Abstract
The involvement of Fbe, a fibrinogen-binding protein of Staphylococcus epidermidis, in the pathogenesis of catheter-associated infection was investigated. An fbe (gene encoding Fbe protein) mutant was constructed by allelic replacement, wherein an erythromycin resistance gene replaced a portion of the A region of fbe. Meanwhile, a rat central venous catheter (CVC) infection model was established to assess the importance of Fbe in the pathogenesis of CVC-associated infection due to S. epidermidis. Fbe-positive S. epidermidis strain HB was significantly more likely to cause a CVC-associated infection resulting in bacteremia and metastatic disease than its isogenic Fbe-deficient mutant (100% versus 20%, P < 0.01). These results confirm the importance of adherence associated with Fbe in the pathogenesis of CVC-associated infection caused by S. epidermidis.
Staphylococcus epidermidis is a commensal inhabitant of human skin that rarely causes disease in healthy persons. In recent years, S. epidermidis has emerged as a major pathogen associated with foreign-body infections, such as prosthetic-joint infection, prosthetic-valve endocarditis, exit site infection in dialysis patients, and cerebrospinal fluid shunt- and catheter-associated infections (5). Unfortunately, these infections are responsible for significant morbidity and mortality.
The propensity of S. epidermidis to cause foreign-body infections can be explained largely by its ability to adhere to and proliferate on polymer surfaces. The adherence of S. epidermidis appears to be a two-step process that requires adhesion of the bacteria to a biomaterial surface, followed by cell-to-cell accumulation and biofilm formation (16).
Shortly after a plastic or metal device is introduced into the body, the surface of the device becomes coated with host proteins (17). Studies of Staphylococcus aureus have shown that bacterial surface proteins binding to host proteins contribute to bacterial infections. Among these surface proteins, clumping factor A and clumping factor B, fibrinogen (Fg)-binding proteins of S. aureus, have been well investigated (7, 8).
In previous study, an Fg-binding protein termed Fbe (also called SdrG) was identified in S. epidermidis strain HB and the sequence of the gene that encodes it (fbe) was determined (9). As a member of the Sdr multigene family of staphylococci, Fbe comprises a 548-residue Fg-binding A domain, two B repeats, a Ser-Asp dipeptide repeated domain, and a cell wall anchoring domain. Sequence comparison with earlier known proteins has revealed that the Fbe protein is related to clumping factor A, the cell wall-bound Fg-binding protein of S. aureus. More studies demonstrated that Fbe and its antibodies could efficiently block the adherence of S. epidermidis to immobilized Fg (11). An assay of adherence to immobilized Fg showed that the Fg-binding ability of an fbe mutant was reduced compared to that of its parental strain (10).
In this study, an fbe mutant was constructed by allelic replacement, wherein an erythromycin (ERM) resistance gene (ermB) replaced a portion of the A region of fbe. A rat model of central venous catheter (CVC)-associated infection was developed with a view to mimicking, to a great extent, the conditions found in the human intravascular system. The rat CVC-associated infection model was then used to compare the mutant and its parental strain for virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. epidermidis cells were grown in Luria-Bertani (LB) medium (Sangon, Shanghai). S. epidermidis HB was an Fbe wild-type strain, and S. aureus RN4220 was a restriction-deficient strain used as a temporary host for plasmids (15). Both were gifts from Jan-Ingmar Flock, Karolinska Institute, Sweden. Plasmid pBT2 was a temperature-sensitive vector with a chloramphenicol (CHL) resistance gene for allelic replacement. Plasmid pEC1 harbored the ermB gene for ERM resistance. They were kindly provided by Cuong Vuong, Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories.
Proteins and reagents.
Human Fg was the product of Calbiochem Co. Ltd., and anti-human Fg mouse monoclonal antibody was obtained from Cedarlane Laboratories Ltd. (Canada). Bovine serum albumin (BSA) was purchased from the Sino-American Biotechnology Company (Shanghai, China), and anti-mouse goat antibody conjugated to horseradish peroxidase (HRP) was the product of Perfect Biotech Co. Ltd. (Shanghai, China). Molecular weight markers used in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and nitrocellulose membranes used for Western blotting were obtained from Pharmacia (Sweden).
Construction of plasmid pBTΔfbe and homologous recombination.
In order to delete the fbe gene in S. epidermidis HB, DNA fragments of 654 bp (upstream, containing EcoRI and BamHI sites) and 1,179 bp (downstream, containing PstI and HindIII sites) of fbe were amplified by PCR assay. Chromosomal DNA from strain HB was used as the template. Both amplifications were done as follows: 94°C for 5 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s; and 72°C for 5 min. The recombination procedure was performed as described by Pei and Flock (10). Two fragments were digested with EcoRI/BamHI and PstI/HindIII, respectively, producing a 527-bp fragment termed fbe1 and a 1,179-bp fbe2 fragment. fbe1 and fbe2 were then inserted into the polylinker region of temperature-sensitive shuttle vector pBT2 together with a 1,450-bp BamHI/PstI-digested fragment containing the ERM resistance gene (ermB) from plasmid pEC1, resulting in a new plasmid, pBTΔfbe. Plasmid pBTΔfbe comprised the fbe::ermB gene. DNA manipulation, isolation of plasmid DNA, and transformation of Escherichia coli were performed by standard procedures (14). A one-step PCR kit and restriction enzymes were obtained from TaKaRa Biotechnology Co., Ltd. (Dalian, China). Constructed plasmid pBTΔfbe was then transformed into restriction-deficient S. aureus RN4220 by electroporation. Plasmid pBTΔfbe, purified from transformed strain RN4220, was transformed into S. epidermidis HB by electroporation, which was performed at room temperature in a 0.2-cm cuvette at settings of 2 kV, 25 μF, and 100 Ω for 2.5 ms (6). The transformants were selected on LB agar plates containing ERM (10 μg/ml) at 30°C. The primers used are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer | Sequence (including restriction enzyme sequence) | Position | Primer combination | Template DNAa | PCR product size (bp) |
|---|---|---|---|---|---|
| P1L | 5′-GGGGGTACCGCTACTACTAAAAAGAAACCTA-3′ (after bp 136 including EcoRI site in fbe) | 59-80 in fbe | P1L-P1R | HB | 654 (fbe1; 527 after digestion) |
| P1R | 5′-CGCGGATCCGGCGGCTGACTTGTTGTT-3′ (BamHI site underlined) | 697-712 in fbe | |||
| P2L | 5′-GAGCTGCAGCGCCGACGCTTAAACATTCA-3′ (PstI site underlined) | 2059-2077 in fbe | P2L-P2R | HB | 1,179 (fbe2) |
| P2R | 5′-CCCAAGCTTGCGCGTGCCTTTAGAGCCATAAT-3′ (HindIII site underlined) | 3218-3238 in fbe | |||
| P3L | 5′-AGGTCATAATGAGGCCAAAGC-3′ | 169-189 in fbe::ermB | P3L-P3R | HBmut | 1,672 |
| P3R | 5′-CTGTGGTATGGCGGGTAAGA-3′ | 1822-1841 in fbe::ermB | HB | 0 (none) | |
| P4L | 5′-ATTGTCCGAGAGTGATTGGTC-3′ | 922-942 in fbe::ermB | P4L-P4R | HBmut | 919 |
| P4R | 5′-CTGTGGTATGGCGGGTAAGA-3′ | 1822-1841 in fbe::ermB | HB | 0 (none) | |
| P5L | 5′-ACTTACCCGCCATACCACAG-3′ | 1822-1841 in fbe::ermB | P5L-P5R | HBmut | 1,574 |
| P5R | 5′-CGTTTCCCTAATAATAACGCTCC-3′ | 3374-3396 in fbe::ermB | P3L-P5R | HB | 0 (none) |
| HBmut | 2,227 (for fbe::ermB sequencing) | ||||
| P6L | 5′-TAAACACCGACGATAATAACCAAA-3′ | 306-329 in fbe | P6L-P6R | HBmut | 0 (none) |
| P6R | 5′-GGTCTAGCCTTATTTTCATATTCA-3′ | 776-800 in fbe | HB | 495 |
HB, wild type; HBmut, mutant.
Identification of the fbe::ermB allele replacement.
Electroporated cells of HB were grown on LB agar plates with ERM (10 μg/ml) at 30°C. An ERM-resistant colony was inoculated into LB broth with ERM (5 μg/ml) and incubated at 40°C for 24 h for 10 generations. The culture was diluted and plated on ERM (5 μg/ml) plates. The same colonies from the ERM plates were inoculated onto both ERM and CHL (10 μg/ml) plates. Colonies that grew only on ERM plates and not CHL plates were selected as candidates for further testing. PCR amplification with different pairs of primers was performed to identify mutants (Table 1). The PCR product obtained with primers P3L and P5R and HBmut template DNA was sequenced by Huanuo Biotech (Shanghai) Co. Ltd.
Bacterial adherence to Fg.
Strains HB and HBmut were grown in LB broth overnight in a 37°C incubator shaker (120 rpm; New Brunswick Scientific). The bacterial cells were washed and resuspended in phosphate-buffered saline (PBS, pH 7.4), and the optical density at 600 nm was adjusted to 0.8. Microtiter wells (Labsystems, Finland) were coated with 100 μl of a bacterial suspension, incubated overnight at 4°C, and blocked with 2% BSA in PBS for 1 h at 37°C. After washing, Fg in PBS at a concentration of 10 μg/ml (100 μl) was added and allowed to adhere for 2 h at room temperature. Following additional washing, anti-Fg mouse antibody, anti-mouse goat antibody with HRP, and substrate were sequentially added and bacterial adherence was determined by optical reading of A492 with a microtiter plate reader (Bio-Rad).
Detection of Fbe expression by SDS-PAGE and Western blotting analysis.
Strains HB and HBmut were grown in LB broth overnight at 37°C in an incubator shaker (120 rpm; New Brunswick Scientific). The bacterial cells were washed and resuspended in PBS (pH 7.0), and then lysostaphin (1 mg/ml) was added and the mixture was incubated for 1 h. After complete sonication and centrifugation (6,000 rpm) at 4°C for 10 min, SDS-PAGE was performed by use of the Phast system with 8 to 25% gradient gels (Pharmacia). To transfer the separated proteins, a nitrocellulose membrane was placed on top of the gel, which was incubated at 65°C. The membrane was then blocked with BSA for 20 min at room temperature. After three washes in PBS-0.05% Tween 20, the membrane was incubated with Fg (5 μg/ml in PBS-0.05% Tween 20) at room temperature for 1 h. Anti-Fg mouse antibody (1:1,000 dilution), anti-mouse goat antibody with HRP (1:1,000 dilution), and the substrate 4-chloronaphthol (Sigma) were then sequentially added (11).
Rat CVC-associated infection model.
A CVC-associated infection rat model was used to compare the pathogenesis of S. epidermidis HB with that of its isogenic mutant S. epidermidis HBmut. Thirty male Sprague-Dawley rats (180 to 200 g) underwent catheterization as previously described, with modification (13). In brief, a polyvinyl chloride (PVC) catheter (inside diameter, 0.5 mm; outside diameter, 1 mm) was inserted into the right jugular vein and advanced into the superior vena cava. The proximal end was cut, sealed, and buried subcutaneously. Every rat was weighed before catheterization. Twenty-four hours after CVC placement, blood was obtained from the tail vein and cultured to ensure sterility and then the rats were divided into two groups and 106 CFU of S. epidermidis HB or HBmut was injected into the tail vein. On day 5, every rat was weighed again and sacrificed. Blood was obtained by cardiac puncture and quantitatively cultured by directly plating 0.1 ml of blood on LB agar. The CVCs were removed aseptically and cut into three parts. The distal end was vigorously vortex washed in PBS, after which the wash fluid was quantitatively cultured. The middle part was cultured in LB broth for 48 h, and the third part was placed in 2.5% glutaraldehyde for later scanning electron microscopy (SEM) to assess the morphological features of S. epidermidis adherent to PVC catheter specimens (3). To ascertain the extent of metastatic disease, the heart, lungs, liver, and kidneys were aseptically harvested, weighed, homogenized in 2 ml of PBS, and quantitatively cultured.
Minimum inoculum size experiments were performed before comparative studies. CVCs were placed as described above. Each group of three rats was challenged with an inoculum of 104, 105, 106, 107, or 108 CFU of S. epidermidis HB, and the animals were evaluated for the presence of infection by blood and CVC culture.
The chi-square test was used to assess whether there was a significant difference in infection rate between the two groups of animals. The Wilcoxon signed-rank test was used to analyze data regarding levels of bacteremia and metastatic disease. Statistical tests were performed with SPSS 10.0.
RESULTS
Deletion of fbe in S. epidermidis strain HB by allele replacement.
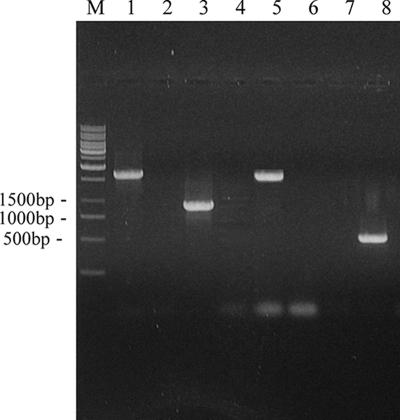
Restriction endonucleases digestion showed that plasmid pBTΔfbe comprising fbe::ermB was constructed successfully. A strain termed HBmut (fbe::ermB) derived from HB by allele replacement of the fbe gene was constructed. PCR analysis of DNAs from strains HB and HBmut was performed with different combinations of primers (Table 1). The PCR product obtained with HBmut template DNA and primers P3L and P3R was 1,672 bp, that obtained with P4L and P4R was 919 bp, and that obtained with P5L and P5R was 1,574 bp, whereas no product was obtained with HB template DNA and the same primers. Likewise, the PCR product obtained with HB template DNA and primers P6L and P6R was 495 bp in size; no product was generated when HBmut template DNA was used in the same way (Fig. 1). fbe::ermB was further demonstrated by sequencing of the PCR-amplified fragment generated with primers P3L and P5R and HBmut template DNA, suggesting that allelic replacement had occurred and the ermB fragment replaced part of fbe between fbe1 and fbe2 (data not shown).
FIG. 1.
PCR analysis of S. epidermidis HBmut and HB. Confirmation of allele replacement of fbe with fbe::ermB in chromosomal DNA. Lanes 1, 3, 5, and 7 are amplified products from strain HBmut. Lanes 2, 4, 6, and 8 are amplified products from strain HB. Lanes 1 and 2, primers P3L and P3R; lanes 3 and 4, primers P4L and P4R; lanes 5 and 6, primers P5L and P5R; lanes 7 and 8, primers P6L and P6R; lane M, markers.
Adherence of the mutant strain to Fg was different from that of its parental strain.
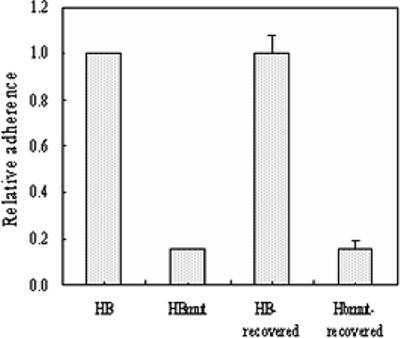
To further demonstrate the function deletion of Fbe protein from strain HBmut, the adherence of the two strains to Fg was tested. The reduction in the adherence of mutant strain HBmut compared with that of wide-type strain HB was significant (Fig. 2). This implied that replacement of fbe affects its Fg affinity and also indicated that the construction of the fbe substitution mutant was successful.
FIG. 2.
Adherence of strains HB and HBmut to immobilized Fg in microtiter wells. Relative adherence was measured by optical reading of A492 with a microtiter plate reader. The relative adherence of strain HB was set to 1. Column HB-recovered shows the mean adherence of 15 strains recovered from rats challenged with strain HB. Column HBmut-recovered shows the mean adherence of three strains recovered from rats challenged with strain HBmut. Mean values and standard deviations are shown.
Detection of Fbe expression by Western blotting.
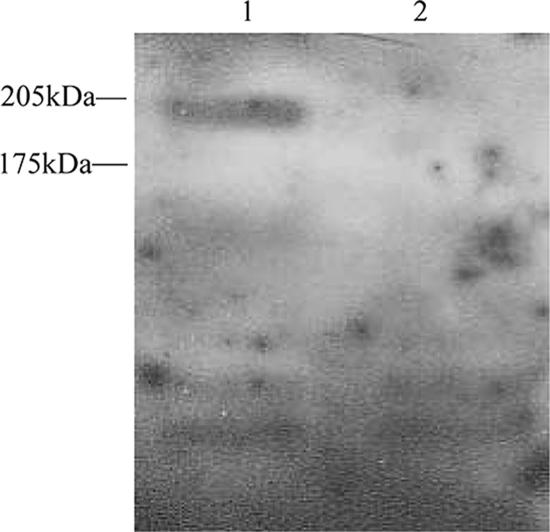
An immunoreactive protein of 190 kDa was expressed by wild-type strain HB but not by HBmut, the fbe mutant (Fig. 3). This implied that HBmut could not express Fbe because it lacked fbe, indicating successful construction of an fbe mutant.
FIG. 3.
Western blotting analysis of Fbe expression. Lane 1, S. epidermidis HB; lane 2, S. epidermidis HBmut.
Minimum-inoculation studies.
Minimum-inoculation studies showed that all of the rats challenged with more than 106 CFU of S. epidermidis HB developed CVC-associated infections, compared with one of three rats challenged with 104 CFU and two of three challenged with 105 CFU. Therefore, the inoculum of 106 CFU, the lowest dose that could reproducibly cause CVC-associated infection, was used in the larger comparative studies.
Comparative studies.
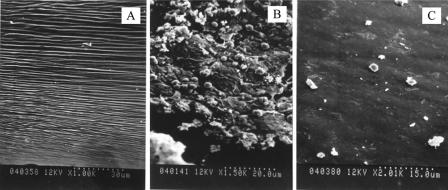
It was found that 93.3% (14/15) of the rats inoculated with S. epidermidis HB had bacteria recovered from the explanted CVC, compared with 13.3% (2/15) of the rats challenged with S. epidermidis HBmut. The median numbers of bacteria recovered from the HB and HBmut groups were 1.4 × 105 and 0 CFU/ml, respectively. There was a significant difference between the two groups of rats by the Wilcoxon signed-rank test (P < 0.001). The morphological features of S. epidermidis colonization of the PVC catheter surface shown by SEM are presented in Fig. 4. Compared with the surface of an unused PVC catheter (Fig. 4A), staphylococcal colonies covered with an amorphous slime matrix were detected on an explanted catheter challenged with 106 CFU of S. epidermidis HB that had bacterial recovery in quantitative and broth cultures (Fig. 4B). In contrast, on the catheter explanted from a rat challenged with S. epidermidis HBmut, neither bacterial cells nor slime formation was visible (Fig. 4C).
FIG. 4.
SEM of an unused sterile PVC catheter surface (A), staphylococcal colonization of the surface of an experimentally infected catheter challenged with S. epidermidis HB (B), and the surface of a catheter explanted from a rat challenged with S. epidermidis HBmut (C).
In addition, 73.3% (11/15) of the rats challenged with S. epidermidis HB had bacteria recovered from the blood, compared with only 20% (3/15) recovery in rats inoculated with S. epidermidis HBmut, and the median numbers of bacteria in the two groups were 50 and 0 CFU/ml, respectively. Wilcoxon signed-rank analysis showed that the difference between the two groups was significant (P < 0.01).
Results from studies defining the burden of metastatic disease in rats challenged with S. epidermidis HB or HBmut through the bloodstream are shown in Table 2. There were more animals with lung, heart, liver, and kidney metastases in the group inoculated with S. epidermidis HB (12/15 to 15/15) than in the group inoculated with S. epidermidis HBmut (1/15 to 3/15). In addition, for all of the organ systems examined, the number of organisms recovered per gram of tissue was greater in the rats challenged with HB (1.9 × 102 to 8.0 × 102 CFU/g tissue) than in those challenged with HBmut (0 CFU/g tissue). There was a significant difference between the two groups by Wilcoxon signed-rank test (P < 0.01).
TABLE 2.
Numbers of S. epidermidis HB and HBmut organisms recovered per gram of tissuea
| Organ | HB
|
HBmut
|
||
|---|---|---|---|---|
| No. of animals infected/total (%) | Median no. of CFU/g tissue (range) | No. of animals infected/total (%) | Median no. of CFU/g tissue (range) | |
| Heart | 12/15 (80) | 1.9 × 102 (0-2.3 × 104) | 3/15 (20) | 0 (0-2.5 × 103) |
| Liver | 15/15 (100) | 3.8 × 102 (30-8.6 × 103) | 3/15 (20) | 0 (0-1.7 × 103) |
| Lung | 14/15 (93.3) | 2.4 × 102 (0-5.1 × 106) | 1/15 (6.7) | 0 (0-1.5 × 104) |
| Kidney | 14/15 (93.3) | 8.0 × 102 (0-7.8 × 104) | 2/15 (13.3) | 0 (0-2.3 × 103) |
All interstrain differences are significant at P < 0.01.
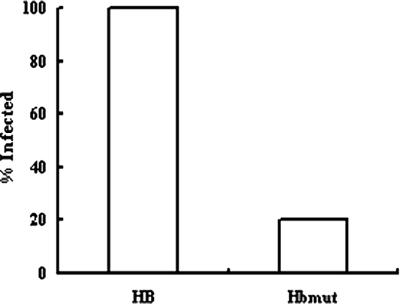
Finally, the overall infection rate, defined as recovery of S. epidermidis from the blood, catheter, or organs at the time of sacrifice, is shown in Fig. 5. The overall infection rate was 100% (15/15) in the HB group, and it was only 20% (3/15) in the HBmut group. There was a significant difference between the two groups by Fisher's exact test (P = 0.000005).
FIG. 5.
Rates of CVC-associated infection of animals challenged with S. epidermidis HB or HBmut.
DISCUSSION
Fbe (also called SdrG), an Fg-binding protein, has been investigated by many researchers in vitro as a member of the subfamily of S. epidermidis bacterial microbial surface components recognizing adhesive matrix molecules. Previous studies showed that Fbe was a major factor involved in the adherence of S. epidermidis to immobilized Fg and appeared to target the thrombin cleavage site in the Fg β chain (2). Nevertheless, the situation in vivo is quite different and whether Fbe mediates S. epidermidis adherence to biomaterials and causes foreign-body infections is unknown. So, the aim of the present study was to evaluate the relative contribution of Fbe to the pathogenesis of intravascular-catheter-associated infection caused by S. epidermidis.
First, we constructed an Fbe-deficient mutant to supply two isogenic background strains. A major problem in performing genetic manipulation of S. epidermidis is the difficulty of introducing plasmids into this organism by transformation. We introduced an efficient system for gene replacement with temperature-sensitive E. coli-Staphylococcus shuttle plasmid PBT2 for delivery, an ERM resistance gene for single-copy selection, and S. aureus RN4220 as a temporary host for the plasmid (1). A mutant termed HBmut (harboring fbe::ermB) was generated by replacing a part of the fbe gene with an ERM resistance gene (ermB) in S. epidermidis strain HB. PCR amplification and sequencing results of HBmut confirmed that fbe::ermB had replaced fbe in the chromosomal DNA.
Bacterial adherence to biomaterials, however, is only one component of a complex process that involves interactions among the host, biomaterial, and microbes. It is difficult to duplicate these conditions in in vitro models. Therefore, it is desirable to do research regarding pathogenesis in in vivo models. Tissue cages in guinea pigs (18) and the mouse subcutaneous-foreign-body infection model (4) have been used to study the interaction between microbes and biomaterials. However, both of these animal models had some weakness in mimicking foreign-body infections in humans. Rupp established a rat CVC-associated infection model to assess the pathogenesis of polysaccharide intercellular adhesin-hemagglutinin in S. epidermidis. This model made use of venous access that allowed long-term venous catheter placement and mimicked the conditions found in surgically implanted CVCs in humans (13). Therefore, we adopted this CVC-associated infection model and modified it a little.
With this model, we demonstrated that Fbe was important in the pathogenesis of CVC-associated infection caused by S. epidermidis. The isogenic, Fbe-deficient mutant produced a significantly lower infection rate and lower bacterial recovery from the implanted catheters and bloodstream and less metastatic disease than the parental strain. These observations might be due to inability of the Fbe-deficient mutant to adhere to the Fg-coated catheter to form macrocolonies and biofilm. We also found that the virulence of strain HB was lower in the absence of medical devices. In a pilot experiment without catheterization, no rat could be infected after inoculation with up to 108 CFU of strain HB (data not shown). These results implied that Fbe protein could promote adherence of bacteria to immobilized Fg substrates and thus contributed to the initiation of foreign-body infection by allowing bacteria to adhere to biomaterial surfaces that had become coated with host proteins after implantation. Bacteria, host proteins, and medical devices were indispensable parts of infection initiation. The reduction in adherence to Fg and lack of 190-kDa protein expression of the mutant in our study also demonstrated the phenotypic alteration in the mutant, which was consistent with the results we had obtained with the in vivo model. Strains HB and HBmut kept their phenotypes after in vivo circulation with similar levels of adherence to Fg (Fig. 2). This in vivo model also could be used to assess the importance of other potential virulence factor in the pathogenesis of CVC-associated S. epidermidis infection.
In conclusion, the present study suggests that Fbe might be an important factor in the pathogenesis of prosthetic-device infection caused by S. epidermidis. A previous study also showed that the severity of systemic infection of mice with S. epidermidis was reduced if the bacteria were preopsonized with anti-Fbe prior to administration (12). Hence, the present findings may provide novel approaches to the prevention and treatment of prosthetic-device infections.
Acknowledgments
We thank Jan-Ingmar Flock, who works at the Karolinska Institutet, Sweden, for providing S. epidermidis HB and S. aureus RN4220 and thank Cuong Vuong, who works at the Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories, for providing plasmids pBT2 and pEC1.
This work was supported by a grant from the National Natural Science Foundation of China (39970040).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 26 March 2007.
REFERENCES
- 1.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Davis, S. L., S. Gurusiddappa, K. W. McCrea, S. Perkins, and M. Hook. 2001. SdrG, a fibrinogen-binding bacterial adhesin of the microbial surface components recognizing adhesive matrix molecules subfamily from Staphylococcus epidermidis, targets the thrombin cleavage site in the Bβ chain. J. Biol. Chem. 276:27799-27805. [DOI] [PubMed] [Google Scholar]
- 3.Franson, T. R., N. K. Sheth, H. D. Rose, and P. G. Sohnle. 1984. Scanning electron microscopy of bacteria adherent to intravascular catheters. J. Clin. Microbiol. 20:500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallimore, B., R. F. Gagnon, R. Subang, and G. K. Richards. 1991. Natural history of chronic Staphylococcus epidermidis foreign body infection in a mouse model. J. Infect. Dis. 164:1220-1223. [DOI] [PubMed] [Google Scholar]
- 5.Huebner, J., and D. A. Goldmann. 1999. Coagulase-negative staphylococci: role as pathogens. Annu. Rev. Med. 50:223-236. [DOI] [PubMed] [Google Scholar]
- 6.Lee, J. C. 1995. Electrotransformation of staphylococci. Methods Mol. Biol. 47:209-216. [DOI] [PubMed] [Google Scholar]
- 7.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 8.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson, M., L. Frykberg, J.-I. Flock, L. Pei, M. Lindberg, and B. Guss. 1998. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 66:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei, L., and J.-I. Flock. 2001. Lack of fbe, the gene for a fibrinogen-binding protein from Staphylococcus epidermidis, reduces its adherence to fibrinogen coated surfaces. Microb. Pathog. 31:185-193. [DOI] [PubMed] [Google Scholar]
- 11.Pei, L., M. Palma, M. Nilsson, B. Guss, and J.-I. Flock. 1999. Functional studies of a fibrinogen binding protein from Staphylococcus epidermidis. Infect. Immun. 67:4525-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rennermalm, A., M. Nilsson, and J.-I. Flock. 2004. The fibrinogen binding protein of Staphylococcus epidermidis is a target for opsonic antibodies. Infect. Immun. 72:3081-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupp, M. E., P. D. Fey, C. Heilmann, and F. Gotz. 2001. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 183:1038-1042. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Tenhami, M., K. Hakkila, and M. Karp. 2001. Measurement of effects of antibiotics in bioluminescent Staphylococcus aureus RN4220. Antimicrob. Agents Chemother. 45:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuong, C., and M. Otto. 2002. Staphylococcus epidermidis infections. Microbes. Infect. 4:481-489. [DOI] [PubMed] [Google Scholar]
- 17.Yu, J., M. N. Montelius, M. Paulsson, I. Gouda, O. Larm, L. Montelius, and A. Ljungh. 1994. Adhesion of coagulase-negative staphylococci and adsorption of plasma proteins to heparinized polymer surfaces. Biomaterials 15:805-814. [DOI] [PubMed] [Google Scholar]
- 18.Zimmerli, W., F. A. Waldvogel, P. Vaudaux, and U. E. Nydegger. 1982. Pathogenesis of foreign body infection: description and characteristics of an animal model. J. Infect. Dis. 146:487-497. [DOI] [PubMed] [Google Scholar]