Abstract
Respiratory pathogens, such as Neisseria meningitidis, secrete site-specific proteases able to cleave human immunoglobulin A1 (IgA1), the first line of defense at mucosal membranes. Bacterial isolates show wide variability in IgA1 protease activity, and those isolated from patients with clinical infection possess the highest levels of activity. A feature of this enzyme is the self-cleavage required for secretion of the mature extracellular form. Known cleavage targets contain a proline-rich consensus recognition sequence, Pro-Pro-Ser-Pro, residing in the variable linker region that connects the protease and translocator domains. Here, we report the sequence of the NMB IgA1 protease and the unexpected self-cleavage and subsequent extracellular release of mature IgA1 protease from mutants lacking the previously defined consensus cleavage site. We investigated the possible link between enzyme secretion and variability in the linker sequence segment using site-directed mutagenesis and linker domain swapping to construct mutated and chimeric forms of the IgA1 protease from N. meningitidis strain NMB. The observed change in secreted activity levels compared to the wild-type clone indicated that the precise amino acid sequence of the intervening region, between mature IgA1 protease and the β-core translocator domain, influences the efficacy of autoproteolytic processing. The broader specificity uncovered for the NMB IgA1 protease suggests that it could cleave a far wider range of human proteins than previously appreciated.
Proteolytic events underpin a vast range of diverse biological processes, including the coagulation cascade (53), apoptosis (54), protein turnover, and sister chromatid separation (17). Many proteinases possess the ability to cleave at a specific sequence, leaving polypeptides lacking the specific recognition site untouched. Examples of this group of proteinases are involved in processes such as the maturation of viral polyproteins (8, 26) and modulation of signaling (4) and in the ontogeny and pathophysiology of the nervous system (41). Bacterial serine-type immunoglobulin A1 (IgA1) proteases have been reported to be site-specific proteinases (45). They are extracellular enzymes able to cleave human IgA1 within its hinge region, resulting in decoupling of the antibody recognition function (Fab fragment mediated) from Fc fragment-mediated effector functions (39). The primary role of human IgA1 is the agglutination of colonizing bacteria, which is greatly enhanced by its polyvalent structure. The resultant bacterium-antibody complexes adhere to mucin and are finally expelled by mucociliary clearance. This essentially mechanical process is defined as immune exclusion and represents the most important defense mechanism operating on mucosal membranes (9, 28).
Neisseria meningitidis and Haemophilus influenzae both produce IgA1 proteinases. They are two of the most common causative agents of bacterial meningitis, septicemia, and a range of other diseases involving mucosal membranes: (24, 27, 44, 49). The iga genes of these species encode extracellular enzymes with the ability to cleave the human IgA1 molecule at a specific site (45). IgA1 proteases specifically cleave the peptide bond distal to the second proline (shown by the vertical line) in the sequence P-P-|-T-P, P-S-P-|-S, or P-T-P-|-S-P. These enzymes have an unusual route of secretion. In contrast to the majority of proteins exported by gram-negative bacteria, the product of the iga gene is relatively self-sufficient in the direction of its own secretion across the bacterial outer membrane, although recent research has uncovered a role for Omp85 in IgA1 protease secretion (67). Van Ulsen et al. reported that another outer membrane autotransporter protein known as NalP could play a role in IgA1 protease maturation, but only in a subset of isolates examined (59, 62).
Detailed analysis of the iga gene and its resultant proteins in Neisseria gonorrhoeae MS11 has generated a model for this type of secretion (25, 46). The precursor of the mature IgA1 protease consists of four functional domains: (i) an amino-terminal signal peptide involved in cytoplasmic membrane transport; (ii) the mature extracellular protease domain; (iii) the α-protein domain, which is secreted in conjunction with the mature protease; and (iv) the β-core domain, essential for transport across the outer membrane. The model proposes that the carboxy-terminal β-core domain integrates into the outer membrane and forms a specific pore through which the mature protease and α-protein are translocated through the periplasm into the extracellular space. In the N. gonorrhoeae MS11 enzyme, autoproteolysis occurs at three target sites, designated a, b, and c. This model was revised in light of evidence suggesting that transport proceeds through the central pore of a ring-shaped complex structure assembled from multiple β-domains (63). A more recently reported crystal structure of a related neisserial autotransporter, NalP, revealed that an N-terminal alpha-helical segment, thought to be part of the passenger domain, was threaded through the β-core. This suggested that the passenger protein domain may indeed be able to translocate through the β-core barrel itself (42), contrary to the previous study. Whatever the details of translocation, the mature IgA1 protease and α-protein are released from the outer-membrane-embedded β-core by autoproteolytic cleavage. The recognition sequences for this self-cleavage are very similar to the target sites in human IgA1: P-A-P-|-S-P, P-P-|-S-P, and P-P-|-A-P (46). Cleavage sites were thought to be restricted to the Yaa-P-Xaa-P consensus sequence, where Yaa may be P (and less frequently P in combination with A, G, or T) and Xaa is T, S, or A (47).
It has been previously shown that the level of IgA1 protease activity varies considerably among individual strains of N. meningitidis and H. influenzae. More importantly, however, levels of enzyme activity were significantly higher in strains associated with disease than in those obtained from asymptomatic carriers (15, 65, 66). The variation in enzyme activities could be explained as the consequence of differences in iga gene sequences among individual strains. Similarly, the suggested failure of N. meningitidis to release an α-protein, in contrast to N. gonorrhoeae strains, could also be explained by the lack of the proteolysis site c in this organism. The aim of this study was to clone, sequence, and investigate the gene determinants involved in autoproteolytic self-processing of IgA1 protease precursor protein in N. meningitidis NMB. We present results showing that autoproteolysis is responsible for the release of mature proteases from wild-type and mutant enzyme precursors lacking canonical self-cleavage consensus sequences. Based on these findings, we suggest that the self-cleavage substrate specificity of IgA1 protease is far wider than previously reported. Furthermore, we present evidence that the amino acid sequence of the intervening region, between the mature IgA1 protease and the β-core translocator domain, contributes to the efficacy of autoproteolytic processing.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
N. meningitidis NMB is a serogroup B strain originally isolated from the cerebrospinal fluid of a patient with invasive meningitis and has been described previously (65). Escherichia coli strain XL1 Blue MRF {Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 Thi-1 recA1 gyrA96 relA1 lac [F′ proAb lacIqZΔM15 Tn10 (Tetr)]} (Stratagene) was used for cloning, stable propagation, and preparation of plasmid DNA. E. coli BL21 (F′ ompT hsdSB gal dcm), an OmpT-negative, protease-deficient strain, was used as a plasmid host strain for all IgA1 protease expression experiments (61). N. meningitidis NMB was grown on chocolate agar plates at 37°C under 5% CO2 in a moist atmosphere. E. coli strains were grown in LB medium supplemented with 50 μg/ml ampicillin.
Cloning of the iga gene from N. meningitidis NMB.
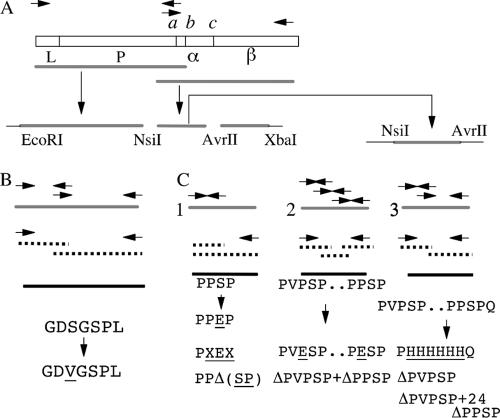
Chromosomal DNA from N. meningitidis NMB was used as the template for PCR amplification; briefly, crude genomic DNA was prepared by emulsifying a loopful of bacteria from a single chocolate agar plate into 0.5 ml of 10 mM Tris-HCl, pH 8, buffer and boiling it for 10 min. All polymerase chain amplification reactions were performed with 1 unit of high-fidelity thermostable Pfu polymerase (34) (Stratagene) and buffer supplied by the manufacturer supplemented with 200 μM of each deoxynucleoside triphosphate, 100 ng of each primer, and 100 ng of template DNA in a total volume of 50 μl. Each cycle consisted of 60 s of denaturation at 94°C, 60 s of annealing at 55°C, and extension at 74°C (2 min per 1,000 bp). The amplifications of distal and proximal parts of the iga gene were performed separately (Fig. 1A). The primers were designed based on the conserved domains of previously cloned iga genes (33, 46). The forward primer for amplification of the proximal part was designed to overlap with the sequence upstream of the N-terminal signal peptide coding sequence and incorporated the N. meningitidis ribosome binding sequence (forward primer MM1255). The reverse primer for the amplification of the distal part, MM1257, was designed to contain an XbaI restriction site. The proximal fragment of the N. meningitidis NMB iga gene was amplified by PCR using oligonucleotides MM1255 and MM1256 (primer sequences are shown in Table 1) (Fig. 1A). The reverse primer was designed to overlap with the conserved distal part of the mature IgA1 protease gene. The forward primer (MM1258) for the amplification of the distal part of the iga gene contained the overlapping sequence of the same region as the reverse primer used for the proximal amplification. The reverse primer (MM1257) was designed to overlap with the conserved DNA uptake signal sequence and also contained an XbaI restriction site. Both sets of primers were designed to amplify fragments that contained the conserved NsiI restriction enzyme site at the point of overlap. After amplification, the proximal fragment was treated with Klenow fragment and then digested with NsiI restriction enzyme. This resulted in the creation of a fragment with one blunt end and an NsiI overlap at the other. The distal fragment was treated with NsiI and XbaI restriction enzymes to create corresponding sticky ends. The plasmid pUC18 was used as a vector after appropriate digestion with Ecl136II and XbaI (68). The proximal 3-kbp and 2-kbp distal fragments were gel purified, treated with the appropriate restriction enzymes, and cloned in a triple ligation reaction into the vector pUC18. The ligation mixture was used to electroporate competent E. coli XL1 Blue MRF cells, and transformants were selected for ampicillin resistance. Plasmid DNA was isolated from individual clones by the alkaline extraction method (7) and analyzed for the presence of an extra 4.8-kbp band corresponding to the NMB iga gene. The representative plasmid was designated pIGAP15. Plasmid pLIT28-09 was constructed by excision of cleavage recognition sequences containing a 0.9-kbp NsiI-AvrII fragment from pIGAP15 and subcloning into the similarly cut plasmid vector pLITMUS 28 (14) (Fig. 1A). All other general DNA manipulations were performed as previously described (55).
FIG. 1.
Cloning and mutagenesis of Neisseria meningitidis NMB IgA1 protease. (A) Physical map of a typical IgA1 protease gene and strategy involved in PCR amplification and cloning. The protease contains a signal peptide, L; a protease domain, P; cleavage sites a, b, and c (flanking the α-protein domain, α); and an autotransporter or β-core domain, β. (B) Schematic showing the approach used for site-directed mutagenesis of the active-site serine residue using the overlap extension method. (C) Site-directed mutagenesis of self-cleavage recognition sequences involved in IgA1 protease precursor autoprocessing using PCR-based megaprimer (1) and overlap extension (2 and 3) methods. Wild-type IgA1 protease sequence, solid gray line; PCR I products, broken line; PCR II products, thick black line; vector sequence, thick black line. PCR primers are represented by arrows. For details, see Materials and Methods.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′) | Position in sequence |
|---|---|---|
| MM1255 | ACCCTTAAAACGGTAAAACCTTATG | 414-438 HF13 igaa |
| MM1256 | AGAGGATCCGGAGCGGTCTTGTACGGATGA | 3241-3261 HF13 iga |
| MM1258 | TCAGGAAGGGCTGAATCTCTTT | 3213-3234 HF13 iga |
| MM1257 | ATGCCGTCTAGAGCCTGAGTTC | 5156-5177 HF13 iga |
| MM1484 | TCACACAGGAAACAGCTATGAC | 2407-2428 Litmus 28 |
| MM1553 | TTGCCTGCGGTTCAGGCGGTGCGA | 3002-3025 NMB igab |
| MM1554 | TTGCCTGCG(ACT)TTCAG(ACT)CGGTGCGA | 3002-3025 NMB iga |
| MM1555 | TTGATTTGCCTGAGGCGGTGCGACGA | 2999-3030 NMB iga |
| MM1482 | AGGGTACCAGAGCTCACCTA | 2572-2591 Litmus 28 |
| MM1622 | TGTGGCAGGAGACTCGTCCGGTTTGACT | 2913-2940 NMB iga |
| MM1623 | AGTCAAACCGGTCGAGTCTCCTGCCA | 2913-2938 NMB iga |
| MM1625 | TGCCTGCGGGCTCTCCGGTGCGACGA | 2999-3024 NMB iga |
| MM1624 | GTCGTCGCACCGGAGAGCCCGCAGGCA | 2998-3024 NMB iga |
| MM1626 | GTGATGGTGATGGTGATGCGGTTTGACTCGGCGGCGGTT | 2902-2922 NMB iga |
| MM1627 | CATCACCATCACCATCACCAGGCAAATCAAGCCGAAGAA | 3019-3039 NMB iga |
| MM1685 | ACGCCGAATTCGAGACCCTTA | |
| MM1750 | TAATGGTGAACCGACATCGCCTAACA | 788-813 NMB iga |
| MM1759 | GTGTTAGGCGATAGCGGTTCACCA | 787-810 NMB iga |
| MM1686 | TGATGCATCAAAGAGATTCAGCCCT | 2754-2778 NMB iga |
| MM1815 | TCAAAATATCGTCGTCGCACAGGCAAATCAAGCCGAAG | 2988-3037 NMB iga |
| MM1816 | TCTTCGGCTTGATTTGCCTGTGCGACGACGATATTT | 2991-3038 NMB iga |
| MM1819 | AACCGCCGCCGAGTCAAAGCCACAAACACGGCTTCTCA | 2902-3054 NMB iga |
| MM1820 | TGAGAAGCCGTGTTTGTGGCTTTGACTCGGCGGCGGTT | 2902-2954 NMB iga |
| MM1821 | AACCGCCGCCGAGTCAAACCGCCTAGCCCGCAGGCA | 2902-3024 NMB iga |
| MM1822 | TGCCTGCGGGCTAGGCGGTTTGACTCGGCGGCGGTT | 2902-3024 NMB iga |
DNA sequence determination.
The 4.8-kbp iga insert in plasmid pIGAP15 was sequenced by the dideoxynucleotide chain termination method (56) using the primer-walking approach. Thirty-two oligonucleotides were synthesized and used for this purpose (data not shown). The identity of each single nucleotide in the sequence was determined by at least two sequencing passes in both directions. The sequences were determined using the dye termination method with an ABI Big Dye sequencing kit (Applied Biosystems, United Kingdom) and analyzed on an ABI automated sequencing machine (Applied Biosystems). DNA sequence analysis was performed using MacVector and AssemblyLIGN (International Biotechnologies, Cambridge, United Kingdom) software packages.
Site-directed mutagenesis.
Inactivation of the active-site serine (267S) was accomplished by replacing it with a valine residue using the overlap extension method (22, 23) (Fig. 1B). Plasmid pIGAP15 was used as a template. The first PCR amplified (PCR I) the proximal 851-bp fragment (primers MM1685 and MM1750) and the 1,993-bp distal fragment (primers MM1759 and MM1686) of the cloned iga gene. The second PCR (PCR II) generated the 2,816-bp fragment using the flanking vector polylinker-specific primer MM1685 and iga-specific NsiI overlapping primer MM1686. PCRs I and II were performed with 15 cycles of amplification only as described above.
Mutants of the cleavage recognition sequence b (1003P-P-S-P) were constructed by site-directed mutagenesis using the PCR-based megaprimer method (30) (Fig. 1C). The mutagenic primers MM1553 (coding for replacement of serine with glutamic acid at position 1005), MM1554 (coding for replacement of serine with glutamic acid at position 1005 and containing degenerate codons instead of proline codons at positions 1004 and 1006), and MM1555 (coding for a deletion of serine and proline at positions 1005 and 1006) were used in combination with the Litmus 28 vector polylinker overlapping primer MM1484 (Table 1). The 330-bp PCR products generated were gel purified, concentrated, and used in PCR II. These products served as large forward primers (megaprimers), together with the reverse vector polylinker overlapping primer MM1482, to amplify the full-length 906-bp fragment. This product was used for cloning replacement of the corresponding wild-type segment in pIGAP15.
Construction of the double proline mutant containing glutamic acid instead of 976P and 1004P residues in recognition sequences a and b, respectively, was performed by the PCR-based overlap extension method (22, 23) (Fig. 1). Primer pairs MM1484-MM1622, MM1623-MM1625, and MM1624-MM1482 were initially used for PCR I to generate three overlapping segments of 245, 114, and 606 bp. Overlaps between MM1622 and MM1623, together with complementary overlaps between MM1625 and MM1624, created the desired mutations. The purified PCR products were used as an overlap template for PCR II, which was performed with the vector polylinker-specific primers MM1484 and MM1482. The mutated 906-bp fragment obtained was used for replacement cloning of the corresponding wild-type segment.
Mutants containing a complete deletion of putative self-cleavage recognition site b or site a, deletion of site a and all 24 amino acids to site b, or simultaneous deletion of sites a and b were constructed using the same overlap extension method (Fig. 1). Deletion of site b alone was created using primer pairs MM1484-MM1816 and MM1815-MM1482 to generate PCR I products. Primers MM1815 and MM1816 both contained central deletions spanning nucleotides 5272 to 5283 (encoding the 1003PPSP segment). In the second reaction, purified PCR I products were used as templates and amplification was completed with the external vector-specific primers MM1484 and MM1482. The PCR II product obtained was finally used to replace the wild-type segment of the cloned NMB iga gene. Removal of site a, including 24 amino acids spanning region 974P to 1002A, was accomplished using an identical strategy and internal primers MM1820 and MM1821. Deletion of both recognition sequences with a preserved intervening 24-amino-acid region was completed using three sets of primers: MM1484-MM1820, MM1819-MM1816, and MM1815-MM1482. Three PCR I products were again used in PCR II, and the final product was used in replacement cloning to generate a mutant lacking recognition sites a and b (Δ974PVPSP plus Δ1003PPSP).
The overlap extension method was used again to construct a deletion mutant IgA1 protease lacking both cleavage recognition sequences and the intervening segment (975V to P1006) (Fig. 1C). The central primers MM1626 and MM1627 were designed to overlap via 18 nucleotides coding for six histidines, thus replacing the deleted region with the histidine tag motif. These primers were used in combination with the above-described MM1484 and MM1482 to amplify the 300-bp and 600-bp fragments of the cloned wild-type 906-bp NsiI-AvrII fragment of the iga gene. The annealed and overlapped products were used as a template for PCR II, which produced the 906-bp mutated fragment.
Linker domain replacement cloning.
Variable linker domain replacement cloning was performed by PCR amplification of this region from two different N. meningitidis strains and subsequent introduction of these amplified segments into the wild-type backbone of a cloned NMB iga gene lacking the natural linker domain. We wanted to use the naturally available restriction sites to perform these manipulations in order to minimize eventual disruption of open reading frames and creation of artificial sequence strings. For this purpose, we randomly selected 10 previously described N. meningitidis strains with reduced IgA1 protease activity. All of these strains were then tested for the presence of NsiI and AvrII restriction enzyme sites flanking the linker domain, as in the case of a cloned iga gene from N. meningitidis NMB. Two sets of primers, MM1892-MM1865 and MM1896-MM954, were used to analyze their potential for generating PCR products. The successfully amplified products were then partially sequenced to determine the availability of the above-mentioned restriction enzyme sites. Based on these criteria, two strains, SVG69 and SVG70, were selected. After amplification using the MM1892-MM1865 primer pair, the PCR products were gel purified, digested with NsiI and AvrII restriction enzymes, and cloned in identically cut pIGAP15 backbones. Three mutants containing SVG69 (M11) and SVG 70 (M12) linker regions were selected for measurement of IgA1 protease activity.
Induction and preparation of bacterial culture supernatants.
Cells from an overnight 3-ml culture (0.1 ml) were used to inoculate a 100-ml flask containing 20 ml of LB medium with 50 μg/ml ampicillin. The culture was grown to an A550 of 0.5 (mid-log phase). At that stage, expression of IgA1 protease was induced by the addition of an appropriate amount of IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration, 0.5 mM). The incubation was continued for 3 h, and the bacterial cells were pelleted by centrifugation. The resultant supernatant was filtered through a 0.22-μm disposable filter unit (Gelman), producing cell-free media for measurement of mature IgA1 protease levels by activity- and enzyme-linked immunosorbent assay (ELISA)-based methods.
Protein preparation.
Induced bacterial cultures were centrifuged to pellet the bacteria. The supernatant, consisting of spent cell-free medium, was filtered through a 0.22-μm filter as described above and transferred to a Vivaspin ultrafiltration device (10,000-molecular-weight cutoff membrane; Vivascience, Hannover, Germany) and concentrated 100-fold. The bacterial cell pellet was lysed by sonication and centrifuged to produce a clarified supernatant (containing soluble cytoplasmic and periplasmic proteins), which was separated from the insoluble fraction by high-speed centrifugation in a Beckman SW50.1 rotor at 20,000 rpm for 1 h. This remaining insoluble fraction was used to obtain outer membrane proteins using the Sarkosyl (BDH Laboratory Supplies) extraction method as described previously (11).
Protein electrophoresis and blotting.
Outer membrane protein fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (29) using 10% polyacrylamide resolving gel and blotted onto BioTrace PVDF membrane (PALL Life Sciences) using a Trans-blot SD semidry transfer cell (Bio-Rad) according to the manufacturer's instruction manual. After Coomassie staining and drying, the excised bands were used for N-terminal amino acid sequencing. N-terminal sequences were determined by J. N. Keen at the Protein Sequencing Facility, School of Biochemistry and Molecular Biology, University of Leeds, Leeds, United Kingdom.
IgA1 protease assays.
The measurement of IgA1 protease activity was performed by a quantitative ELISA as described previously (51, 65). One unit of enzyme activity was defined as the amount of enzyme able to effect a change in optical density of 1 absorbance unit (at 490 nm) in 1 h at 37°C. A second independent alternative method to monitor the expression of IgA1 protease in culture supernatants was developed based on the ability of mouse monoclonal antibody AH207 to react with bacterial IgA1 protease (36). We used this antibody in a separate ELISA to measure the amount of IgA1 protease produced, in parallel with the determination of IgA1 protease activity. Bacterial cell supernatants prepared as described above (50 μl) were mixed with 50 μl of carbonate-bicarbonate coating buffer, pH 9, and the mixture was used to cover microtiter plate wells (Immulon 2; Dynex). The plates were incubated overnight at 37°C with continuous shaking. The wells were washed using the same buffer and protocol described for the IgA1 protease activity ELISA. In the second step, the anti-IgA1 protease antibody AH207 (1:250 dilution; 0.1 ml per well) was added, and incubation continued for 2 h at 37°C. Finally, after being washed, the plates were incubated with secondary antibody (peroxidase-conjugated rabbit anti-mouse antibody; 1:500 dilution; 0.1 ml per well; DAKO). The plates were developed using the chromogenic substrate o-phenylenediamine dihydrochloride (Sigma), and the intensity of color was recorded at 490 nm. The amount of IgA1 protease was calculated as the difference in the signal intensity between the recorded value for each culture supernatant sample and the control (wells coated with LB-coating-buffer mixture only). Assays were performed in triplicate.
RESULTS
Sequence analysis of the NMB iga gene.
Analysis of the nucleotide sequence of the cloned iga gene in plasmid pIGA15 revealed a 4,659-bp open reading frame encoding a protein of 1,552 amino acids containing typical attributes of IgA1 protease precursor proteins (GenBank accession number, AF235032): the 27-amino-acid amino-terminal leader peptide, the mature protease domain, and the carboxyl-terminal β helper domain. The mature protease and β-core domains of the cloned iga gene proved to be highly homologous to the previously described Neisseria IgA1 protease genes. Several complete Neisseria iga gene sequences were available for comparison with our NMB iga gene, as well as their encoded proteins (NC_003116 encoding NP_283693, X82474 encoding CAA57857, and NC_003112 encoding NP_273742 from meningococcal strains Z2491, HF13, and MC58, respectively, and X04835 encoding P09790 from a gonococcus MS11 isolate). The overall nucleotide sequence identity was high, e.g., 95% for N. meningitidis HF13 (33) and 94% for N. gonorrhoeae MS11 (46) (data not shown). High levels of homology (96 to 99% identity at the nucleotide level) were identified when the available mature protease gene fragment sequences from nine N. meningitidis strains (52) were used for comparison (data not shown). Ninety-seven to 98% identity was apparent compared with published β-core sequences of three Neisseria strains (19) (data not shown). The most variable region of the NMB iga gene corresponds to residues 1007 to 1225, containing the α-protein domain, a region linking the highly conserved protease and β-core domains (Fig. 2).
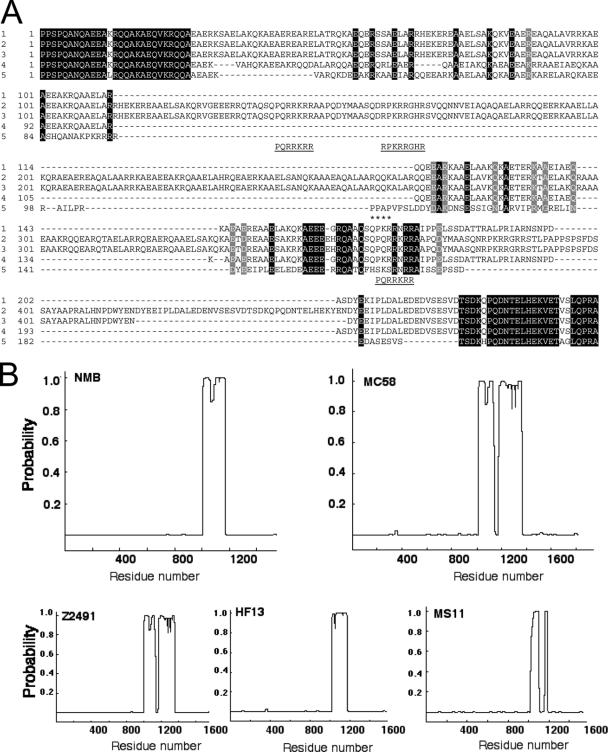
FIG. 2.
Sequence analysis of α-protein regions from N. meningitidis IgA1 protease sequences. (A) Multiple-sequence alignment of IgA1 protease sequences from N. meningitidis (1) NMB (AAK15023), (2) MC58 (NP-273742), (3) Z2491 (NP-283693), and (4) HF13 (CAA57857) and from (5) N. gonorrhoeae MS11 (P09790). The numbering of the α-proteins is relative to recognition site b (1003 in AAK15023). The position of the autoproteolytic site c in the MS11 sequence is also shown (marked by asterisks). Invariant amino acids sequences are highlighted in black and similar residues in gray. Putative nuclear localization sequences are shown underlined beneath the corresponding section of the alignment. (B) Coiled-coil prediction was carried out for the IgA1 proteases encoded by the indicated strains of N. meningitidis aligned in panel A using the Paircoil server (see Materials and Methods). The probability of each part of the amino acid sequences forming a coiled coil is represented.
The conserved segment, G-D-S-G-S-P-L, characteristic of bacterial serine-type IgA1 protease (3), was also identified as corresponding to residues 265 to 271. The typical cleavage recognition sequence involved in autoprocessing was identified as P-P-S-P (residues 1003 to 1006). This sequence corresponds to the previously described cleavage site b of N. gonorrhoeae MS11 (46). A second putative recognition sequence, P-V-P-S-P (residues 974 to 978), was also present in the NMB sequence (cleavage site a according to the N. gonorrhoeae MS11 labeling). The NMB enzyme lacked an obvious cleavage site c, located downstream of a and b, as in the MS11 IgA1 protease. Protein secondary-structure analysis using the Paircoil server (http://paircoil.lcs.mit.edu/cgi-bin/paircoil) (6) predicted the existence of a 165-amino-acid α-helical coiled-coil domain corresponding to residues 1007 to 1171 in the NMB sequence (Fig. 2B). This predicted helical segment was followed by a potential nuclear localization signal, P-K-R-R-N-R-R, at positions 1172 to 1178 (12). It was shown previously that the α-helical structure and the existence of nuclear localization signals are characteristic of N. gonorrhoeae α-proteins (48). It was also suggested that these α-helices might form coiled-coil structures. The same type of structure analysis indeed predicted the existence of an 87-amino-acid coiled-coil structure between positions 1021 and 1107 of the N. gonorrhoeae MS11 α-protein (Fig. 2). In previously examined N. gonorrhoeae strains, these peptides were released together with the mature IgA1 protease as a result of precursor autoprocessing. This was possible due to the presence of cleavage sites b and c flanking the α-helical region in those isolates. Previously, it had been reported that N. meningitidis strain HF13 does not contain a released α-domain equivalent to the gonococcal α-protein, as it lacks the autoproteolytic cleavage site c (33). Secondary-structure prediction analysis again suggested the existence of an α-helical coiled-coil region spanning 156 amino acids (positions 1016 to 1172) in N. meningitidis HF 13 IgA1 protease (Fig. 2), followed by a nuclear localization signal.
Expression of the cloned NMB iga gene.
To examine the selected clones for IgA1 protease production, we used two different experimental strategies. The first approach was based on a sensitive quantitative ELISA in which the supernatants of liquid cultures were tested for IgA1 protease activity using human IgA1 as a substrate (51, 65). The second strategy used an ELISA in which the amount of IgA1 protease in the supernatant was measured by titration with mouse monoclonal anti-IgA1 protease antibody (see Materials and Methods). One of the clones producing IgA1 protease verified by both criteria, E. coli XL1 Blue MRF(pIGA15), was used for detailed investigations. Plasmid DNA was isolated from this clone and used to transform E. coli BL21. E. coli BL21(pIGA15) was grown in liquid culture under different induction regimes, and IgA1 protease expression was measured by both methods. Readily detectable levels of IgA1 protease were obtained following induction with 0.5 mM IPTG for 3 h (data not shown). SDS-PAGE analysis of the supernatant revealed that mature IgA1 protease was present, and N-terminal sequence analysis of the membrane-embedded β-core showed that processing had occurred within the b site (P-P- -S-P) after residue 1004.
Effect of point mutations in the IgA1 protease autoproteolytic recognition sequence PPSP.
To investigate whether the recognition sequence 1003P-P-S-P1006 is indeed the target for autoproteolytic processing, we constructed a range of IgA1 protease mutants with alterations in this region. These mutations included replacing Ser1005 with Glu, a relatively gross change to the site in terms of both steric and charge considerations, in order to assess how tolerant the protease is of changes in its recognition sequence. We also altered the −1 and +2 proline residues and deleted the +1 and +2 residues to determine whether these changes would affect autoproteolysis. The PCR-based site-directed mutagenesis megaprimer method was used (30). Three classes of mutants were constructed: (i) M1, a mutant containing glutamic acid instead of wild-type serine at position 1005; (ii) M2, a mutant containing glutamic acid instead of wild-type serine at position 1005 and threonine and alanine substitutions for prolines at positions 1004 and 1006; and (iii) M3, a mutant with residues 1005S-P1006 deleted (Fig. 1). A double mutant (M4) of IgA1 protease containing glutamic acid instead of proline at the −1 position in the potential recognition sequence (Pro976Glu) and the confirmed site of processesing (Pro1004Glu) was constructed using the PCR-based overlap extension method (22, 23) (Fig. 1). The presence of all the desired mutations was confirmed by DNA sequencing.
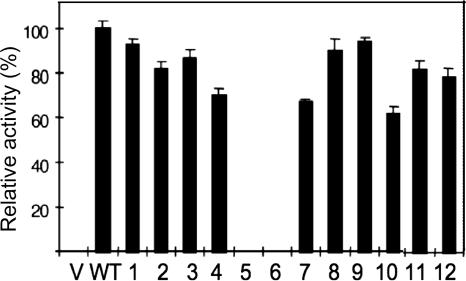
In order to examine IgA1 protease expression in clones containing the mutants of the cloned iga gene, we performed all experiments in parallel with two control strains. The host strain, E. coli BL21, containing vector plasmid, was used as a negative control, and E. coli BL21(pIGA15), producing wild-type enzyme, was used as a positive control. All strains were grown in parallel until the cultures reached an A550 of 0.5, at which point the lac promoter was induced with 0.5 mM IPTG for 3 h. The supernatants were harvested and tested for the presence of IgA1 protease using the activity assay. The results of these experiments are presented in Fig. 3. To our surprise, mutants M1, M2, and M3 containing modified consensus recognition sites showed only slight reductions in protease secretion. Even more surprisingly, the M4 mutant (with glutamic acid rather than proline at the −1 position in both sites a and b) was still found in the mature, secreted form. However, this mutant showed the biggest drop in secreted protease levels (ca. 30% lower than the wild type).
FIG. 3.
Expression of N. meningitidis NMB IgA1 protease wild type (WT) and mutants (1 to 12, corresponding to mutants M1 to M12). Enzyme activity was measured by quantitative ELISA with human IgA1 as a substrate, as described in Materials and Methods. V, supernatants from E. coli BL21 containing vector plasmid only (no iga gene) as negative controls showed no activity. The error bars indicate standard errors of the means.
The consequences of deleting the putative a and confirmed b self-cleavage recognition sites.
As our initial mutations failed to abolish secretion of the mature protease, we undertook more drastic surgery on the target sites. Mutants lacking the confirmed self-cleavage recognition site b (Δb; M7), lacking putative site a (Δa; M8), or lacking site a and the intervening residues between sites a and b (Δa27; M9) and a double mutant lacking both sites (Δa/Δb; M10) were constructed using the overlap extension PCR method and replacement cloning. Details of the strategies involved in their construction are described in Materials and Methods and in Fig. 1C. When we examined the supernatants of induced cultures of BL21 carrying the plasmids pIGAPM7-M10, we found a surprising amount of secreted IgA1 protease in the media (Fig. 3 and 4). Secretion was reduced by only 40% in the double-deletion mutant M10 (Δa/Δb), with M7 to M9 showing less reduction of secreted protease. The only recognition site mutant that failed to produce detectable secreted protease in culture supernatants was M5, the mutant in which the entire region spanning sites a and b was replaced with a hexahistidine linker. This mutant did produce the IgA1 preprotein, as could be seen from the outer membrane preparation from the construct (Fig. 4).
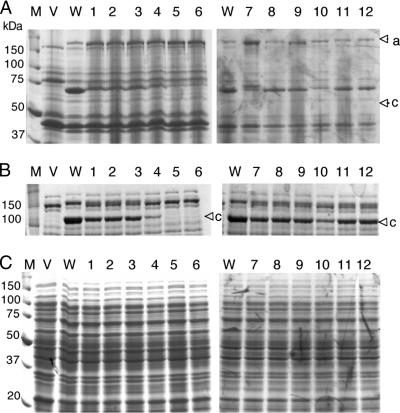
FIG. 4.
SDS-PAGE analysis of self-cleavage processing of wild-type and mutant IgA1 proteases of N. meningitidis NMB. (A) Accumulation of β-core fragments in outer membrane fractions as an indication of IgA1 protease self-cleavage activity in recombinant E. coli expressing IgA1 protease and its mutants. The upper arrowhead (a) denotes the position of the preprotein, while the lower arrowhead (b) shows the membrane-associated β-core fragment after self-cleavage. (B) Analysis of spent cell-free medium fractions concentrated 100 times. The arrowhead (c) denotes the mature processed IgA1 protease. (C) Analysis of cytoplasmic and periplasmic fractions. Lanes: M, molecular mass standards; V, E. coli BL21 host strain containing vector plasmid only; WT, clone producing wild-type IgA1 protease; 1 to 12, clones producing mutant variants of IgA1 protease enzyme. Table 1 shows details of mutant M1 to M12 sequences, and Fig. 3 shows quantitative determination of secreted IgA1 protease activity.
An IgA1 protease mutant with a disabled active site.
It has been shown that mutagenesis of the active-site serine in the H. influenzae protease resulted in the loss of IgA1 protease release due to a block in protein autoprocessing (unpublished result quoted previously in references 3 and 50). The deduced amino acid sequence of the cloned iga gene of N. meningitidis NMB contains the conserved active-site serine (italicized) in the characteristic sequence GDSGSPL (3). Thus, the Ser267Val mutant (M6) was constructed to investigate whether the active-site serine is required for self-cleavage and the subsequent extracellular release of mature neisserial IgA1 protease. It also allowed us to determine whether endogenous E. coli proteases might be able to cleave the IgA1 protease precursor. We used the PCR-based overlap extension method to replace the active-site serine 267 with a valine residue (Fig. 1B).
No free extracellular Ser267Val mutant protein was detectable by SDS-PAGE analysis, indicating that this mutant failed to undergo self-cleavage. However, it could be clearly seen in the high-molecular-weight, membrane-bound precursor form in outer membrane preparations and was absent from the cell supernatant of BL21(pIGAPM6), as shown by SDS-PAGE analysis (Fig. 4). Thus, it appears that successful secretion of the IgA1 protease does indeed occur via an autoproteolytic mechanism, as expected.
Determination of autoproteolytic self-cleavage sites for wild-type and mutant enzymes.
The exact site of self-cleavage for each mutant preprotein was determined by N-terminal amino acid sequencing of the β-core fragment, which remains embedded in the bacterial outer membrane after self-cleavage and release of the mature form of IgA1 protease. The protein preparation for each mutant strain, along with the experimentally determined self-cleavage site, is shown in Fig. 4 and Table 2. Figure 4 shows accumulation of processed β-core fragments (top), mature IgA1 proteases (middle), and combined cytoplasmic and periplasmic fractions (bottom) of the wild-type and all mutant NMB enzymes. Under the same conditions of induction and with similar levels of total protein loading, it is apparent that wild-type protease is the most efficiently produced (see the quantitative analysis in Fig. 3 and in Fig. 4A and B, lanes W). The levels of processed β-core in mutant strains M1 to M4 and M7 to M12 are lower than in the wild type, with reciprocal increases in levels of unprocessed preprotein apparent in these strains. These results are in good correlation with the accumulation of free, mature IgA1 protease in the media (Fig. 4B), as well as with the quantitative results described above (Fig. 3). The efficacy of autoprocessing is most affected in mutants M4, M7, and M10, again in agreement with enzyme assay data (Fig. 3). The cytoplasmic fraction of all strains tested appeared indistinguishable from one another, ruling out the possibility that some of the mutant proteases had accumulated in the cytoplasm. The exact site of self-cleavage in mutants M1 to M4, M8, and M9 (with alterations in sites a and b or deletion of site a) resides in site b, the same site that is the target for wild-type enzyme (Table 1). Deletion of site b results in cleavage at three new points. Two of those are in the target site a, and the third, after 996P, between sites a and b. The further removal of site a in M10 (a mutant with both sites deleted) results in the single cleavage point after 996P in the sequence 994A-K-P-|-Q-N-I. This target clearly deviates from all previously identified IgA1 protease cleavage sites.
TABLE 2.
N. meningitidis NMB wild-type and mutant IgA1 proteases, their putative self-cleavage targets, and determined sites of self cleavage
| Protease | Putative wild-type and mutated self-cleavage sitesa | N-terminal sequence of outer membrane embedded β-core | Context of deduced cleavage site (*) |
|---|---|---|---|
| Wild type | 1003P-P-S-P | 1005S-P-Q-A-N | 1001V-A-P-P-*-S-P-Q-A |
| M1 | 1003P-P-E-P | 1005E-P-Q-A-N | 1001V-A-P-P-*-E-P-Q-A |
| M2 | 1003P-T-E-A | 1004T-E-A-Q-A | 1001V-A-P-*-T-E-A-Q-A |
| M3 | 1003P-P-(S-P deleted) | 1007Q-A-N-Q-A | 1001V-A-P-P-*-Q-A-N-A |
| M4 | 974P-V-E-S-P 1003P-E-S-P | 1004E-S-P-Q-A | 1001V-A-P-*-E-S-P-Q-A |
| M5 | 974P-H-H-H-H-H-H-Q | NDc | |
| M6b | 1003P-P-S-P | ND | |
| M7 | Δ1003P-P-S-P (Δ b) | 977S-P-A-T-N | 974P-V-P-*-S-P-A-T |
| 979A-T-N-T-A | 976P-S-P-*-A-T-N-A | ||
| 997Q-N-I-V-V | 994A-K-P-*-Q-N-I-V | ||
| M8 | Δ974P-V-P-S-P (Δ a) | 1005S-P-Q-A-N | 1001V-A-P-P-*-S-P-Q-A |
| M9 | Δ974P-1002A (Δ a+24) | 1005S-P-Q-A-N | 1003P-P-*-S-P-Q-A |
| Δ974P-V-P-S-P (Δ a) | |||
| M10 | + | 997Q-N-I-V-V | 994A-K-P-*-Q-N-I-V-V |
| Δ1003P-P-S-P (Δ b) |
Δ, deleted site.
Active-site serine (267S) mutant.
ND, not detected.
Construction of mutant IgA1 proteases containing heterologous linker domains.
Mutant IgA1 proteases containing linker domain swaps and encoding the variable α-protein domains from the two N. meningitidis strains SVG69 and SVG70 were created (65). They were subsequently introduced into the wild-type backbone of the cloned NMB iga gene instead of its natural linker domain. This was facilitated by the presence of naturally available NsiI and AvrII restriction sites flanking the linker regions in these strains. Thus, the construction of domain-swapped mutants containing no extraneous residues or frameshifts was possible. These chimeric IgA1 protease variants were characterized by expression levels mirroring their wild-type parental IgA1 proteases. For example, the levels of activity in M11 (SVG69 linker) and M12 (SVG70 linker) were reduced by 18% and 22%, respectively, compared to the cloned wild-type enzyme. This trend reflects the observed reductions in activity levels for the parental N. meningitidis strains SVG69 (27%) and SVG70 (49%) compared to N. meningitidis strain NMB (65).
DISCUSSION
In this study, we present data demonstrating that the IgA1 protease of N. meningitidis NMB possesses a far broader self-cleaving specificity than the initially defined consensus recognition sequence. This was illustrated by expression, autoproteolytic processing, and release of the mature enzyme into the growth medium by mutant strains with altered wild-type recognition sequences involved in autoproteolytic cleavage.
We created a range of iga gene mutants with altered internal cleavage recognition sequences to confirm that the identified sequences do indeed represent the sites of autoproteolytic processing necessary for the final release of mature IgA1 protease. The first set of mutants contained the mutated 1005S, deletion 1005S-P1006, or mutations of three adjacent amino acids (1004P-S-P1006) in recognition sequence b. We were able to detect IgA1 protease in the culture supernatants of all these mutants, based on IgA1 protease activity. However, the level of enzyme production was only slightly lower than in the wild-type clone. We also identified a second potential recognition sequence a (974P-V-P-S-P978) 24 amino acids upstream of the first. Although this sequence fulfils the basic rule of the consensus recognition sequence (prolines at positions −1, −3, and +2) (47), the cleavage of a sequence with valine at position −2 has not been reported previously. Creation of the double proline mutant (−1 position prolines in both recognition sequences replaced with glutamic acid) again did not abolish the release of IgA1 protease into the culture supernatants, although the level of enzyme activity was further reduced compared to the mutants examined earlier.
We created more extensive mutations to further investigate N. meningitidis NMB IgA1 protease autoprocessing after the apparent and unsuspected detection of self-cleavage susceptibility in the first set of mutants. Mutants containing the complete deletion of the putative self-cleavage recognition sites were constructed. Again, these mutants were able to undergo successful autoprocessing and to release mature IgA1 protease. The level of detected activity was again reduced in comparison to the wild type, with Δb (M7) and Δa plus Δb (M10) mutants being the most affected. This cleavage could be explained either by the presence of alternative IgA1 protease-susceptible sites in the precursor molecule or by proteolysis with endogenous E. coli outer membrane proteases (13). The lack of secreted IgA1 protease in the supernatant of the active-site serine mutant eliminates the possibility that release of IgA1 protease from our mutants is a consequence of endogenous E. coli outer membrane protease action. Furthermore, all our expression experiments were performed in the outer membrane protease-deficient BL21 strain (OmpT−) of E. coli. However, an additional mutant was created to investigate whether IgA1 protease secretion in the mutant clones is indeed the consequence of authentic enzymatic autolysis from the outer-membrane-embedded β-core domain. The iga mutant M5 containing the 32-amino-acid deletion and lacking all potential cleavage recognition sequences (975V-P1006) was produced. This mutant failed to produce any detectable level of mature IgA1 protease expression in the culture supernatants. This suggests that the fragment 975V-P1006 contains all potential cleavage sites for N. meningitidis NMB IgA1 protease autoprocessing. This mutant was indistinguishable with regard to its IgA1 protease expression from the mutant created by the replacement of the active-site serine residue with valine (M6). These results indicate that self-cleavage is an inherent property of several of the mutant IgA1 protease precursor molecules and that experiments in the E. coli BL21 background represent an appropriate alternative to N. meningitidis as the natural host for studying autoprocessing.
This is the first report of IgA1 protease activity acting on substrates lacking the previously defined consensus recognition sequence. IgA1 protease has been reported to act on chimeric substrates containing the previously reported IgA1 protease cleavage recognition sequence inserted into recombinant proteins. Cleavage of the N-terminal part of the bacteriophage MS2 polymerase-β-domain-IgA1 protease fusion protein at the recognition sequence linking the two domains has been demonstrated, using purified IgA1 protease (47). Cleavage was also demonstrated on a tripartite fusion protein consisting of the N-terminal part of ms2 polymerase, the cholera toxin B subunit, and the IgA1 protease β-domain. The cholera toxin B domain in this fusion was flanked by two IgA1 protease recognition sequences. Cleavage of human CD8 protein at the naturally occurring IgA1 protease recognition sequence was also demonstrated when this protein was part of the fusion protein. In addition, successful IgA1 protease-mediated cleavage was obtained using mutant endonuclease A from Cellulomonas fimi, in which the natural linker connecting the two domains was replaced with the hinge sequence of human IgA1 (35). In all these examples, cleavage has been demonstrated on purified proteins. Attempts to use purified IgA1 protease to induce cleavage of the CD8 protein, which is naturally exposed on the T-cell surface, was not successful (47). This suggests that the recognition sequence alone is not sufficient for IgA1 protease cleavage and that other factors, such as the three-dimensional conformation within and around the recognition sequence, may also play important roles. This has also been illustrated by simultaneous cleavage at two different naturally occurring recognition sequences in the hinge region of human IgA1 when substituted for the linker region in a chimeric C. fimi endonuclease A construct (35). This cleavage pattern was detected in spite of IgA1 proteases being previously characterized by a single recognition sequence specificity for human IgA1 (37, 38). In experiments with recombinant IgA, in which the hinge regions of IgA1 and IgA2 were swapped, it was found that IgA1(hinge2) was not susceptible to cleavage by N. gonorrhoae IgA1 protease whereas IgA2(hinge1) was cleaved (10). In similar experiments, it was shown that bacterial IgA proteases are capable of cleaving mutated, synthetic forms of the hinge region of recombinant human IgA molecules (57, 58).
Instead of introducing the same recognition sequence into different protein backbones (as shown with the recognition sequence containing fusion proteins mentioned above), we have applied a different strategy and created a situation in which a novel target sequence, i.e., a mutated recognition sequence, was introduced into the same enzyme backbone. Similarly, we also performed linker domain swaps involving the insertion of two different N. meningitidis strain linker domains into the same NMB IgA1 protease background. E. coli cells producing chimeras with linkers from N. meningitidis SVG69 and SVG70 substituted (65) displayed reduced levels of secreted protease compared with E. coli cells expressing wild-type NMB IgA1 protease. This correlates with the IgA1 protease levels seen in the neisserial isolates themselves (NMB > SVG69 > SVG70).
This suggests the possibility that IgA1 protease secretion levels could be influenced at the level of posttranslational autoprocessing. The precise amino acid composition and the three-dimensional structure of the intervening region between the mature IgA1 protease and the conserved β-core translocator domain could contribute to the efficacy of autoproteolytic processing. This region is the most polymorphic of iga gene domains (50). Polymorphism arises during the process of natural transformation of Neisseria strains (60) and subsequent homologous recombination of DNA segments carrying the IgA1 protease gene (19, 32, 33).
Apart from regulating the amount of mature enzyme released, the processing pattern could also regulate the fate of the coexpressed α-protein. It has been shown that α-protein from N. gonorrhoeae MS11 has the ability to migrate into the nucleus of human epithelial cells (48). Neisserial α-proteins possess functional nuclear localization signals and extended regions of the predicted α-helical structure, including hydrophobic heptad repeats resembling leucine zippers. Such features are found in some eukaryotic transcription factors (20, 43). These findings suggest that α-proteins may act as regulators of host cell functions or as carriers for IgA1 protease nuclear transport (48). In the latter case, the proteases could be involved in enzymatic cleavage of target nuclear proteins. According to our results, the autoprocessing pattern of the IgA1 protease of N. meningitidis NMB appears to differ from that previously reported in N. gonorrhoeae MS11 (46). Instead of being released as a free protein, the α-protein remains associated with the cell membrane due to lack of the corresponding c recognition site. This lack of cleavage site c was also reported for N. meningitidis HF13 IgA1 protease (33). This suggests an autoprocessing pattern similar to that of our IgA1 protease.
Two groups have demonstrated that N. gonorrhoeae IgA1 protease is able to cleave the human lysosome-associated membrane protein LAMP-1 that forms a protective lining enclosing terminal phagolysosomes (21, 31). It is important to note that human LAMP-1 contains a proline/serine-rich region similar to the hinge region of human IgA1 (including potential IgA1 protease cleavage sites) (64). The destruction of this membrane could result in bacterial escape from phagolysosomes (1). Once in the cell cytoplasm, bacteria could start interfering with normal nucleocytoplasmic transport mediated by NLS on the surface-exposed α-proteins. What could be the potential consequence of surface expression of α-peptides in Neisseria meningitidis? It is tempting to speculate that surface-exposed nuclear localization signal sequences could act as a decoy device for the integrin family of cytoplasmic receptors necessary for normal transport into the cell nucleus (18, 40). An example of a similar type of interference has been reported for a viral protein (16). It has been shown that the human immunodeficiency virus type 1 auxiliary protein Vif contains the sequence 90RKKR93, similar to that of the prototypic nuclear localization signal, which is directly involved in inhibition of nuclear transport via the importin pathway. An alternative role of these surface-exposed α-proteins could be a more specific interaction with particular eukaryotic cellular components, with the same ultimate goal—enhanced survival of the bacterium.
The work presented here shows that neither the −2 nor the +2 proline is essential for autocleavage and that both the +1 and +2 positions will tolerate quite major amino acid substitutions. This suggests the possibility that there may be a far wider range of potential IgA1 protease targets than previously suspected. The search for novel substrate candidates has until now been restricted to the homology search approach to discover proteins containing the same cleavage recognition sequence (48, 65). However, two proteins lacking a classical IgA1 protease recognition site are known to be cleaved by this proteinase: the type II tumor necrosis factor receptor (5) and human lysosome-associated membrane protein 2 (2). The cleavage of these proteins by IgA1 protease may be readily understood by taking into account the results presented in our present study showing that IgA1 protease has a wider substrate specificity than previously appreciated. Thus, it may be that IgA1 protease has a number of as-yet-undiscovered targets, implying a more extensive role for this enzyme at the host-pathogen interface than hitherto suspected.
Acknowledgments
We thank Mark Achtman for supplying hybridoma-producing anti-IgA1 protease antibody.
We thank the Meningitis Research Foundation and Interleukin Genetics Inc., Waltham, MA, for financial support.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Apicella, M. A., M. Ketterer, F. K. Lee, D. Zhou, P. A. Rice, and M. S. Blake. 1996. The pathogenesis of gonococcal urethritis in men: confocal and immunoelectron microscopic analysis of urethral exudates from men infected with Neisseria gonorrhoeae. J. Infect. Dis. 173:636-646. [DOI] [PubMed] [Google Scholar]
- 2.Ayala, P., L. Lin, S. Hopper, M. Fukuda, and M. So. 1998. Infection of epithelial cells by pathogenic Neisseriae reduces the levels of multiple lysosomal constituents. Infect. Immun. 66:5001-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachovchin, W. W., A. G. Plaut, G. R. Flentke, M. Lynch, and C. A. Kettner. 1990. Inhibition of IgA1 proteinases from Neisseria gonorrhoeae and Haemophilus influenzae by peptide prolyl boronic acids. J. Biol. Chem. 265:3738-3743. [PubMed] [Google Scholar]
- 4.Beck, S., J. A. Le Good, M. Guzman, N. Ben Haim, K. Roy, F. Beermann, and D. B. Constam. 2002. Extraembryonic proteases regulate nodal signalling during gastrulation. Nat. Cell. Biol. 4:981-985. [DOI] [PubMed] [Google Scholar]
- 5.Beck, S. C., and T. F. Meyer. 2000. IgA1 protease from Neisseria gonorrhoeae inhibits TNF alpha-mediated apoptosis of human monocytic cells. FEBS Lett. 472:287-292. [DOI] [PubMed] [Google Scholar]
- 6.Berger, B., D. B. Wilson, E. Wolf, T. Tonchev, M. Milla, and P. S. Kim. 1995. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrion, R., Jr., Y. T. Ro, and J. L. Patterson. 2003. Purification, identification, and biochemical characterization of a host-encoded cysteine protease that cleaves a leishmaniavirus Gag-Pol polyprotein. J. Virol. 77:10448-10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Childers, N. K., M. G. Bruce, and J. R. McGhee. 1989. Molecular mechanisms of immunoglobulin A defense. Annu. Rev. Microbiol. 43:503-536. [DOI] [PubMed] [Google Scholar]
- 10.Chintalacharuvu, K. R., P. D. Chuang, A. Dragoman, C. Z. Fernandez, J. Qiu, A. G. Plaut, K. R. Trinh, F. A. Gala, and S. L. Morrison. 2003. Cleavage of the human immunoglobulin A1 (IgA1) hinge region by IgA1 proteases requires structures in the Fc region of IgA. Infect. Immun. 71:2563-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, R. L., R. A. Wall, and S. P. Borriello. 1990. Comparison of methods for the analysis of outer-membrane antigens of Neisseria meningitidis by Western blotting. J. Immunol. Methods 134:215-225. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 13.Enfors, S. O. 1992. Control of in vivo proteolysis in the production of recombinant proteins. Trends Biotechnol. 10:310-315. [DOI] [PubMed] [Google Scholar]
- 14.Evans, P. D., S. N. Cook, P. D. Riggs, and C. J. Noren. 1995. LITMUS: multipurpose cloning vectors with a novel system for bidirectional in vitro transcription. BioTechniques 19:130-135. [PubMed] [Google Scholar]
- 15.Fernaays, M. M., A. J. Lesse, X. Cai, and T. F. Murphy. 2006. Characterization of igaB, a second immunoglobulin A1 protease gene in nontypeable Haemophilus influenzae. Infect. Immun. 74:5860-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedler, A., N. Zakai, O. Karni, D. Friedler, C. Gilon, and A. Loyter. 1999. Identification of a nuclear transport inhibitory signal (NTIS) in the basic domain of HIV-1 Vif protein. J. Mol. Biol. 289:431-437. [DOI] [PubMed] [Google Scholar]
- 17.Funabiki, H., H. Yamano, K. Kumada, K. Nagao, T. Hunt, and M. Yanagida. 1996. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature 381:438-441. [DOI] [PubMed] [Google Scholar]
- 18.Gorlich, D., and I. W. Mattaj. 1996. Nucleocytoplasmic transport. Science 271:1513-1518. [DOI] [PubMed] [Google Scholar]
- 19.Halter, R., J. Pohlner, and T. F. Meyer. 1989. Mosaic-like organization of IgA protease genes in Neisseria gonorrhoeae generated by horizontal genetic exchange in vivo. EMBO J. 8:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison, S. C. 1991. A structural taxonomy of DNA-binding domains. Nature 353:715-719. [DOI] [PubMed] [Google Scholar]
- 21.Hauck, C. R., and T. F. Meyer. 1997. The lysosomal/phagosomal membrane protein h-lamp-1 is a target of the IgA1 protease of Neisseria gonorrhoeae. FEBS Lett. 405:86-90. [DOI] [PubMed] [Google Scholar]
- 22.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 24.Kilian, M., J. Reinholdt, H. Lomholt, K. Poulsen, and E. V. G. Frandsen. 1996. Biological significance of IgA1 proteases in bacterial colonization and pathogenesis: critical evaluation of experimental evidence. APMIS 104:321-338. [DOI] [PubMed] [Google Scholar]
- 25.Klauser, T., J. Kramer, K. Otzelberger, J. Pohlner, and T. F. Meyer. 1993. Characterization of the Neisseria Iga (Beta)-core—the essential unit for outer-membrane targeting and extracellular protein secretion. J. Mol. Biol. 234:579-593. [DOI] [PubMed] [Google Scholar]
- 26.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornfeld, S. J., and A. G. Plaut. 1981. Secretory immunity and the bacterial IgA proteases. Rev. Infect. Dis. 3:521-534. [DOI] [PubMed] [Google Scholar]
- 28.Kraehenbuhl, J. P., and M. R. Neutra. 1992. Molecular and cellular basis of immune protection of mucosal surfaces. Physiol. Rev. 72:853-879. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Landt, O., H. P. Grunert, and U. Hahn. 1990. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96:125-128. [DOI] [PubMed] [Google Scholar]
- 31.Lin, L., P. Ayala, J. Larson, M. Mulks, M. Fukuda, S. R. Carlsson, C. Enns, and M. So. 1997. The Neisseria type 2 IgA1 protease cleaves LAMP1 and promotes survival of bacteria within epithelial cells. Mol. Microbiol. 24:1083-1094. [DOI] [PubMed] [Google Scholar]
- 32.Lomholt, H., K. Poulsen, D. A. Caugant, and M. Kilian. 1992. Molecular polymorphism and epidemiology of Neisseria meningitidis immunoglobulin A1 proteases. Proc. Natl. Acad. Sci. USA 89:2120-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomholt, H., K. Poulsen, and M. Kilian. 1995. Comparative characterization of the iga gene encoding IgA1 protease in Neisseria meningitidis, Neisseria gonorrhoeae and Haemophilus influenzae. Mol. Microbiol. 15:495-506. [DOI] [PubMed] [Google Scholar]
- 34.Lundberg, K. S., D. D. Shoemaker, M. W. Adams, J. M. Short, J. A. Sorge, and E. J. Mathur. 1991. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene 108:1-6. [DOI] [PubMed] [Google Scholar]
- 35.Miller, P. B., H. Shen, N. R. Gilkes, D. G. Kilburn, R. C. Miller, A. G. Plaut, and R. A. J. Warren. 1992. Endoglucanase-a from Cellulomonas fimi in which the hinge sequence of human IgA1 is substituted for the linker connecting its 2 domains is hydrolyzed by IgA proteases from Neisseria gonorrhoeae. FEMS Microbiol. Lett. 92:199-203. [DOI] [PubMed] [Google Scholar]
- 36.Morelli, G., B. Malorny, K. Muller, A. Seiler, J. F. Wang, J. delValle, and M. Achtman. 1997. Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol. Microbiol. 25:1047-1064. [DOI] [PubMed] [Google Scholar]
- 37.Mulks, M. H., A. G. Plaut, H. A. Feldman, and B. Frangione. 1980. IgA proteases of two distinct specificities are released by Neisseria meningitidis. J. Exp. Med. 152:1442-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulks, M. H., and J. S. Knapp. 1987. Immunoglobulin-A1 protease types of Neisseria gonorrhoeae and their relationship to auxotype and serovar. Infect. Immun. 55:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulks, M. H., and R. J. Shoberg. 1994. Bacterial immunoglobulin A1 proteases. Methods Enzymol. 235:543-554. [DOI] [PubMed] [Google Scholar]
- 40.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 41.Noorbakhsh, F., N. Vergnolle, M. D. Hollenberg, and C. Power. 2003. Proteinase-activated receptors in the nervous system. Nat. Rev. Neurosci. 4:981-990. [DOI] [PubMed] [Google Scholar]
- 42.Oomen, C. J., P. Van Ulsen, P. Van Gelder, M. Feijen, J. Tommassen, and P. Gros. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23:1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254:539-544. [DOI] [PubMed] [Google Scholar]
- 44.Plaut, A. G., J. V. Gilbert, M. S. Artenstain, and J. D. Capra. 1975. Neisseria gonorrhoeae and Neisseria meningitidis: extracellular enzyme cleaves human immunoglobulin A. Science 193:1103-1105. [DOI] [PubMed] [Google Scholar]
- 45.Plaut, A. G., and W. W. Bachovchin. 1994. IgA-specific prolyl endopeptidases—serine type. Methods Enzymol. 244:137-151. [DOI] [PubMed] [Google Scholar]
- 46.Pohlner, J., R. Halter, K. Beyreuther, and T. F. Meyer. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325:458-462. [DOI] [PubMed] [Google Scholar]
- 47.Pohlner, J., T. Klauser, E. Kuttler, and R. Halter. 1992. Sequence-specific cleavage of protein fusions using a recombinant Neisseria type-2 IgA protease. Bio-Technology 10:799-804. [DOI] [PubMed] [Google Scholar]
- 48.Pohlner, J., U. Langenberg, U. Wolk, S. C. Beck, and T. F. Meyer. 1995. Uptake and nuclear transport of Neisseria IgA1 protease-associated alpha-proteins in human cells. Mol. Microbiol. 17:1073-1083. [DOI] [PubMed] [Google Scholar]
- 49.Poulsen, K., J. Brandt, J. P. Hjorth, H. C. Thogersen, and M. Kilian. 1989. Cloning and sequencing of the immunoglobulin-A1 protease gene (iga) of Hemophilus influenzae serotype B. Infect. Immun. 57:3097-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poulsen, K., J. Reinholdt, and M. Kilian. 1992. A comparative genetic study of serologically distinct Haemophilus influenzae type 1 immunoglobulin A1 proteases. J. Bacteriol. 174:2913-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinholdt, J., and M. Kilian. 1983. A sensitive enzyme-linked immunosorbent-assay for IgA protease activity. J. Immunol. Methods 63:367-376. [DOI] [PubMed] [Google Scholar]
- 52.Reinholdt, J., and M. Kilian. 1997. Comparative analysis of immunoglobulin A1 protease activity among bacteria representing different genera, species, and strains. Infect. Immun. 65:4452-4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rezende, S. M., R. E. Simmonds, and D. A. Lane. 2004. Coagulation, inflammation, and apoptosis: different roles for protein S and the protein S-C4b binding protein complex. Blood 103:1192-1201. [DOI] [PubMed] [Google Scholar]
- 54.Salvesen, G. S., and V. M. Dixit. 1997. Caspases: intracellular signaling by proteolysis. Cell 91:443-446. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Manniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY.
- 56.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senior, B. W., and J. M. Woof. 2005. The influences of hinge length and composition on the susceptibility of human IgA to cleavage by diverse bacterial IgA1 proteases. J. Immunol. 174:7792-7799. [DOI] [PubMed] [Google Scholar]
- 58.Senior, B. W., and J. M. Woof. 2005. Effect of mutations in the human immunoglobulin A1 (IgA1) hinge on its susceptibility to cleavage by diverse bacterial IgA1 proteases. Infect. Immun. 73:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shoberg, R. J., and M. H. Mulks. 1991. Proteolysis of bacterial-membrane proteins by Neisseria gonorrhoeae type-2 immunoglobulin-A1 protease. Infect. Immun. 59:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sparling, P. F. 1966. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J. Bacteriol. 92:1364-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 62.van Ulsen, P., L. van Alphen, J. ten Hove, F. Fransen, P. van der Ley, and J. Tommassen. 2003. A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 50:1017-1030. [DOI] [PubMed] [Google Scholar]
- 63.Veiga, E., E. Sugawara, H. Nikaido, V. de Lorenzo, and L. A. Fernandez. 2002. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 21:2122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Viitala, J., S. R. Carlsson, P. D. Siebert, and M. Fukuda. 1988. Molecular cloning of cDNAs encoding lamp A, a human lysosomal membrane glycoprotein with apparent Mr approximately equal to 120,000. Proc. Natl. Acad. Sci. USA 85:3743-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vitovski, S., R. C. Read, and J. R. Sayers. 1999. Invasive isolates of Neisseria meningitidis possess enhanced immunoglobulin A1 protease activity compared to colonizing strains. FASEB J. 13:331-337. [DOI] [PubMed] [Google Scholar]
- 66.Vitovski, S., K. T. Dunkin, A. J. Howard, and J. R. Sayers. 2002. Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels. JAMA 287:1699-1705. [DOI] [PubMed] [Google Scholar]
- 67.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 68.Yanish-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]