Abstract
Staphylococcus aureus can stimulate activation and aggregation of platelets, which are thought to be factors in the development of infective endocarditis. Previous studies have identified clumping factor A (ClfA) and fibronectin binding proteins A and B (FnBPA and FnBPB) as potent platelet aggregators. These proteins are able to stimulate rapid platelet aggregation by either a fibrinogen- or a fibronectin-dependent process which also requires antibodies specific to each protein. Slower aggregation has been seen in other systems where specific fibrinogen binding ligands are absent and platelet aggregation is mediated by complement and specific antibodies. Bacteria expressing ClfB aggregate platelets with a longer lag time than ClfA or FnBPA and FnBPB. In order to investigate whether ClfB causes platelet aggregation in a complement- or fibrinogen-dependent manner, a non-fibrinogen-binding mutant of ClfB (ClfB Q235A) was constructed. Lactococcus lactis expressing ClfB Q235A was able to stimulate platelet aggregation in platelet-rich plasma without a significant increase in lag time. The requirements for platelet aggregation were investigated using gel-filtered platelets. Fibrinogen and specific anti-ClfB antibodies were found to be sufficient to allow platelet aggregation mediated by the wild-type ClfB protein. It seems that ClfB causes platelet aggregation by a fibrinogen-dependent mechanism. The non-fibrinogen-binding ClfB mutant was unable to stimulate platelet aggregation under these conditions. However, bacteria expressing ClfB Q235A caused platelet aggregation in a complement-dependent manner which required specific anti-ClfB antibodies.
Staphylococcus aureus is a commensal of the human anterior nares which is commonly associated with nosocomial disease. As well as causing superficial infections, S. aureus can cause serious invasive conditions such as septic arthritis and infective endocarditis (IE). IE is characterized by the buildup of vegetative bodies on heart valve surfaces which consist of bacteria and platelet thrombi. It is though that the initial coaggregation between bacteria and platelets can trigger activation of nearby platelets, leading to thrombus formation. This condition is caused predominantly by S. aureus and has a high mortality rate. Treatment has become more difficult due to the recent emergence of multidrug-resistant strains. Risk factors for infection include rheumatic heart conditions and the use of prosthetic heart valves, although S. aureus can colonize previously undamaged heart valves (21).
The ability to cause platelet aggregation is thought to contribute to the development of IE (21, 27, 31). Several surface-expressed proteins of S. aureus have been shown to stimulate platelet activation and aggregation. These include the fibrinogen binding proteins clumping factor A (ClfA) and ClfB and the bifunctional fibronectin-fibrinogen binding proteins A and B (FnBPA and FnBPB) (11, 23). Thus, the interaction between S. aureus and platelets is multifactorial.
Bacteria that cause platelet aggregation interact directly or indirectly with receptors on the platelet surface. This initial interaction results in the upregulation of the active form of platelet integrin GPIIb/IIIa (10, 17). In its active form GPIIb/IIIa can bind avidly to fibrinogen and fibronectin in solution (2, 3, 29). Subsequent aggregation occurs when neighboring platelets interact via bound fibrinogen (21). Aggregation occurs after a variable period of time referred to as the lag time. This time reflects the time taken for activation and aggregation to occur after the bacteria and platelets come into contact.
Bacteria expressing ClfA and FnBPA cause rapid aggregation with short lag times (1 to 2 min) (11, 18, 27). ClfA interacts with platelets in a fibrinogen-dependent manner (11). The initial adhesion between the bacterium and the resting form of GPIIb/IIIa occurs via a fibrinogen bridge. Resting GPIIb/IIIa is able to bind fibrinogen coating the bacterium, as it resembles fibrinogen bound to a surface. One end of the bivalent fibrinogen molecule is bound at the γ chain by ClfA, while the other γ chain is free to interact with GPIIb/IIIa (9, 16, 18). Previous studies have shown that the level of ClfA protein on the bacterial surface is crucial in this process. A “threshold” level of protein expression is required for platelet activation to occur. ClfA-specific antibodies are also required to interact with platelet FcγRIIa receptors which cluster to trigger activation and intracellular signaling (18).
ClfA is expressed predominantly in the stationary phase of growth and is the main mediator of platelet aggregation for stationary-phase cells (18). In the exponential phase of growth, rapid platelet activation is caused by FnBPA and FnBPB. FnBPA causes platelet aggregation in a manner similar to that of ClfA (11). Fibrinogen bound by the A domain or fibronectin bound by the BCD domains of the protein can interact with GPIIb/IIIa on the surface of platelets. Specific antibodies to FnBPA are also required to trigger activation and subsequent aggregation (11).
Longer lag times to aggregation occur with bacteria lacking potent proaggregatory surface components. Instead, a slower process involving complement assembly coupled with specific antibody binding is required for aggregation. Platelet aggregation mediated by Streptococcus sanguis requires complement and immunoglobulin G (IgG) (12, 13). This was also found to be the case for a non-fibrinogen-binding mutant of ClfA (ClfA PY). Bacteria expressing ClfA PY caused platelet aggregation in a complement-dependent manner with a longer lag time of 8 to 10 min. This suggests that assembly of complement proteins and IgG binding are required to cross-link bacteria to a platelet complement receptor and FcγRIIa (18).
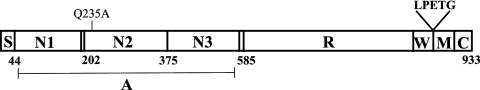
ClfB belongs to a family of surface expressed proteins known as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules). It is most highly expressed in the early exponential phase of growth (19). ClfB plays an important role in nasal colonization by binding cytokeratin 10 in the desquamated nasal epithelium (24, 33). Recent studies have shown that immunizing mice with ClfB reduces nasal colonization (30). ClfB can also bind to fibrinogen, which is assumed to be significant in platelet activation and aggregation. Unlike ClfA, FnBPA, and FnBPB, which bind the γ chain of fibrinogen, ClfB binds to the α chain (22, 25). This interaction is presumed to occur by the “dock-lock-latch” mechanism in a hydrophobic trench located between domains N2 and N3 of the ClfB molecule (26) (Fig. 1).
FIG. 1.
Domain organization of ClfB. The signal sequence (S) is followed by the ligand binding A domain, which is projected from the cell surface by the repeat region (R). The C terminus consists of wall (W), membrane (M), and cytoplasmic (C) spanning domains. The LPETG motif is requited for cross-linking ClfB to the cell wall by sortase.
Although ClfB is known to cause platelet aggregation and has been shown to contribute to the pathogenesis of experimental endocarditis in rats, little is known about the mechanism by which platelet aggregation occurs (8, 23). Despite its ability to bind fibrinogen, Lactococcus lactis expressing ClfB caused aggregation with longer lag times (6 to 8 min) than ClfA and FnBPA. The aims of this study were to establish whether ClfB causes platelet aggregation by a fibrinogen- or complement-dependent mechanism or by a combination of both. The role of ClfB in platelet aggregation was studied in S. aureus strain Newman and in a surrogate host, Lactococcus lactis. In order to establish the role of fibrinogen in ClfB-mediated aggregation, a non-fibrinogen-binding mutant of ClfB was constructed. The involvement of individual plasma proteins was studied using gel-filtered platelets (GFP).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. L. lactis was grown statically at 28°C in M17 (Difco) agar or broth with 0.5% (wt/vol) glucose. S. aureus was grown on tryptic soy agar or broth at 37°C with shaking (200 rpm). The following antibiotics (Sigma) were added to the media as required: chloramphenicol at 10 μg/ml, erythromycin at 10 μg/ml, and tetracycline at 2 μg/ml.
TABLE 1.
Bacterial strains and vectors
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| L. lactis strain NZ9800 | Host for expression plasmids; ΔnisA | 4 |
| S. aureus strains | ||
| Newman | Strain expressing high levels of ClfA and coagulase | 7 |
| DU5999 | Newman clfA5; frameshift mutation in clfA | 11 |
| DU6000 | Newman clfA5 clfB::lacZ (Emr); transduced clfB mutation into DU5999 | 11 |
| DU5854 | Newman strain with coagulase mutation (Δcoa::Tcr) | 20 |
| DU6016 | Newman clfA5 coa; transduction of coa::Tcr into DU5999 | This study |
| DU6017 | Newman clfA5 coa::TcrclfB::lacZ (Emr); transduction of coa::Tcr into DU6000 | This study |
| L. lactis plasmids | ||
| pNZ8037 | Expression vector with nisin-inducible promoter (Cmr) | 5 |
| pNZ8037clfA | pNZ8037 with full-length ClfA (Cmr) | 18 |
| pNZ8037clfB | pNZ8037 with full-length ClfB (Cmr) | This study |
| pNZ8037clfB Q235A | pNZ8037 with full-length clfB Q235A | This study |
Staphylococcus aureus mutants.
A deletion-substitution coagulase mutation (Δcoa::Tcr) was transduced into strains Newman, DU5999, and DU6009 by using phage 85 (1). Transductants were validated by the coagulase tube test.
Construction of L. lactis(pNZ8037clfB).
The clfB gene was amplified from genomic DNA of S. aureus Newman by using the forward primer CTTGCCATGGAAAAAAGAATTGATTATTTGTCG and the reverse primer TGCTCTAGATTCTTCCGGTAAAATGACTG. Following amplification, PCR products were digested with NcoI and XbaI (sites are underlined) and ligated with pNZ8037 plasmid digested with the same enzymes. Ligated DNA was electroporated into competent L. lactis cells (34).
Site-directed mutagenesis.
The protein homology/analogy recognition engine (PHYRE) was used to predict the three-dimensional structure of ClfB based on other homologous proteins. Resulting structures were further analyzed using a computer modeling program (Chimera) to select residues in the putative ligand binding trench of ClfB. Mutagenesis of plasmid pNZ8037clfB was carried out using the QuikChange protocol (Stratagene). Complementary forward (CATTTGACCCTAATGCAAGTGGTAACACATTTATGG) and reverse (CCATAAATGTGTTACCACTTGCATTAGGGTCAAATG) primers incorporating a mutation (mutated nucleotides are in bold) were used to amplify the circular template. The PCR products were digested with DpnI to eliminate parental DNA and transformed into Escherichia coli XL-1Blue. Mutations were verified by DNA sequencing. The PCR conditions used were as outlined by the QuikChange protocol.
Fibrinogen binding assays.
L. lactis strains were grown to stationary phase, diluted 1/100 in fresh GM17, and allowed to reach exponential phase. Cultures were induced with nisin and grown to stationary phase. Bacterial cells were then washed twice in phosphate-buffered saline (PBS) and resuspended to an OD600 of 1. Cells were added to 96-well plates (Sarstedt) coated with 10 μg/ml of fibrinogen (Calbiochem). Crystal violet staining was used to measure adherent bacteria (15).
Whole-cell immunoblots.
L. lactis strains were grown and induced as described above. Doubling dilutions of washed cells (5 μl) were dotted onto a nitrocellulose membrane (Protran). The membrane was blocked for 1 h with 10% (wt/vol) milk powder (Marvel) in TS buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl) and then probed with rabbit anti-ClfB polyclonal antibody (19). Bound antibody was detected using goat anti-rabbit antibody conjugated to horseradish peroxidase (HRP) (18).
Preparation of PRP.
Platelet-rich plasma (PRP) was prepared as described previously. Briefly, blood was drawn into a syringe containing 3.2% (wt/vol) Na citrate. Whole blood was then centrifuged at 150 × g for 10 min. The top platelet layer was removed and the red blood cells centrifuged again to obtain platelet-poor plasma (18).
Preparation of GFP.
Blood was drawn into a syringe containing acid-citrate-dextrose. Following preparation of PRP (as described above), the pH of the platelets was adjusted to 6.5 and prostaglandin E1 (1 μM) was added to prevent activation during preparation of GFP. PRP was centrifuged at 720 × g for 10 min. Platelets were resuspended in 1 ml of JNL buffer (6 mM dextrose, 130 mM NaCl, 9 mM NaCl2, 10 mM Na citrate, 10 mM Tris, 3 mM KCl, 0.8 mM KH2PO4, and 0.9 mM MgCl2 [pH 7.4]) and passed through a 10-ml Sepharose 2B column (Sigma). The original volume of platelets was restored with JNL and supplemented with 2 mM CaCl2 (18).
Depletion of IgG from fibrinogen and human serum.
Fibrinogen was purified of contaminating IgG by passage through a column of protein A coupled to Sepharose (Amersham Biosciences). The flowthrough was collected and concentrated back to the original volume in a centrifugal filter device (Amicon). Human serum was depleted of IgG in the same manner. Enzyme-linked immunosorbent assay were carried out to confirm that fibrinogen had been depleted of antibodies, and serum samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to confirm loss of IgG.
Depletion of ClfB-specific antibodies from pooled human IgG.
Cultures of L. lactis(pNZ8037) and L. lactis(pNZ8037clfB) induced with 3.2 ng/ml of nisin were pelleted and washed in PBS. Cells were resuspended in 1 ml of pooled human IgG (Gammmagard; Baxter) at 5% (wt/vol) and incubated with rotation at 4°C for 1 h. Bacterial cells were pelleted, and the absorbed IgG was collected and filtered using a 0.45-μm filter (Sarstedt). Depletion of anti-ClfB antibodies was confirmed by testing reactivity with recombinant ClfB by enzyme-linked immunosorbent assay (28).
Inactivation of complement proteins in human serum.
Human serum was depleted of complement by heating at 56°C for 30 min or by treatment with zymosan (100 mg/ml) as described previously (13).
Platelet aggregation.
L. lactis cells were induced and grown to stationary phase as described above. S. aureus cells were grown to mid-exponential phase. Cells were washed twice with M17 broth or PBS and resuspended to an OD600 of 1.6. Bacterial cells (25 μl) were added to 225 μl of PRP in glass cuvettes. PRP and bacteria were incubated with stirring in an aggregometer (Bio-Data) for up to 25 min at 37°C, and light transmission was monitored. Aggregation was inhibited by anti-GPIIb/IIIa (Abciximab, Eli Lily) antibodies at a 1/100 dilution and anti-FcγRIIa antibodies (IV3; kindly provided by R. Klimkowski, Medarex) at a 1/50 dilution. GFP were supplemented with purified fibrinogen at a final concentration of 0.5 mg/ml. Pooled IgG and depleted IgG were added to a final concentration of 2 μg/ml. Twenty-five microliters of human serum was added to a final volume of 225 μl GFP.
Statistical analysis.
The data presented for this study represent the means from three experiments ± standard errors of the means unless otherwise stated. The unpaired t test was used to determine the significance of differences in aggregation between strains, with significance defined as a P value of <0.05.
RESULTS
Contribution of ClfB to strain Newman-mediated platelet aggregation.
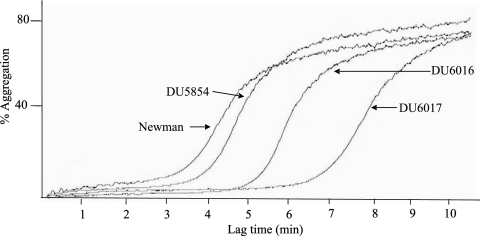
S. aureus strain Newman was selected for this study because it does not have FnBPA and FnBPB attached to its cell surface due to mutations that result in secretion of truncated forms of each protein (14). It was therefore possible to study the effect of ClfB expressed in the exponential phase of growth without interference from the FnBPA and FnBPB proteins. Strain Newman expresses high levels of coagulase, which causes formation of fibrin clots in plasma. A coagulase-defective mutation was therefore introduced into strains DU5999 and DU6000 by transduction to prevent interference in light transmission in the aggregometer caused by clot formation.
Figure 2 shows a typical aggregation trace. Strains Newman and DU5854 (Newman Δcoa::Tcr) had equal lag times to aggregation of approximately 3 min. Comparing strains DU5854 and DU6016 (Newman coa clfA5), an increase in lag time was observed, from 3 to 4.67 min. This difference was found to be statistically significant (P = 0.0123). Thus, the low level of ClfA on exponential-phase cells contributes to aggregation.
FIG. 2.
Representative aggregation trace. Strains Newman, DU5854 (Newman coa), DU6016 (Newman coa clfA), and DU6017 (Newman coa clfA clfB) were grown to exponential phase, washed, and added to PRP in an aggregometer.
Strain DU6017 (Newman coa clfA clfB) had a mean lag time of 6.5 min in two of the donors tested. In the third donor no aggregation occurred. When the 6.5-min lag time was compared to the lag time caused by strain DU6016, the difference was statistically significant (P = 0.0191). These data show that both ClfA and ClfB play a role in the ability of S. aureus from the exponential phase of growth to interact with platelets.
Controlled expression of ClfB from L. lactis.
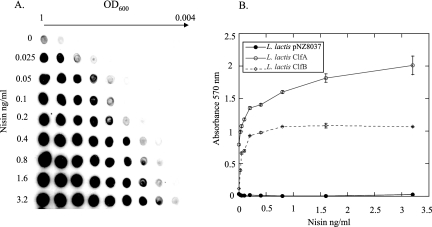
In order to express ClfB on the surface of L. lactis cells in a controlled fashion, the clfB gene was cloned into the nisin-inducible pNZ8037 vector. Whole-cell immunoblotting was carried out to study ClfB protein expression by L. lactis(pNZ8037clfB), using nisin concentrations ranging from 0.025 ng/ml to 3.2 ng/ml. A constant number of cells were dotted onto a nitrocellulose membrane and probed with anti-ClfB antibodies. An approximate 64-fold increase in ClfB expression was seen in cells induced with 3.2 ng/ml nisin compared to uninduced cells (Fig. 3A).
FIG. 3.
Expression of ClfB from L. lactis and fibrinogen binding. (A) Whole-cell immunoblot detecting ClfB expression. Doubling dilutions of bacterial cells induced with nisin from 0 to 3.2 ng/ml were dotted onto nitrocellulose membranes. The membranes were probed with rabbit anti-ClfB antibody, and bound antibody was detected using goat anti-rabbit antibody conjugated to HRP. (B) L. lactis ClfA, L. lactis ClfB, and L. lactis(pNZ8037) were induced with nisin at from 0 to 3.2 ng/ml and grown to stationary phase. Adherence of the cells to immobilized fibrinogen was assessed by crystal violet staining.
The ability of L. lactis expressing ClfB (L. lactis ClfB) to adhere to immobilized fibrinogen was assessed. L. lactis ClfB, L. lactis ClfA, and a strain containing the empty pNZ8037 vector were induced with increasing nisin concentrations, and their abilities to bind to microtiter wells coated with fibrinogen were compared. Increasing the level of ClfB on the cell surface correlated with a sharp increase in fibrinogen binding up to nisin concentrations of 0.4 ng/ml. Increasing the inducer concentration above this level did not increase adherence to fibrinogen. L. lactis ClfB adhered to fibrinogen less avidly than L. lactis ClfA across the range of inducer concentrations used (Fig. 3B). This could be due to a lower level of ClfB expression compared to ClfA or to a lower affinity for fibrinogen. L. lactis(pNZ8037) did not bind detectably to fibrinogen.
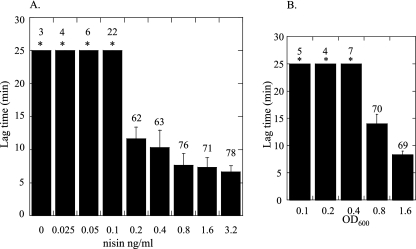
Threshold level of ClfB necessary for platelet activation and aggregation.
Previous studies have shown that a minimum level of ClfA is required for platelet activation and aggregation to occur (18). To test the level of ClfB required, L. lactis(pNZ8037clfB) was induced with increasing concentrations of nisin. Induced cells were added to PRP, and the lag time to aggregation was measured. It was found that 0.2 ng/ml of nisin was sufficient to cause platelet aggregation. Below this level of inducer, no aggregation was seen (Fig. 4A). The cells induced with the highest nisin concentrations produced the shortest lag times (6.7 min at 3.2 ng/ml nisin, compared to 11.7 min at 0.2 ng/ml nisin [P < 0.01]). These results indicate that the lag time to aggregation is dependent on the level of ClfB protein expressed on the bacterial cell surface. There seems to be a decrease in total percent aggregation caused by cells induced with lower levels of nisin (0.2 and 0.4 ng/ml) compared to fully induced cells (78%). This trend was not statistically significant, although this would probably change if a larger number of samples were tested.
FIG. 4.
(A) Threshold level of ClfB expression required for platelet activation. L. lactis ClfB cultures induced with increasing concentrations of nisin were incubated with PRP, and the lag time to activation was measured. (B) Threshold level of bacterial cells required for platelet activation. Doubling dilutions of fully induced L. lactis ClfB (OD600 of 1.6) were added to platelets, and the lag time to activation was measured. Asterisks indicate no activation after 25 min. The percent aggregation is shown above the error bars.
Experiments were also carried out to determine the threshold number of bacterial cells necessary to cause ClfB-mediated aggregation. Doubling dilutions of fully induced L. lactis ClfB (OD600 of 1.6) were added to platelets. It was found that an OD600 of 0.8 was necessary to cause platelet aggregation (Fig. 4B). The lag time to aggregation increased from 8 min at an OD600 of 1.6 to 14 min at an OD600 of 0.8 (P = 0.0061). (Fig. 4B). In contrast, when 20 μM ADP was added to platelets, aggregation occurred immediately and reached 82% (data not shown).
Inhibition of aggregation by anti-GPIIb/IIa and anti-FcγRIIa antibodies.
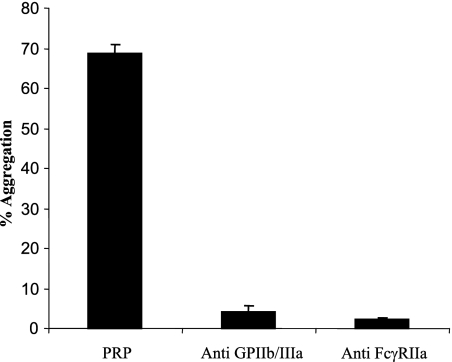
Antibodies to platelet receptors GPIIb/IIIa and FcγRIIa were used to inhibit ClfB-mediated aggregation. Anti-GPIIB/IIIa reduced aggregation from 69% to 4% (Fig. 5). Anti-FcγRIIa reduced aggregation from 69% to 2%, showing that both receptors are absolutely required in ClfB-mediated platelet aggregation.
FIG. 5.
Inhibition of platelet aggregation using anti-GPIIb/IIIa and anti-FcγRIIa antibodies. PRP was incubated with the inhibitory antibodies for 15 min at 37°C prior to the addition of L. lactis ClfB.
Non-fibrinogen-binding mutant of ClfB.
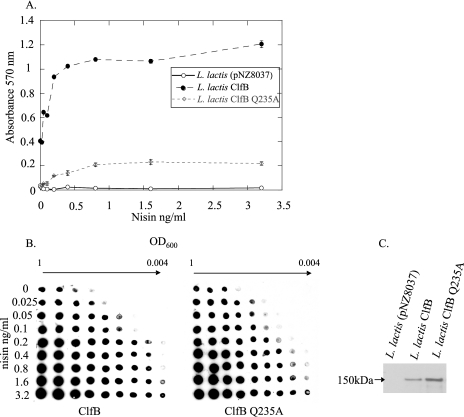
A three-dimensional molecular model of domains N2 and N3 of ClfB was constructed based on the known structures of ClfA and SdrG. The structures were analyzed to select residues in the putative ligand binding trench of ClfB that could play a role in ligand binding. Thus, amino acid residue Q235 was selected for mutagenesis. The pNZ0837clfB plasmid was used as a template for mutagenesis, and ClfB Q235A was expressed on L. lactis. The abilities of L. lactis expressing wild-type ClfB and ClfB Q235A to adhere to immobilized fibrinogen were compared. ClfB Q235A showed a dramatic decrease in adherence to fibrinogen (Fig. 6A). Whole-cell immunoblotting was carried out to compare the expression of wild-type ClfB and ClfB Q235A on L. lactis at different concentrations of the inducer nisin. There was no noticeable difference in the levels of expression detected with rabbit anti-ClfB antibody (Fig. 6B). Whole-cell immunoblotting was also carried out with pooled human IgG which contains anti-ClfB antibodies. No difference was detected in IgG binding between L. lactis expressing ClfB and ClfB Q235A. This strongly suggests that the proteins were expressed at the same level (data not shown). Western immunoblotting showed that ClfB and ClfB Q235A migrated as bands of 150 kDa, which shows that the mutant protein is intact (Fig. 6C).
FIG. 6.
(A) Fibrinogen binding abilities of L. lactis ClfB and L. lactis ClfB Q235A. L. lactis ClfB, L. lactis ClfB Q235A, and L. lactis (pNZ8037) were induced with increasing nisin concentrations and grown to stationary phase. Adherence of washed cultures to 10 μg/ml of fibrinogen was assessed by crystal violet staining. (B) Expression of ClfB and ClfB Q235A from L. lactis. Expression of ClfB Q235A from L. lactis was compared to that of wild-type ClfB. Doubling dilutions of bacterial cells induced with nisin at from 0 to 3.2 ng/ml were dotted onto nitrocellulose membranes. The membranes were probed with rabbit anti-ClfB antibody, and bound antibody was detected using goat anti-rabbit antibody conjugated to HRP. (C) Western immunoblot of protoplasts of L. lactis, L. lactis ClfB, and L. lactis ClfB Q235A.
Fibrinogen and ClfB-specific antibodies are required for ClfB-mediated platelet activation and aggregation.
The lag times and percent aggregation in PRP were very similar for L. lactis ClfB and L. lactis ClfB Q235A. L. lactis ClfB caused aggregation after a lag time of 5.7 min, compared to 6 min for L. lactis ClfB Q235A. The percent aggregation was slightly lower for L. lactis ClfB Q235A (70.3%, compared to 80.3% for L. lactis ClfB wild type), but this was not found to be statistically significant (P = 0.1621) (Fig. 7).
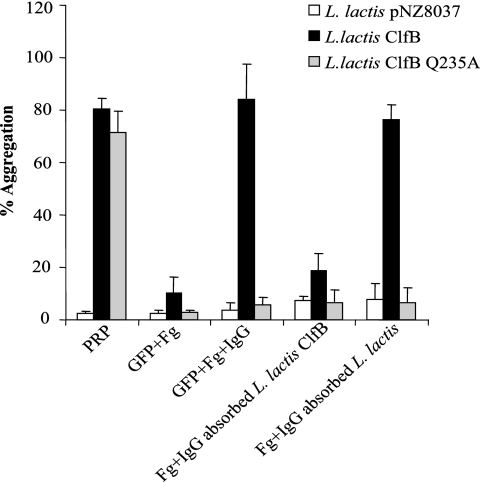
FIG. 7.
Activation of GFP. L. lactis(pNZ8037), L. lactis ClfB, and L. lactis ClfB Q235A were added to GFP supplemented with fibrinogen, pooled antibodies, and pooled antibodies absorbed with L. lactis(pNZ8037) or L. lactis ClfB. Percent aggregation was recorded after 25 min.
To determine which plasma proteins were required for ClfB-mediated platelet activation, studies were performed with GFP. When fibrinogen alone was added to GFP, no activation occurred. The further addition of pooled human antibodies restored the ability of L. lactis ClfB to cause platelet aggregation. However, fibrinogen and pooled IgG were not able to restore the ability of ClfB Q235A to aggregate platelets, which indicates that additional plasma proteins are required (Fig. 7).
In order to determine if anti-ClfB antibodies in pooled human IgG were required to stimulate platelet activation, the IgG was absorbed with fully induced L. lactis(pNZ8037clfB) and L. lactis(pNZ8037) cells. When IgG was depleted of specific ClfB antibodies, aggregation by L. lactis ClfB was reduced to 18.7%. IgG absorbed with L. lactis(pNZ8037) still supported activation and subsequent aggregation (Fig. 7).
Complement fixation is required for platelet aggregation mediated by ClfB Q235A.
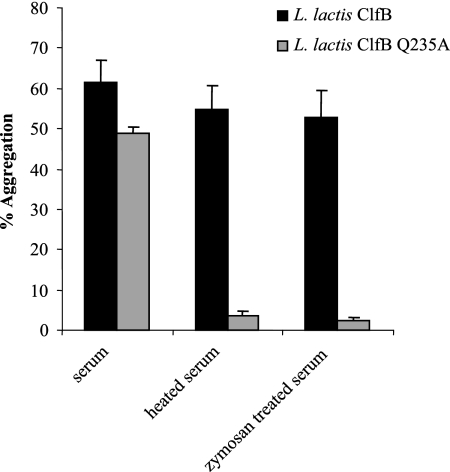
L. lactis ClfB Q235A was unable to cause platelet activation in GFP supplemented with fibrinogen and pooled antibody (Fig. 7). Upon the addition of serum and fibrinogen (required for aggregation following activation) to GFP, the non-fibrinogen-binding mutant was able to cause aggregation (Fig. 8). The apparent difference in percent aggregation caused by the mutant and wild-type proteins (49% compared to 62%, respectively) was not statistically significant (P = 0.7325). Serum was treated with zymosan or heated at 56°C for 30 min to inactivate complement. In both cases activation by L. lactis ClfB was not affected, whereas L. lactis ClfB Q235A did not stimulate activation. This shows that the non-fibrinogen-binding mutant of ClfB could promote platelet activation only by a complement-dependent mechanism (Fig. 8).
FIG. 8.
Role of complement proteins in aggregation mediated by ClfB and ClfB Q235A. L. lactis ClfB and L. lactis ClfB Q235A were added to GFP to which fibrinogen and serum, serum treated with zymosan, or serum heated at 56°C was added.
Specific anti-ClfB antibody is required for platelet aggregation mediated by ClfB Q235A.
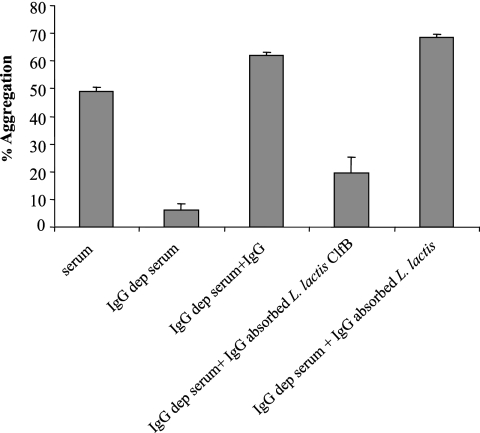
In order to determine if anti-ClfB antibodies were required for L. lactis ClfB Q235A-mediated aggregation, serum was depleted of IgG by absorption with protein A-Sepharose. Serum depleted of IgG did not support platelet activation (P = 0.0001). Addition of pooled human IgG restored the ability of ClfB Q235A to cause activation (62% aggregation), as did the addition of pooled human IgG absorbed with L. lactis(pNZ8037). When pooled human IgG absorbed with L. lactis(pNZ8037clfB) was added to IgG-depleted serum, aggregation was reduced to 19% (P = 0.0080). Thus, specific anti-ClfB antibodies are required for L. lactis ClfB Q235A-promoted activation of platelets (Fig. 9).
FIG. 9.
Requirement for specific ClfB antibodies in ClfB Q235A-mediated platelet activation. Serum depleted of IgG was added to GFP supplemented with fibrinogen. Pooled IgG and absorbed IgG were added to samples.
DISCUSSION
Several surface proteins of S. aureus have been shown to be involved in the process of platelet activation and aggregation. A common mechanism for activation by ClfA and FnBPA involves an initial interaction between the bacterium and the platelet receptor GPIIb/IIIa. Bound fibrinogen and fibronectin coat the bacterial surface and cross-link the bacterium to GPIIb/IIIa on the platelet surface. The abilities of ClfA to bind avidly to fibrinogen and of FnBPA to bind both fibrinogen and fibronectin allow for short lag times to aggregation (11, 18). When specific binding proteins are absent, extended lag times for aggregation are seen, such as were described for the non-fibrinogen-binding mutant of ClfA and for platelet aggregation mediated by Streptococcus sanguis (13, 18). These longer lag times are reflective of the time taken for complement assembly to occur on the bacterial surface. ClfB is known to bind to the α chain of fibrinogen. It was therefore surprising that longer lag times to aggregation (6 min) were caused by L. lactis expressing ClfB. The mean lag time was marginally shorter than that of the non-fibrinogen-binding mutant of ClfA (8 to 10 min) (18).
The results of this study show that ClfB expressed from L. lactis can cause activation and aggregation of platelets in a fibrinogen-dependent manner, although binding fibrinogen alone is not sufficient to stimulate activation. When GFP were supplemented with fibrinogen and pooled human IgG, L. lactis ClfB was able to stimulate platelet aggregation with a lag time of approximately 6 min. Further experiments showed that anti-ClfB antibodies present in pooled human IgG were necessary for activation to occur (Fig. 7). It seems that ClfB is able to cause platelet aggregation in the same manner as ClfA and FnBPA. Bound fibrinogen acts as a bridge between the platelet and the bacterium, while specific anti-ClfB antibody can bind the Fc receptor on platelets to cause activation through receptor clustering and intracellular signaling. The fact that inhibitors of GPIIb/IIIa (Abciximab) and FcγRIIa (IV3) prevented aggregation mediated by L. lactis ClfB confirms the role of these receptors in the interaction between platelets and bacteria expressing ClfB (Fig. 5).
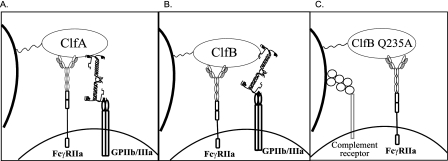
Since ClfB can cause aggregation in a fibrinogen-dependent manner, it seems strange that the lag time to aggregation is longer than that seen for ClfA and other fibrinogen binding molecules (11, 18). There are several possibilities to explain this difference. Fibrinogen is a symmetrical molecule consisting of two γ chains, two α chains, and two β chains (16). Unlike ClfA, FnBPA, and FnBPB, ClfB binds the α chain of fibrinogen in a flexible region located between residues 240 and 410 (32). In the case of ClfA and the FnBPs, the interaction with platelets occurs via a fibrinogen molecule bound at the γ chain protruding from the end of domain D by GPIIb/IIIa and ClfA. This may orient fibrinogen in a manner that is more suitable for binding GPIIb/IIIa than when it is bound at the α chain (Fig. 10).
FIG. 10.
(A) Fibrinogen-dependent platelet aggregation by ClfA. ClfA requires both fibrinogen and IgG to interact with GPIIb/IIIa and FcγRIIa platelet receptors. Both GPIIb/IIIa and ClfA bind the γ chain of fibrinogen. (B) Fibrinogen-dependent platelet aggregation by ClfB. ClfB requires fibrinogen and IgG to cause platelet activation. ClfB interacts with the α chain of fibrinogen, while GPIIb/IIIa binds the γ chain. (C) Complement-dependent platelet activation mediated by ClfB Q235A. Assembly of complement proteins and IgG binding are required to engage the platelet complement receptor and FcγRIIa, respectively.
The affinities of ClfB and ClfA for fibrinogen have not been compared directly, and it is possible that ClfB binds with a lower affinity. The α chain of fibrinogen has also been shown to be susceptible to cleavage by proteases (6, 32). These two possibilities would also explain the difference seen between L. lactis ClfB and L. lactis ClfA in fibrinogen binding assays. Across the range of inducer concentrations tested, cells expressing ClfB bound at consistently lower levels than cells expressing ClfA (Fig. 3). Any of these factors could contribute to the longer lag time to aggregation caused by ClfB.
Platelet activation and aggregation mediated by ClfB require a minimum level of ClfB protein to be expressed on the surface of cells. The threshold for platelet activation stimulated by L. lactis ClfB occurred at 0.2 ng/ml of nisin (Fig. 4A). Below this level, no activation was be seen. Increasing the number of ClfB molecules on the surface of the cell decreases the lag time to aggregation. The initial adhesion of bacteria to platelets via fibrinogen seems to be important in determining the lag time. Similar results have been seen with bacteria expressing ClfA (18). This study also found that a threshold number of bacterial cells are needed to cause platelet activation. At lower concentrations of bacteria, the lag time to activation increased. Decreasing the level of ClfB on the surface of L. lactis seemed to decrease percent aggregation, although the difference was not statistically significant.
Bacteria expressing the non-fibrinogen-binding mutant ClfB Q235A were still able to cause aggregation in PRP. The lag time to aggregation was not significantly different from that caused by the wild-type protein. However, experiments carried out with GFP demonstrated that ClfB Q235A did not cause aggregation in a fibrinogen-dependent manner. Instead, aggregation was dependent on the presence of complement. When serum was heated to inactivate complement and added to GFP, only cells expressing the wild-type ClfB protein could cause aggregation. The same result was seen with zymosan-treated serum (Fig. 8). Platelet aggregation could be restored by adding pooled IgG or IgG absorbed against L. lactis(pNZ8037). Adding IgG absorbed against L. lactis(pNZ8037clfB) did not restore activation (Fig. 9). We can therefore conclude that specific anti-ClfB antibodies are required for L. lactis ClfB Q235A-mediated platelet activation and aggregation. These results suggest that ClfB Q235A causes aggregation in the same way as the non-fibrinogen-binding mutant of ClfA (ClfA PY). Complement proteins bound to the bacterium can presumably interact with a complement receptor on the surface of the platelets. Bound anti-ClfB antibodies form another link to the platelet via the FcγRIIa receptor. The fact that the fibrinogen- and complement-dependent mechanisms of platelet aggregation identified in this study have similar lag times suggests that both mechanisms can contribute to overall activation and aggregation caused by ClfB.
This study also investigated the role that ClfB plays in platelet aggregation mediated by S. aureus strain Newman. This strain produces a high level of coagulase, which caused fibrin clot formation when cells were incubated with PRP for long periods of time. For this reason a coagulase-defective mutant of strain Newman (DU5854) was used. Strain DU5854 had the same lag time as wild-type Newman (approximately 3 min). Eliminating ClfA increased the lag time from 3 min to 4.67 min. Normally, in the exponential phase of growth bacterial cells express high levels of FnBPA and FnBPB, which cause rapid aggregation (11). However, Newman is defective in FnBP expression (14). ClfA is expressed predominantly in the stationary phase of growth. Previous studies using stationary-phase cells have shown that ClfA causes rapid activation with a lag time of 1 min (18). ClfA is not expressed at high levels on exponential-phase cells, and therefore an extended lag time was seen in this study.
ClfB contributes to platelet activation and aggregation by exponentially growing cells. Strain DU6017 lacks both ClfA and ClfB. In two of the donors tested, DU6017 activated platelets with an average lag time of 6.5 min, compared to the 4.67-min lag times caused by DU6016. In the third donor, DU6017 failed to activate platelets. This suggests that variable host factors, such as antibody levels and the presence of platelet receptors, contribute to the ability of bacteria expressing ClfB to cause activation. Overall, the presence of ClfB on S. aureus did shorten the lag time to aggregation. We propose that ClfB can contribute to and amplify the platelet aggregation caused by FnBPA and FnBPB in the exponential phase of growth.
In conclusion, this study found that ClfB expressed on the surface of bacteria can aggregate platelets in a fibrinogen-dependent manner. Bacteria expressing a non-fibrinogen-binding mutant, ClfB Q235A, were still able to cause aggregation by a complement-dependent mechanism. Both mechanisms of platelet aggregation required specific anti-ClfB antibodies to cross-link to the platelet FcγRIIa receptor and trigger activation. The two mechanisms have similar lag times, and it is feasible that both contribute to the overall activation and aggregation caused by ClfB.
Acknowledgments
We acknowledge support from the Health Research Board of Ireland (RPO09/2002).
Editor: A. Camilli
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Asheshov, E. H. 1966. Loss of antibiotic resistance in Staphylococcus aureus resulting from growth at high temperature. J. Gen. Microbiol. 42:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Calvete, J. J. 1995. On the structure and function of platelet integrin alpha IIb beta 3, the fibrinogen receptor. Proc. Soc. Exp. Biol Med. 208:346-360. [DOI] [PubMed] [Google Scholar]
- 3.Calvete, J. J. 1999. Platelet integrin GPIIb/IIIa: structure-function correlations. An update and lessons from other integrins. Proc. Soc. Exp. Biol Med. 222:29-38. [DOI] [PubMed] [Google Scholar]
- 4.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doolittle, R. F. 1984. Fibrinogen and fibrin. Annu. Rev. Biochem. 53:195-229. [DOI] [PubMed] [Google Scholar]
- 7.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 8.Entenza, J. M., T. J. Foster, D. Ni Eidhin, P. Vaudaux, P. Francioli, and P. Moreillon. 2000. Contribution of clumping factor B to pathogenesis of experimental endocarditis due to Staphylococcus aureus. Infect. Immun. 68:5443-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell, D. H., P. Thiagarajan, D. W. Chung, and E. W. Davie. 1992. Role of fibrinogen alpha and gamma chain sites in platelet aggregation. Proc. Natl. Acad. Sci. USA 89:10729-10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald, J. R., T. J. Foster, and D. Cox. 2006. The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 4:445-457. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald, J. R., A. Loughman, F. Keane, M. Brennan, M. Knobel, J. Higgins, L. Visai, P. Speziale, D. Cox, and T. J. Foster. 2006. Fibronectin-binding proteins of Staphylococcus aureus mediate activation of human platelets via fibrinogen and fibronectin bridges to integrin GPIIb/IIIa and IgG binding to the FcgammaRIIa receptor. Mol. Microbiol. 59:212-230. [DOI] [PubMed] [Google Scholar]
- 12.Ford, I., C. W. Douglas, D. Cox, D. G. Rees, J. Heath, and F. E. Preston. 1997. The role of immunoglobulin G and fibrinogen in platelet aggregation by Streptococcus sanguis. Br. J. Haematol. 97:737-746. [DOI] [PubMed] [Google Scholar]
- 13.Ford, I., C. W. Douglas, J. Heath, C. Rees, and F. E. Preston. 1996. Evidence for the involvement of complement proteins in platelet aggregation by Streptococcus sanguis NCTC 7863. Br. J. Haematol. 94:729-739. [DOI] [PubMed] [Google Scholar]
- 14.Grundmeier, M., M. Hussain, P. Becker, C. Heilmann, G. Peters, and B. Sinha. 2004. Truncation of fibronectin-binding proteins in Staphylococcus aureus strain Newman leads to deficient adherence and host cell invasion due to loss of the cell wall anchor function. Infect. Immun. 72:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartford, O., P. Francois, P. Vaudaux, and T. J. Foster. 1997. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol. Microbiol. 25:1065-1076. [DOI] [PubMed] [Google Scholar]
- 16.Herrick, S., O. Blanc-Brude, A. Gray, and G. Laurent. 1999. Fibrinogen. Int. J. Biochem. Cell. Biol. 31:741-746. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, S. P., W. S. Nesbitt, and S. Kulkarni. 2003. Signaling events underlying thrombus formation. J. Thromb. Haemost. 1:1602-1612. [DOI] [PubMed] [Google Scholar]
- 18.Loughman, A., J. R. Fitzgerald, M. P. Brennan, J. Higgins, R. Downer, D. Cox, and T. J. Foster. 2005. Roles for fibrinogen, immunoglobulin and complement in platelet activation promoted by Staphylococcus aureus clumping factor A. Mol. Microbiol. 57:804-818. [DOI] [PubMed] [Google Scholar]
- 19.McAleese, F. M., E. J. Walsh, M. Sieprawska, J. Potempa, and T. J. Foster. 2001. Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 276:29969-29978. [DOI] [PubMed] [Google Scholar]
- 20.McDevitt, D., P. Vaudaux, and T. J. Foster. 1992. Genetic evidence that bound coagulase of Staphylococcus aureus is not clumping factor. Infect. Immun. 60:1514-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreillon, P., and Y. A. Que. 2004. Infective endocarditis. Lancet 363:139-149. [DOI] [PubMed] [Google Scholar]
- 22.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien, L., S. W. Kerrigan, G. Kaw, M. Hogan, J. Penades, D. Litt, D. J. Fitzgerald, T. J. Foster, and D. Cox. 2002. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol. Microbiol. 44:1033-1044. [DOI] [PubMed] [Google Scholar]
- 24.O'Brien, L. M., E. J. Walsh, R. C. Massey, S. J. Peacock, and T. J. Foster. 2002. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell. Microbiol. 4:759-770. [DOI] [PubMed] [Google Scholar]
- 25.Perkins, S., E. J. Walsh, C. C. Deivanayagam, S. V. Narayana, T. J. Foster, and M. Hook. 2001. Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J. Biol. Chem. 276:44721-44728. [DOI] [PubMed] [Google Scholar]
- 26.Ponnuraj, K., M. G. Bowden, S. Davis, S. Gurusiddappa, D. Moore, D. Choe, Y. Xu, M. Hook, and S. V. Narayana. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115:217-228. [DOI] [PubMed] [Google Scholar]
- 27.Que, Y. A., J. A. Haefliger, L. Piroth, P. Francois, E. Widmer, J. M. Entenza, B. Sinha, M. Herrmann, P. Francioli, P. Vaudaux, and P. Moreillon. 2005. Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J. Exp. Med. 201:1627-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roche, F. M., R. Massey, S. J. Peacock, N. P. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 29.Rosado, J. A., E. M. Meijer, K. Hamulyak, I. Novakova, J. W. Heemskerk, and S. O. Sage. 2001. Fibrinogen binding to the integrin alpha(IIb)beta(3) modulates store-mediated calcium entry in human platelets. Blood 97:2648-2656. [DOI] [PubMed] [Google Scholar]
- 30.Schaffer, A. C., R. M. Solinga, J. Cocchiaro, M. Portoles, K. B. Kiser, A. Risley, S. M. Randall, V. Valtulina, P. Speziale, E. Walsh, T. Foster, and J. C. Lee. 2006. Immunization with Staphylococcus aureus clumping factor B, a major determinant in nasal carriage, reduces nasal colonization in a murine model. Infect. Immun. 74:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullam, P. M., A. S. Bayer, W. M. Foss, and A. L. Cheung. 1996. Diminished platelet binding in vitro by Staphylococcus aureus is associated with reduced virulence in a rabbit model of infective endocarditis. Infect. Immun. 64:4915-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsurupa, G., L. Tsonev, and L. Medved. 2002. Structural organization of the fibrin(ogen) alpha C-domain. Biochemistry 41:6449-6459. [DOI] [PubMed] [Google Scholar]
- 33.Walsh, E. J., L. M. O'Brien, X. Liang, M. Hook, and T. J. Foster. 2004. Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J. Biol. Chem. 279:50691-50699. [DOI] [PubMed] [Google Scholar]
- 34.Wells, J. M., P. W. Wilson, and R. W. Le Page. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629-636. [DOI] [PubMed] [Google Scholar]