McaP as an indicator of protein loads in these experiments.
As shown in lane 2 of Fig. 2B, disrupting mhaC (strain O35E.C) resulted in the absence of a band reacting with MhaC-specific antibodies. In the WT strain, this band migrated with an apparent mass of ∼65 kDa, which is smaller than predicted by sequence analysis of the O35E mhaC gene (i.e., 78,589). MhaC is present in greater quantities in the OM of strains with mutations in mhaB1 (O35E.B1) (lane 3 in Fig. 2B), mhaB2 (O35E.B2) (lane 4 in Fig. 2B), or both (O35E.B1B2) (Fig. 2B, lane 5). Western blot analysis with antibodies against His-MhaB indicated that the WT isolate O35E (which should express both MhaB1 and MhaB2) and the mhaB1 isogenic mutant O35E.B1 (which should express only MhaB2) both yielded MhaB-reactive bands migrating with a similar apparent size of ∼120 kDa (Fig. 2A, lanes 1 and 3, respectively). These immunoblot experiments also revealed that the mhaB2 mutant O35E.B2 (presumably expressing only MhaB1) produces a protein that reacts with the MhaB antibodies and appears slightly larger than the MhaB-reactive band observed in O35E and O35.B1 (compare lane 4 to lanes 1 and 3 in Fig. 2A). Of note, this difference in size between MhaB1 and MhaB2 is reproducible and has been observed in several experiments involving electrophoresis of OM preparations for varying periods of time. These observations suggest that MhaB2 is primarily expressed in the parent strain O35E and that the slightly larger MhaB1 protein is expressed at detectable levels only in the absence of MhaB2. As expected, the His-MhaB antibodies no longer reacted with an ∼120 kDa band in the OM of the mhaB1 mhaB2 double mutant O35E.B1B2 (Fig. 2A, lane 5). Furthermore, it was discovered that the His-MhaB antibodies did not react with this ∼120-kDa band in the OM of the mhaC mutant O35E.C (Fig. 2A, lane 2), although the antibodies clearly recognized the antigen in whole-cell preparations of the mutant (data not shown). These results support the hypothesis that these proteins are OM located and comprise a TPS system where MhaC functions as a transporter which is necessary for the secretion and localization of MhaB1 and MhaB2 in the OM of M. catarrhalis strain O35E.
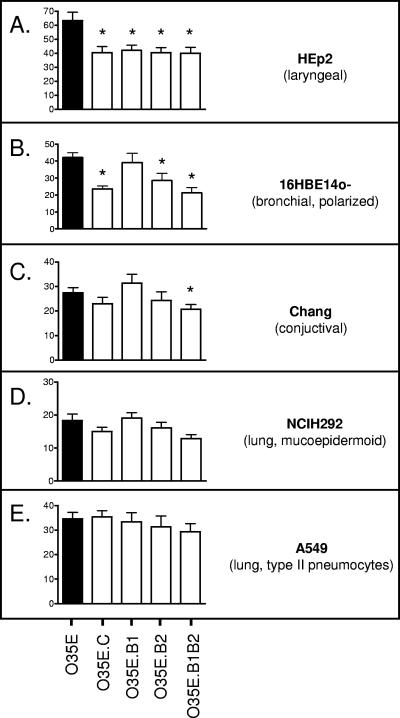
When the mha mutants were tested in attachment assays, it was discovered that all mutations significantly decreased binding to HEp2 monolayers (Fig. 3A). The absence of MhaC (O35E.C) or MhaB2 (alone in O35E.B2, or together with MhaB1 in O35E.B1B2) in the OM of mutants also resulted in a statistically significant decrease in adherence to 16HBE14o− cells, whereas the mhaB1 mutant (O35E.B1) attached at WT levels to these monolayers (Fig. 3B). Binding to Chang monolayers was found to be reduced only in the isogenic mutant strain lacking expression of both MhaB1 and MhaB2 (i.e., O35E.B1B2) (Fig. 3C). Disruption of the mha genes did not have a statistically significant effect on the attachment of M. catarrhalis O35E to NCIH292 or A549 cells (Fig. 3D and 3E). Of note, the reduction in adherence of the O35E mha mutants was not due to lower expression of other known M. catarrhalis adhesins, as the presence of Hag, McaP, OMPCD, and UspA1 was verified by Western blot analysis (data not shown). These results demonstrate that the mhaC and mhaB2 gene products are involved in the adherence of M. catarrhalis strain O35E to HEp2 and 16HBE14o− cells and that MhaB1 participates in the binding of this isolate to HEp2 cells. Expression of both MhaB1 and MhaB2 also appears to contribute to adherence to Chang monolayers.
FIG. 3.
Adherence of M. catarrhalis strain O35E (black bars) and mha mutants (open bars) to human epithelial cells in vitro. Adherence is expressed as the percentage (±standard error) of inoculated bacteria bound to epithelial cells following a 5-min incubation (shown on y axes). The Mann-Whitney test was used to determine whether decreases in adherence were statistically significant (*, P < 0.05).
The mhaB1CB2 locus is conserved among M. catarrhalis isolates.
To determine whether these gene products are expressed by strains other than O35E, we obtained OM preparations from 15 M. catarrhalis isolates of various clinical and geographical origins (Table 1). These preparations were then tested by Western blotting using MhaC- and MhaB-reactive murine sera as well as antibodies against McaP as a loading indicator. As shown in Fig. 4, nine strains (O12E, 7169, McGHS1, McGH, Mc34F, V1171, 13P18B1, 32P11B1, and O46E) expressed readily detectable levels of MhaC. These nine strains were also found to express MhaB-reactive antigens in their OM which were similar in size to that detected in O35E. Upon longer exposure, MhaB-reactive bands of ∼120 kDa were also detected in the OM preparations of 7P94B1, 11P29B1, and TTA24 (data not shown).
The mhaC genes from six isolates (O12E, McGHS1, Mc34F, TTA37, O46E, and V1171) were amplified by PCR and sequenced to assess conservation. The mhaC gene products from these strains (and O35E) were found to be highly conserved (98 to 99% identity). The mhaC ORFs from O12E, O35E, McGHS1, Mc34F, and O46E were each 705 aa in length, whereas those from V1171, TTA37, and the previously published ATCC 43617 strain (nt 58937 to 61048 of GenBank accession no. AX067458) were lacking a phenylalanine at codon 140, resulting in the smaller ORFs of 704 aa. Of note, the mhaC gene was amplified from strain TTA37 which did not demonstrate expression of either MhaC- or MhaB-reactive proteins in its OM (Fig. 4).
The nucleotide sequences of the mhaB1 ORFs from strains O12E and McGHS1 were also determined, and their gene products were found to be highly conserved (90% identity) (Table 2). The MhaB1 proteins of strains O12E and O35E were virtually identical (98%), whereas the McGHS1 gene product shared 90% identity with the O12E and O35E MhaB1 proteins (Table 2). The dissimilarity among these molecules was localized to their C termini, as residues 1 to 1,552 were 99.2% identical (data not shown). Nucleotide sequence analyses of mhaB2 from strains O12E and O35E revealed that the gene products also diverged toward their C termini. Amino acids 1 to 1,200 were found to be 99.5% identical, while the remainder of these two MhaB2 molecules only exhibit 19.3% identity (data not shown); O12E and O35E MhaB2 were well conserved with an overall identity of 68.8% (Table 2). It should be noted that even though strain McGHS1 appears to contain a mhaB2 gene based on PCR analysis, several attempts at amplifying and sequencing this gene in its entirety were unsuccessful. Comparison of O12E-MhaB1 and O12E-MhaB2 indicated that these proteins were better conserved than their O35E counterparts (82.4% versus 69.5%) (Table 2). This higher level of identity between the O12E MhaB proteins was due to the fact that their first 1,441 residues, as opposed to 1,200 in the O35E molecules, were perfectly conserved; the remainder of the O12E MhaB proteins only displayed 10.8% identity (data not shown).
TABLE 2.
Comparison of predicted amino acid sequences of the mhaB1 and mhaB2 genes of M. catarrhalis strains O35E, O12E, and McGHS1a
| Gene product (aa) | % Identity with gene product (aa):
|
||||
|---|---|---|---|---|---|
| O35E MhaB1 (1,727) | O35E MhaB2 (1,888) | O12E MhaB1 (1,726) | O12E MhaB2 (1,795) | McGHS1 MhaB1 (1,722) | |
| O35E MhaB1 (1,727) | 100 | ||||
| O35E MhaB2 (1,888) | 69.5 | 100 | |||
| O12E MhaB1 (1,726) | 98.0 | 69.4 | 100 | ||
| O12E MhaB2 (1,795) | 82.1 | 68.8 | 82.4 | 100 | |
| McGHS1 MhaB1 (1,722) | 90.1 | 69.5 | 90.3 | 81.9 | 100 |
Numbers in parentheses indicate the number of amino acid residues specified by each gene product.
The role of the mha gene products in adherence is conserved among M. catarrhalis isolates.
To test whether MhaB1 and MhaB2 are involved in the adherence of other M. catarrhalis isolates, the mhaC genes of strains O12E and McGHS1 were disrupted with a kanamycin resistance cassette. As shown in Fig. 5, these mutations abolished the appearance of both MhaC- and MhaB-reactive bands in the OM of strains O12E.C and McGHS1.C. These results are consistent with the analysis of our panel of O35E mutants (Fig. 2) in that a lack of the transporter MhaC results in the absence of MhaB1 and MhaB2 in the OM.
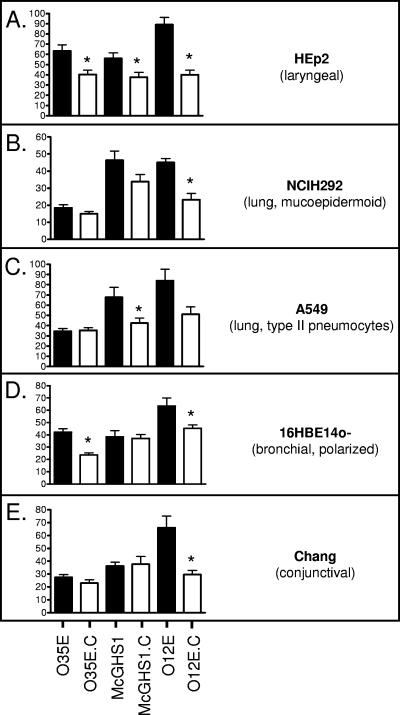
When the adherence of the mhaC mutants was compared to that of their respective parent strains, substantial decreases in adherence were observed (Fig. 6). Disruption of the mhaC gene in McGHS1 significantly reduced binding to HEp2 and A549 but not NCIH292, 16HBE14o−, or Chang cells. The absence of the Mha proteins in the OM of the mutant O12E.C resulted in decreased adherence to HEp2, NCIH292, 16HBE14o−, and Chang monolayers. Although the adherence of the O12E mhaC mutant to A549 cells appeared lower, this difference was not statistically significant. Interestingly, the contribution of the Mha proteins to epithelial cell binding varies among the isolates tested. For example, disruption of mhaC affects adherence of strains O35E and O12E to 16HBE14o− cells but not that of McGHS1, whereas McGHS1.C is the only mutant exhibiting significantly lower binding to A549 pneumocytes (Fig. 6). Overall, our data clearly demonstrate that the role of these TPS proteins in adherence is conserved among M. catarrhalis isolates.
FIG. 6.
Adherence of M. catarrhalis WT (black bars) and mhaC mutant (open bars) strains to human epithelial cells in vitro. Adherence is expressed as the percentage (±standard error) of inoculated bacteria bound to epithelial cells following a 5-min incubation (shown on y axes). The Mann-Whitney test was used to determine whether decreases in adherence were statistically significant (*, P < 0.05).
Only three plasmids, namely pWW102B (51), pWW115 (52), and pEMCJH04 (19), have been described for use in M. catarrhalis. These vectors are not, however, amenable for complementing the various mha mutations due to the incompatibility of antibiotic resistance markers and replication in a restricted number of M. catarrhalis isolates. To strengthen the hypothesis that the Mha proteins form a TPS system contributing to adherence, the WT mhaC gene was reintroduced in the chromosome of mutants O35E.C, O12E.C, and McGHS1.C. As shown in Fig. 5 and Table 3, all three repaired strains (i.e., O35E.CR1, O12E.CR1, and McGHS1.CR1) express MhaC- and MhaB-reactive bands in their OM and adhere at WT levels to HEp2 monolayers.
TABLE 3.
Adherence of M. catarrhalis strains to HEp2 cells
| Strain | Description | Expression of:
|
Adherence ± SD | |
|---|---|---|---|---|
| MhaCa | MhaBb | |||
| O35E | WT strain | + | + | 10.7 ± 7.1d |
| O35E.C | mhaC mutant | − | − | 4.8 ± 0.9 |
| O35E.CR1c | Repaired mhaC mutant | + | + | 10.6 ± 2.1d |
| O12E | WT strain | + | + | 70.3 ± 4.5d |
| O12E.C | mhaC mutant | − | − | 47.8 ± 4.5 |
| O12E.CR1c | Repaired mhaC mutant | + | + | 74.2 ± 8.2d |
| McGHS1 | WT strain | + | + | 42.9 ± 1.8d |
| McGHS1.C | mhaC mutant | − | − | 25.5 ± 3.3 |
| McGHS1.CR1c | Repaired mhaC mutant | + | + | 40.1 ± 2.8d |
Expression of MhaC in the OM was measured by Western blotting.
Expression of MhaB-reactive antigens in OM was measured by Western blotting.
Repaired strains were found to express WT levels of other known adhesins (UspA1, Hag, McaP, and OMPCD) by Western blotting (data not shown).
The difference in adherence between the strain and corresponding mhaC mutant is statistically significant.
The absence of MhaC, MhaB1, and MhaB2 in the OM of our panel of mutants may affect the proper surface display of other adhesins, thereby indirectly reducing adherence to epithelial cells. To test whether these proteins are directly involved in adherence, recombinant E. coli bacteria expressing the MhaC and MhaB1 proteins from strain O12E were tested in adherence assays. To achieve this, the mhaC and mhaB1 genes were isolated from a plasmid-based library of large M. catarrhalis DNA fragments (see Materials and Methods) and introduced into a nonadherent cloning strain of E. coli. As shown in Fig. 7A, these recombinant bacteria were found to express MhaB1 and MhaC in their OM and to bind to HEp2 cells at levels substantially higher than those of the negative control (Fig. 7B). Expression of O12E-MhaC and O12E-MhaB1 was also found to substantially increase the adherence of E. coli cells to Chang and 16HBE14o− monolayers (data not shown). Of note, control experiments demonstrated that growth of E. coli is minimal during incubation with monolayers and that the replication of control and test strains is comparable (data not shown). These data conclusively demonstrate that MhaB1 and MhaC are directly involved in adherence to human epithelial cells. Several attempts at expressing both MhaC and MhaB2 in recombinant E. coli were unsuccessful.
DISCUSSION
The identification of TPS systems in gram-negative bacteria is becoming a common occurrence (20, 26, 27). Several well-characterized (5, 11, 31, 40, 45-47) and recently identified (12, 41) TPS systems have been shown to be involved in adherence. Though very similar in function, the specifics of these systems are continually being elucidated and the extent of variation is becoming evident. This makes it difficult to hypothesize on the particulars of how MhaC, MhaB1, and MhaB2 function, but the most highly conserved characteristics of TPS systems are found in this newly characterized TPS system of M. catarrhalis.
First, MhaB1, MhaB2, and MhaC were identified on the basis of the similarity of their sequences, as well as predicted structural features, to the well-characterized FHA proteins of B. pertussis. Secondly, we discovered that, like FHA and other TPS exoproteins such as H. influenzae HMW1 and HMW2, MhaB1 and MhaB2 are involved in adherence to certain human epithelial cell types. Upon closer examination of the attachment assays with our panel of O35E mutants to Chang, NCIH292, and 16HBE14o− cells (Fig. 3), we noted a conserved pattern of adherence. The strains exhibiting the lowest level of binding were consistently O35E.B1B2 and O35E.C, both of which lack MhaB1 and MhaB2 in their OM (Fig. 2A). The mhaB2 mutant O35E.B2, which only expresses MhaB1 in its OM, also displayed reduced adherence to epithelial cells, though not to the same extent as O35E.C and O35E.B1B2. The mutant O35E.B1, which only expresses MhaB2 in its OM, attached to monolayers at levels undistinguishable from that of the WT strain O35E. Although some of the decreases in adherence to Chang and NCIH292 cells were not statistically significant (Fig. 3), the aforementioned attachment pattern of the mutants is clearly distinguishable. These observations indicate that expression of MhaB2 in the OM of strain O35E is necessary for WT levels of adherence, which is consistent with the Western blot analysis of our panel of mutants in which MhaB2 appears to be the TPS exoprotein that is primarily expressed by strain O35E (Fig. 2A), and thus, one might expect MhaB2's contribution to adherence by this WT isolate to be greater than that of MhaB1.
This adherence pattern of O35E mha mutants to NCIH292, 16HBE14o−, and Chang cells suggests that MhaB1 plays little or no role in adherence by M. catarrhalis strain O35E. This interpretation, however, is contradicted by the results of attachment assays using HEp2 cells in which comparable decreases in adherence were seen for O35E.B1, O35E.B2, O35E.C, and O35E.B1B2 (Fig. 3A). MhaB1 thus appears to be involved in M. catarrhalis adherence to HEp2 monolayers, and this hypothesis is supported by the fact that expression of MhaB1 and MhaC in the OM of a nonadherent cloning strain of E. coli leads to a gain of adherence to these laryngeal cells (Fig. 7). Clearly, the adhesive properties of the MhaB1 and MhaB2 proteins are complex. Studying their expression under conditions resembling those in attachment assays, using methods such as Western blotting with monoclonal antibodies specific for each adhesin or quantitative real-time PCR, may help in elucidating their respective contributions to M. catarrhalis adherence.
Because we were not able to express MhaB2 in recombinant E. coli, it is not clear whether this protein directly mediates adherence to epithelial cells or if MhaB2 expression is necessary for proper display of MhaB1 (or other adhesins) on the surface of M. catarrhalis cells. However, our data support a direct involvement for MhaB2. Our panel of mutants were found to express WT levels of Hag and UspA1 (data not shown), which have been previously shown to play major roles in the binding of strain O35E to Chang (1) and A549 cells (22), respectively. Based on these reports, one might expect that improper surface display of UspA1 and/or Hag to result in 60-fold (for Chang cells) and 10-fold (for A549 cells) reductions in attachment. As shown in Fig. 3C and E, the lack of expression of MhaB2 in the mutant O35E.B2 did not result in such decreases. Furthermore, expression of MhaB1 by recombinant E. coli increased binding to HEp-2 cells (Fig. 7), thus demonstrating that MhaB2 expression is not necessary for MhaB1 to function as an adhesin.
Our Western blot analysis of the O35E mutants suggests that MhaB1 migrates with a different mass than, and is present at detectable levels in the OM only in the absence of, MhaB2 (Fig. 2A). Interestingly, a similar occurrence has been reported by Ward and colleagues (53) for the H. ducreyi TPS system specifying the large supernatant proteins LspA1 and LspA2. Using monoclonal antibodies, these investigators demonstrated that culture supernatants of the H. ducreyi WT strain 3500HP contain LspA1 but barely detectable levels of LspA2. It was also discovered that an lspA1 mutant expressed greater levels of LspA2 than the WT strain 3500HP. Whether MhaB1 and MhaB2 are coordinately expressed in a similar manner remains to be determined by the use of monoclonal antibodies specifically recognizing each molecule.
The predicted amino acid sequences of MhaB1 from strains O35E, O12E, and McGHS1 were 90 to 98% identical, indicating a high level of conservation. The mhaB2 gene products, however, were more divergent (68.8% identity), particularly in the last one-third of the proteins (19.3% identity). It is tempting to speculate that this higher level of sequence divergence in the MhaB2 proteins might be the result of selective pressure for antigenic variation in vivo. This hypothesis would be consistent with our Western blot analysis, suggesting that MhaB2 is primarily expressed (relative to MhaB1) in the wild-type isolate O35E. The high level of sequence homology between these proteins makes them attractive as vaccine targets. In addition, our Western blots indicate that at least 63% of isolates express MhaB-reactive antigens in their OM. Of note, these preparations were obtained from cultures grown at 37°C in TH broth which may not represent the optimal conditions for expression of these TPS proteins. This hypothesis is supported by our finding that strain TTA37 specifies a mhaC gene product which does not appear to be expressed in the OM of this isolate (Fig. 4).
MhaC appears to comply with what is known of the TPS transporter family. These molecules are well-conserved among various organisms at the amino acid level as well as structurally (26, 27) and are considered members of the Omp85 family of proteins (15, 50). As observed with other Omp85-like proteins, the secondary structure of MhaC is predicted to be rich in β-strands, which is suggestive of a β-barrel porin-like conformation in the OM. Another characteristic of these β-barrel proteins is aberrant migration when resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. As shown in Fig. 2B, the mature MhaC protein, predicted to be 78 kDa, resolves at ∼65 kDa, which is substantially smaller than its predicted size. Whether MhaC forms a β-barrel structure that permits the specific passage of MhaB1 and MhaB2 across the OM of M. catarrhalis remains to be elucidated. Fleckenstein and colleagues recently reported data suggesting an adhesive role for the transporter of a TPS system expressed by uropathogenic E. coli (12), a finding which had not been previously reported for other TPS systems. Our results showing that mhaC mutants of strains O35E, O12E, and McGHS1 all exhibit reduced binding to epithelial cells (Fig. 7) raises the possibility that the M. catarrhalis MhaC protein might directly mediate adherence. The fact that the O35E.B1B2 mutant, which lacks expression of MhaB1 and MhaB2 but expresses MhaC in its OM (Fig. 2), exhibits the same defect in adherence as the O35E mhaC strain O35E.C, which lacks OM expression of all three proteins, argues against this possibility. Furthermore, while attempting to identify recombinant E. coli bacteria expressing both the M. catarrhalis MhaC and MhaB2 proteins, we generated several clones that expressed MhaC in their OM. When tested in attachment assays, these constructs did not bind to epithelial cells (data not shown). Thus, it is unlikely that the M. catarrhalis transporter MhaC itself possesses adhesive properties.
In summary, our data show that MhaC, MhaB1, and MhaB2 are highly conserved OM proteins expressed by several M. catarrhalis isolates. In strain O35E, MhaB1 and MhaB2 function as adhesins, though their specific contribution to human cell attachment remains to be elucidated. The role of MhaB1 and MhaB2 in adherence appears to be conserved, as isogenic mutants of strains O12E and McGHS1 lacking expression of MhaB-reactive antigens in their OM also have reduced binding to human epithelial cells. Furthermore, our results demonstrate that MhaB1 and MhaB2 rely on the transporter protein MhaC to reach the OM of M. catarrhalis, and as such, these three molecules constitute a novel TPS system. Future studies will be aimed at determining the cell-binding specificities of the MhaB proteins as well as to explore the vaccinogenic potential of these surface-located antigens.
Acknowledgments
This study was supported by a grant from NIH/NIAID (AI051477) to E.R.L.
We thank Anthony Campagnari and Pascale Plamondon at the State University of New York at Buffalo for providing strain 7169. We also thank Eric Hansen at the University of Texas Southwestern Medical Center in Dallas and Tim Murphy at the State University New York at Buffalo for providing M. catarrhalis strains and antibodies. We also express our thanks to Brian Bullard, Christine Akimana, William Grose, Robert Blumenthal, Mark Wooten, and Randall Worth for their helpful comments on the manuscript.
Editor: D. L. Burns
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson, W., J. Hamborsky, L. McIntyre, and S. Wolfe (ed.). 2007. Epidemiology and prevention of vaccine-preventable diseases, 10th ed., p. 81-100. Public Health Foundation, Washington, DC. http://www.cdc.gov/nip/publications/pink/pert.pdf.
- 4.Attia, A. S., E. R. Lafontaine, J. L. Latimer, C. Aebi, G. A. Syrogiannopoulos, and E. J. Hansen. 2005. The UspA2 protein of Moraxella catarrhalis is directly involved in the expression of serum resistance. Infect. Immun. 73:2400-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., and J. W. St Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 6.Beachey, E. H. 1981. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surface. J. Infect. Dis. 143:325-345. [DOI] [PubMed] [Google Scholar]
- 7.Bullard, B., S. L. Lipski, and E. R. Lafontaine. 2005. Hag directly mediates the adherence of Moraxella catarrhalis to human middle ear cells. Infect. Immun. 73:5127-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlone, G. M., M. L. Thomas, H. S. Rumschlag, and F. O. Sottnek. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J. Clin. Microbiol. 24:330-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, G. H. McCracken, Jr., and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cripps, A. W., D. C. Otczyk, and J. M. Kyd. 2005. Bacterial otitis media: a vaccine preventable disease? Vaccine 23:2304-2310. [DOI] [PubMed] [Google Scholar]
- 11.Dawid, S., S. Grass, and J. W. St Geme III. 2001. Mapping of binding domains of nontypeable Haemophilus influenzae HMW1 and HMW2 adhesins. Infect. Immun. 69:307-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleckenstein, J. M., K. Roy, J. F. Fischer, and M. Burkitt. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect. Immun. 74:2245-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsgren, A., M. Brant, M. Karamehmedovic, and K. Riesbeck. 2003. The immunoglobulin D-binding protein MID from Moraxella catarrhalis is also an adhesin. Infect. Immun. 71:3302-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsgren, A., M. Brant, A. Mollenkvist, A. Muyombwe, H. Janson, N. Woin, and K. Riesbeck. 2001. Isolation and characterization of a novel IgD-binding protein from Moraxella catarrhalis. J. Immunol. 167:2112-2120. [DOI] [PubMed] [Google Scholar]
- 15.Gentle, I. E., L. Burri, and T. Lithgow. 2005. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 58:1216-1225. [DOI] [PubMed] [Google Scholar]
- 16.Giebink, G. S., Y. Kurono, L. O. Bakaletz, J. M. Kyd, S. J. Barenkamp, T. F. Murphy, B. Green, P. L. Ogra, X. X. Gu, J. A. Patel, T. Heikkinen, S. I. Pelton, M. Hotomi, and P. Karma. 2005. Recent advances in otitis media. 6. Vaccine. Ann. Otol. Rhinol. Laryngol. Suppl. 194:86-103. [PubMed] [Google Scholar]
- 17.Guedin, S., E. Willery, C. Locht, and F. Jacob-Dubuisson. 1998. Evidence that a globular conformation is not compatible with FhaC-mediated secretion of the Bordetella pertussis filamentous haemagglutinin. Mol. Microbiol. 29:763-774. [DOI] [PubMed] [Google Scholar]
- 18.Guedin, S., E. Willery, J. Tommassen, E. Fort, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2000. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 275:30202-30210. [DOI] [PubMed] [Google Scholar]
- 19.Hays, J. P., K. Eadie, C. M. Verduin, H. Verbrugh, and A. van Belkum. 2005. A novel plasmid (pEMCJH03) isolated from moraxella catarrhalis possibly useful as a cloning and expression vector within this species. Plasmid 53:263-268. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holm, M. M., S. L. Vanlerberg, I. M. Foley, D. D. Sledjeski, and E. R. Lafontaine. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 72:1906-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm, M. M., S. L. Vanlerberg, D. D. Sledjeski, and E. R. Lafontaine. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao, C. B., S. Sethi, and T. F. Murphy. 1995. Outer membrane protein CD of Branhamella catarrhalis: sequence conservation in strains recovered from the human respiratory tract. Microb. Pathog. 19:215-225. [DOI] [PubMed] [Google Scholar]
- 24.Hu, W. G., J. Chen, J. C. McMichael, and X. X. Gu. 2001. Functional characteristics of a protective monoclonal antibody against serotype A and C lipooligosaccharides from Moraxella catarrhalis. Infect. Immun. 69:1358-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacob-Dubuisson, F., C. El-Hamel, N. Saint, S. Guedin, E. Willery, G. Molle, and C. Locht. 1999. Channel formation by FhaC, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 274:37731-37735. [DOI] [PubMed] [Google Scholar]
- 26.Jacob-Dubuisson, F., R. Fernandez, and L. Coutte. 2004. Protein secretion through autotransporter and two-partner pathways. Biochim. Biophys. Acta 1694:235-257. [DOI] [PubMed] [Google Scholar]
- 27.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 28.Klein, J. O. 2000. The burden of otitis media. Vaccine 19(Suppl. 1):S2-S8. [DOI] [PubMed] [Google Scholar]
- 29.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locht, C., P. Bertin, F. D. Menozzi, and G. Renauld. 1993. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol. Microbiol. 9:653-660. [DOI] [PubMed] [Google Scholar]
- 32.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 20:1754-1760. [DOI] [PubMed] [Google Scholar]
- 33.Mollenkvist, A., T. Nordstrom, C. Hallden, J. J. Christensen, A. Forsgren, and K. Riesbeck. 2003. The Moraxella catarrhalis immunoglobulin D-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J. Bacteriol. 185:2285-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172:195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, T. F., C. Kirkham, E. DeNardin, and S. Sethi. 1999. Analysis of antigenic structure and human immune response to outer membrane protein CD of Moraxella catarrhalis. Infect. Immun. 67:4578-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy, T. F., C. Kirkham, D. F. Liu, and S. Sethi. 2003. Human immune response to outer membrane protein CD of Moraxella catarrhalis in adults with chronic obstructive pulmonary disease. Infect. Immun. 71:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy, T. F., and M. R. Loeb. 1989. Isolation of the outer membrane of Branhamella catarrhalis. Microb. Pathog. 6:159-174. [DOI] [PubMed] [Google Scholar]
- 38.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichichero, M. E., and J. R. Casey. 2002. Otitis media. Expert Opin. Pharmacother. 3:1073-1090. [DOI] [PubMed] [Google Scholar]
- 40.Relman, D. A., M. Domenighini, E. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. USA 86:2637-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rojas, C. M., J. H. Ham, W. L. Deng, J. J. Doyle, and A. Collmer. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. USA 99:13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schonherr, R., R. Tsolis, T. Focareta, and V. Braun. 1993. Amino acid replacements in the Serratia marcescens haemolysin ShIA define sites involved in activation and secretion. Mol. Microbiol. 9:1229-1237. [DOI] [PubMed] [Google Scholar]
- 44.St Geme, J. W., III. 1997. Bacterial adhesins: determinants of microbial colonization and pathogenicity. Adv. Pediatr. 44:43-72. [PubMed] [Google Scholar]
- 45.St Geme, J. W., III. 1994. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun. 62:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.St Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 47.St Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, T. T., A. Forsgren, and K. Riesbeck. 2006. The respiratory pathogen Moraxella catarrhalis binds to laminin via ubiquitous surface proteins A1 and A2. J. Infect. Dis. 194:493-497. [DOI] [PubMed] [Google Scholar]
- 49.Timpe, J. M., M. M. Holm, S. L. Vanlerberg, V. Basrur, and E. R. Lafontaine. 2003. Identification of a Moraxella catarrhalis outer membrane protein exhibiting both adhesin and lipolytic activities. Infect. Immun. 71:4341-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voulhoux, R., and J. Tommassen. 2004. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155:129-135. [DOI] [PubMed] [Google Scholar]
- 51.Wang, W., A. S. Attia, L. Liu, T. Rosche, N. J. Wagner, and E. J. Hansen. 2006. Development of a shuttle vector for Moraxella catarrhalis. Plasmid 55:50-57. [DOI] [PubMed] [Google Scholar]
- 52.Wang, W., and E. J. Hansen. 2006. Plasmid pWW115, a cloning vector for use with Moraxella catarrhalis. Plasmid 56:133-137. [DOI] [PubMed] [Google Scholar]
- 53.Ward, C. K., J. L. Latimer, J. Nika, M. Vakevainen, J. R. Mock, K. Deng, R. J. Blick, and E. J. Hansen. 2003. Mutations in the lspA1 and lspA2 genes of Haemophilus ducreyi affect the virulence of this pathogen in an animal model system. Infect. Immun. 71:2478-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward, C. K., S. R. Lumbley, J. L. Latimer, L. D. Cope, and E. J. Hansen. 1998. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J. Bacteriol. 180:6013-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward, C. K., J. R. Mock, and E. J. Hansen. 2004. The LspB protein is involved in the secretion of the LspA1 and LspA2 proteins by Haemophilus ducreyi. Infect. Immun. 72:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitby, P. W., D. J. Morton, and T. L. Stull. 1998. Construction of antibiotic resistance cassettes with multiple paired restriction sites for insertional mutagenesis of Haemophilus influenzae. FEMS Microbiol. Lett. 158:57-60. [DOI] [PubMed] [Google Scholar]
- 57.Willems, R. J., C. Geuijen, H. G. van der Heide, G. Renauld, P. Bertin, W. M. van den Akker, C. Locht, and F. R. Mooi. 1994. Mutational analysis of the Bordetella pertussis fim/fha gene cluster: identification of a gene with sequence similarities to haemolysin accessory genes involved in export of FHA. Mol. Microbiol. 11:337-347. [DOI] [PubMed] [Google Scholar]