Abstract
FimH is the tip adhesin of mannose-specific type 1 fimbriae of Escherichia coli, which are critical to the pathogenesis of urinary tract infections. Point FimH mutations increasing monomannose (1M)-specific uroepithelial adhesion are commonly found in uropathogenic strains of E. coli. Here, we demonstrate the emergence of a mixed population of clonally identical E. coli strains in the urine of a patient with acute cystitis, where half of the isolates carried a glycine-to-arginine substitution at position 66 of the mature FimH. The R66 mutation induced an unusually strong 1M-binding phenotype and a 20-fold advantage in mouse bladder colonization. However, E. coli strains carrying FimH-R66, but not the parental FimH-G66, had disappeared from the patient's rectal and urine samples collected from 29 to 44 days later, demonstrating within-host instability of the R66 mutation. No FimH variants with R66 were identified in a large (>600 strains) sequence database of fimH-positive E. coli strains. However, several strains carrying genes encoding FimH with either S66 or C66 mutations appeared to be relatively stable in the E. coli population. Relative to FimH-R66, the FimH-S66 and FimH-C66 variants mediated only moderate increases in 1M binding but preserved the ability to enhance binding under flow-induced shear conditions. In contrast, FimH-R66 completely lost shear-enhanced binding properties, with bacterial adhesion being inhibited by shear forces and lacking a rolling mode of binding. These functional trade-offs may determine the natural populational instability of this mutation or other pathoadaptive FimH mutations that confer dramatic increases in 1M binding strength.
Most Escherichia coli strains express type 1 fimbriae, hair-like adhesive organelles that mediate mannose-sensitive binding to host glycoproteins. The tip subunit, adhesin FimH, mediates shear-enhanced adhesion to monomannose (1M) receptors; the strength of binding is relatively weak under static conditions but is dramatically increased at high flow (21, 22). Binding properties of FimH can be dramatically altered by single point mutations that occur throughout the protein and are responsible for up to 15-fold increases in the ability of E. coli strains to bind 1M under static conditions (4, 14, 15, 17, 18). Uropathogenic strains tend to bind 1M significantly better than strains of intestinal origin, and 1M binding is correlated with the ability to bind uroepithelial cells and with colonization of the urinary bladder in the mouse model of urinary tract infection (15). Thus, FimH mutations resulting in enhanced 1M binding are pathogenicity adaptive (pathoadaptive) in nature.
Though FimH variants are 99% identical at the protein level, evolutionary analysis indicates that mutations occur in the adhesin more frequently than in even the structurally diverse major subunit of type 1 fimbriae, FimA (23). At the same time, mutations that enhance 1M binding do not persist in evolutionary terms due to the trade-off effects of pathoadaptive mutations (15). However, the nature of trade-off effects and the relative stability of clones carrying different pathoadaptive FimH variants have not been evaluated.
Here, we studied FimH mutation dynamics in a single patient, as well as at the E. coli species level. We show that a mutant FimH variant with powerful functional effects can be observed within a single patient but then disappears in less than 2 months, without entering stable species-level circulation among E. coli strains. At the same time, FimH mutations with more moderate functional effects do circulate within the species. We also show that loss of the shear-enhanced mode of FimH adhesion may represent an important functional trade-off for mutations that enhance 1M binding.
MATERIALS AND METHODS
Strain collection.
Isolates TOP17-AC-G66 and -R66, TOP17-AC-R1 and -R2, and TOP17-ABU and -RC (recurrent cystitis), along with TOP5-RC, PUMA1007-AC, HH36-R, and TOP41-RC, were obtained from the University of Washington UTI Strain Repository. All strains in the Repository were isolated from urine and rectal samples obtained from adult females during and between episodes of acute cystitis (AC), under protocols approved by Human Subjects Research Committee at University of Washington. To identify additional strains with position 66 polymorphisms, we queried our laboratory database, which consists of fimH sequences for more than 600 E. coli isolates of diverse origin with regard to geography, host species, health status, and anatomic site of recovery. The strains used for fimH analysis here are as follows. Reference strains Ecor10, Ecor25, Ecor38, Ecor39, Ecor41, Ecor51, Ecor54, and Ecor55, were originally obtained by Howard Ochman (12). Extraintestinal pathogenic strains PAP1071, Py114, VA1010, U1, and V20, as well as fecal strain G1040, were provided by James Johnson (University of Minnesota) and previously described regarding virulence characteristics and analyzed for fimH diversity (18). Netherlands newborn bacterial meningitis strain 90-1634 was generously provided by J. R. Johnson (5). Strains bearing the respective parent (G66 coding) alleles for the C66 and S66 variants are as follows. Strain F-18 (7) carries the gene encoding g259/G66 but is otherwise identical to FimH-C66b; likewise, strain Ed1e (4) carries the gene encoding g259/G66 but is otherwise identical to FimH-S66a.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was carried out as previously published (2). Briefly, bacteria from single colonies were grown overnight on solid media. The entire cells were embedded into agarose and lysed. After washing, the agarose blocks containing bacterial DNA were digested with XbaI and loaded onto agarose gels and the DNA fragments were separated by pulsed-field electrophoresis. After staining with ethidium bromide, digital images were obtained and analyzed using the BioNumerics software package (Applied Maths, Inc., Austin, TX).
Sequencing.
Sequencing primers for seven housekeeping loci and for fimH have been previously published (23, 25). Internal fragments of the seven housekeeping genes were concatenated. For descriptive purposes, housekeeping gene alleles and sequence types were assigned according to the publicly available, web-based multiple locus sequence type (MLST) website (25) curated by Mark Achtman (http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli). Nucleotide sequences for fimH and concatenated housekeeping gene haplotypes were aligned and FimH amino acid sequences were predicted using MacVector software (Accelrys, Inc., San Diego, CA). Phylogenetic relatedness among the concatenated housekeeping gene haplotypes was determined using the neighbor-joining algorithm.
Cloning.
fimH alleles of interest were cloned into pGB17 and transformed into the K-12 laboratory strain background, as previously described (17). Briefly, fimH genes were amplified from wild-type strains by PCR, subcloned into pACYC184-based plasmid pGB17 (Cmr), and introduced into fimH-null E. coli strain AAEC191A(pPKL114) for substrate binding and tissue adhesion assays.
Substrate binding assay.
Purified receptor compounds (yeast mannan and bovine RNase B) were obtained from Sigma Chemical Co. (St. Louis, MO). Glycoproteins were dissolved at 20 μg/ml, and collagen was dissolved at 50 μg/ml, in 0.02 M bicarbonate buffer, and 100-μl aliquots were incubated in 96-well microtiter plates for 75 min at 37°C, under static (nonshaking) conditions. The wells were then washed three times with phosphate-buffered saline (pH 7.2) and quenched with 0.1% bovine serum albumin in the same buffer. [3H]thymidine-labeled bacteria (5 × 107 CFU) were added with or without 50 mM methyl α-d-mannopyranoside inhibitor (Sigma, St. Louis, MO) and incubated for 45 min at 37°C without shaking to achieve saturation, and the wells were then washed to remove unbound bacteria. The number of bound bacteria in each well was determined by measuring the radioactivity level using scintillation counting. Experiments were carried out in duplicate, and reported results represent mean values with standard errors of the mean.
Bladder epithelial cell binding assay.
Confluent monolayers of T24 bladder cells were flask grown in minimum essential medium containing fetal bovine serum with penicillin and streptomycin. Tissue cells were briefly dissociated with trypsin-EDTA, seeded onto Lab-Tek chamber slides (Nalge Nunc Int., Naperville, IL), and grown in 5% CO2 at 37°C for 24 h to confluence. After washing with Hanks’ balanced salt solution (HBSS), cells were incubated at 37°C for 1 h in medium. Bacteria were adjusted in HBSS to an optical density at 600 nm of 0.6 and then diluted twofold with or without methyl α-d-mannopyranoside inhibitor. Bacteria were added to cells, incubated at 37°C for 1 h, and then removed. Wells were washed six times with 0.5 ml HBSS and then fixed and stained with Diff-Quik B4132-1 stain set (Dade Behring Inc., Newark, DE). After wells were washed with phosphate-buffered saline, slides were prepared for microscopic examination. The number of bacteria attached to five cells per high-power field was counted, examining four fields per coverslip. Experiments were performed in triplicate wells, and reported results represent mean values with standard errors of the mean.
Mouse urinary tract infection model.
We utilized the previously described ascending newborn mouse model of urinary tract infection (9). Briefly, following urethral catheterization of a female newborn C3H/HeN mouse (Charles River Labs, Wilmington, MA), a 50-μl volume containing 5 × 107 CFU of bacterial suspension was instilled into the bladder. Five mice each were infected with either TOP17-AC-G66 or TOP17-AC-R66. Mice were sacrificed at 24 h. The urinary bladders were homogenized and plated by serial dilution on agar for quantification. Reported results represent mean values with standard errors of the mean.
Flow chamber studies.
Binding assays under shear conditions were carried out in a flow chamber as previously published (11). Briefly, bacteria in buffer are passed over the surface of 1M-bovine serum albumin-coated dishes in a parallel-plate flow chamber (GlycoTech, Gaithersburg, MD). Digital images of the bacterial binding behavior were acquired by a Nikon TE200 or Nikon Diaphot inverted microscope with a ×10 phase-contrast objective, a Roper Scientific high-resolution Cascade camera, and MetaMorph video acquisition software (Molecular Devices Corp., Downington, PA) and analyzed using the MetaMorph software. Rolling and stationary bacteria were counted as described previously (21).
Genomic polymorphism detection.
Comparative analysis of genomic polymorphisms by differential hybridization of DNA from two clinical isolates was performed by NimbleGen (Madison, WI). A custom microarray was designed, based on the published CFT073 sequence (PubMed accession no. AE014075). In total, approximately 1 Mb throughout the chromosome was selected, including several major pathogenicity islands, the fim cluster, and the seven housekeeping genes (see supplemental material). Chromosomal DNA for TOP17 isolates from day 0 (AC; urine, FimH-R66) and day +44 (RC; urine, FimH-G66) was obtained. Comparative analysis identified putative nucleotide polymorphism signals in the window containing fimH nucleotide 259, as well as two additional windows in which direct sequencing did not confirm true point mutations.
RESULTS AND DISCUSSION
Mutational split in fimH in a patient with acute cystitis.
To investigate the populational dynamics of FimH mutations, we first analyzed within-host E. coli populations in urine samples obtained from six patients with AC. The urine samples were streaked on agar plates, and fimH sequencing was performed for eight colonies per plate. In samples from five patients, the within-patient population of AC bacteria carried identical fimH alleles. However, in the sample from one patient (TOP17), an even split in the AC population was detected, with four colonies carrying genes encoding g259 in fimH and four colonies carrying genes encoding c259, with the sequences otherwise identical. The nucleotide polymorphism resulted in an amino acid difference between the two encoded FimH proteins, with either glycine (G) or arginine (R), respectively, in position 66 of the mature protein.
The strains expressing FimH-G66 or FimH-R66 variants (TOP17-AC-G66 and TOP17-AC-R66, respectively) were of identical MLST and PFGE profiles (Fig. 1), indicating that they derived from the same parental strain and that the allelic split occurred very recently, possibly within the patient. According to the publicly available MLST database, the strains belong to the large ST73 clonal complex (25), which includes model uropathogenic E. coli strain CFT073 (24). ST73, in turn, resides in phylogenetic group B2, one of four major groups of the E. coli species and one from which most human uropathogenic strains originate (6).
FIG. 1.
Phylogenetic tree and PFGE profiles. To the left is a phylogenetic tree (dendrogram) built based on MLST haplotypes concatenated from the integral fragments of seven housekeeping genes, with distances indicating absolute numbers of nucleotide differences distributed proportionally among branches. To the right are PFGE profiles obtained using XbaI digestion. ST, sequence type. Abbreviations for TOP17 strains further identified by clinical setting: AC, acute cystitis; R, rectal; ABU, asymptomatic bacteriuria; RC, recurrent cystitis. Abbreviations for site of isolation: U, urine; S, stool; R, respiratory secretions; B, blood. Abbreviations for place of isolation: WA, Washington (Seattle); IA, Iowa; MN, Minnesota (Minneapolis); SE, Sweden; NY, New York; TO, Tonga.
Based on analysis of our database of fimH sequences from over 600 E. coli strains, g259 and thus G66 represent the consensus nucleotide and amino acid, respectively, appearing in the vast majority of alleles, including all variants of 11 fully sequenced model E. coli strains (Fig. 2A). No other FimH variant in our database had an R66 mutation. Thus, the FimH-G66 variant from patient TOP17 is the parent, wild-type protein, while FimH-R66 is the mutational derivative.
FIG. 2.
Amino acid and DNA sequences for FimH adhesin variants. Consensus residues are indicated below each position number. For sequenced strains, only polymorphisms relative to the consensus are shown, with identical positions indicated by hyphens. Published genomes compared to TOP17 isolates (A) and S66 and C66 FimH variants identified from our clinical fimH sequence database (B).
R66 is a mutation with pathogenicity-adaptive functional effect.
To determine whether the R66 mutation has a functional effect, we compared FimH-mediated binding of the TOP17-AC-G66 and TOP17-AC-R66 isolates. To ensure expression of the type 1 fimbriae, both isolates were passed three times through broth culture before the binding assay was performed. Both strains bound well to trimannose (3M) substrate RNase B (Fig. 3A), a model glycoprotein that possesses high-mannose, Man5-type oligosaccharides and is bound well by all functional FimH variants via a trisaccharide-specific mechanism (3, 15). However, the TOP17-AC-R66 strain bound more than four times better than the TOP17-AC-G66 strain to yeast mannan, a model 1M substrate to which different FimH variants demonstrate more than 15-fold variability in binding level (15). The 1M/3M binding ratios were 0.96 for FimH-R66 bacteria and 0.29 for FimH-G66 bacteria.
FIG. 3.
(A) 1M-specific (filled bars) and 3M-specific (open bars) binding of wild-type isolates under static conditions. (B) 1M-specific (filled bars) and 3M-specific (open bars) binding of isogenic strain pairs under static conditions: (i) FimH-G66 versus FimH-R66 variants from the TOP17 isolates. (ii) FimH-C66 and its parent G66 variant. (iii) FimH-S66 and its parent G66 variant. (C) T24 bladder cell binding of the isogenic strain pair expressing the TOP-17 FimH-R66 and parent FimH-G66 variants. (D) Mouse bladder colonization at 24 h of TOP17-AC-R66 and TOP17-AC-G66 strains. (E) Fifty percent inhibitory concentrations (IC50) of α-methylmannoside for the isogenic strain pairs. Results represent mean values with standard errors of the mean.
The same 1M and 3M binding pattern was observed when the wild-type and mutant FimH variants were compared in a recombinant isogenic background (Fig. 3B, panel i), with the FimH-R66 strain binding more than twofold better than the isogenic FimH-G66 strain. The less-pronounced difference in the 1M binding of recombinant strains versus wild-type isolates is unlikely to be due to additional non-type 1 fimbria-related differences between the latter variants, because the 1M binding was inhibited more than 90% by soluble mannose. Instead, we believe that the use of a multicopy-plasmid system for fimbrial expression in the isogenic strains results in a much higher level of FimH expression on the surface of both strains, leading to binding saturation effects in the adhesion test.
In addition, the recombinant FimH-R66 strain adhered to T24 bladder epithelial cells at a greater than threefold higher rate than the FimH-G66 strain (Fig. 3C), in accordance with previous studies demonstrating that FimH-mediated E. coli binding to uroepithelial cells occurs via 1M-specific mechanisms (10, 26). (Wild-type isolates could not be used in the cell-binding assay due to their pronounced cytotoxicity.)
Thus, R66 belongs to the category of pathoadaptive, 1M-enhancing FimH mutations that have been shown to provide an advantage in urinary tract colonization (15). Indeed, the TOP17-AC-R66 strain colonized mouse bladders approximately 20 times better than TOP17-AC-G66 bacteria (Fig. 3D). Thus, the R66 mutation likely provided a selective advantage in bladder colonization during the course of acute cystitis in patient TOP17.
Clones carrying the FimH R66 mutation are relatively unstable.
We further investigated within-host populational dynamics of the FimH R66 mutation by analyzing strain diversity in additional samples from patient TOP17: (i) a rectal swab obtained at the same time as the AC urine sample (day 0); (ii) a rectal swab obtained during healthy surveillance on day +29; (iii) urine obtained on day +35, when the patient had asymptomatic bacteriuria; and (iv) urine obtained on day +44, when the patient was diagnosed with recurrent cystitis. Both rectal E. coli populations were mixed (containing a second, non-ST73 strain [not shown]) but did contain a strain with MLST/PFGE genotypes matching those of TOP17-AC-G66/R66 strains: isolates TOP17-AC-R1 and TOP17-AC-R2 (Fig. 1). The two subsequent urinary samples (represented by isolates TOP17-ABU and TOP17-RC) demonstrated homogeneous populations, with MLST/PFGE profiles identical to those of the TOP17-AC-G66/R66 strains. Thus, the subsequent infections were caused by the same E. coli strain found in the AC and concurrent rectal samples. To determine fimH variability of the subsequent sample populations, sequencing of fimH was performed for 30 to 45 bacterial colonies from each urine or rectal sample, with an expanded number of colonies screened from the original AC urine sample as well.
As in the AC population, a mixture of FimH-G66 and -R66 strains was detected among the isoclonal bacteria from the day 0 rectal sample. However, FimH-R66 carriers were not as common in the rectal population (12 of 45 colonies) as in the AC population (17 of 34 colonies; P < 0.05). The concurrent recovery of R66-FimH strains in the day 0 rectal and urine cultures obscures the true site of origin of the mutant strain. It is possible that FimH-R66 was selected originally in the intestine (or other nonurinary compartment) and then introduced into the bladder alongside FimH-G66 in a mixed population, where it was selected further. It is also possible that FimH-R66 originated in the bladder compartment during the acute infection and spread back into the perianal and/or rectal compartments.
In contrast to the FimH-variable populations seen on day 0, only FimH-G66 bacteria were detected in the subsequent rectal and urinary samples and the lack of FimH-R66 colonies was statistically significant compared to the day 0 urinary and rectal populations (P < 0.05 and P < 0.01, respectively). This drastic reduction (if not outright disappearance) of FimH-R66 E. coli from the colonizing and infecting populations indicates within-host instability of the FimH-R66 clone.
It is uncertain in which host compartment the R66 clone was selected against and whether the R66 mutation itself was subject to negative selection. We attempted to confirm a FimH-specific selection process by seeking additional mutations in approximately 1 Mb of chromosomal sequence by using a custom gene-tiling microarray (NimbleGen) to compare genomic DNA of TOP17-AC-R66 (day 0) and TOP17-RC (day +44). For the microarray design, we selected pathogenicity island-associated and backbone genes from the sequenced genome of E. coli strain CFT073, which belongs to the same clonal complex (ST73) as the TOP17 isolates. If mutations occurred at high frequency in this strain background, additional mutations would be expected to accumulate around the bacterial chromosome over time. However, no mutations other than FimH-R66 were detected between the two strains. Thus, the strain TOP17 background did not demonstrate hypermutability, leaving the fimH locus among a likely few primary targets of positive selection and, presumably, of negative selection later on.
The critical role of FimH mutation R66 in the natural instability of the carrier clone is also supported by E. coli species-level population analysis. As mentioned above, no FimH-R66 mutations were found in our database of naturally occurring fimH allelic (entire coding region) variants, compiled from over 600 E. coli strains, of which one-third were of urinary tract origin. In the database, however, we identified three unique fimH alleles that encoded FimH with C66 instead of the consensus G66 and four unique alleles encoding FimH with S66 (Fig. 2B). None of these seven alleles was phylogenetically linked; that is, they differed by various silent and, in some cases, amino acid changes. The latter indicates that C66 or S66 mutations have occurred independently multiple times in E. coli fimH, suggesting that position 66 is a FimH mutational hot spot, which is consistent with the R66 mutation emerging under positive selection (that is, by providing competitive advantage over G66-bearing strains). In contrast to the FimH-R66 allele, some FimH-C66/S66 alleles were found in multiple strains: FimH-C66a and FimH-C66b alleles were found in five and two strains, respectively, while FimH-S66a and FimH-S66b alleles were found in five and four strains, respectively (Fig. 1). Strains bearing certain of these fimH alleles have been recovered from different hosts (and some even from different species) at different times—spanning up to 10 years—and/or in different geographical locations. Moreover, such strains were genetically diverse. Though FimH-C66a, FimH-C66b, or FimH-S66b strains were either completely or nearly identical to each other by MLST, the PFGE profiles for corresponding strains were distinctive, indicating genotypic diversification of the clones subsequent to acquisition of the respective FimH mutations. Furthermore, some strains carrying FimH-S66a belonged to different MLST haplotypes and even to distantly related major clonal groups of E. coli, indicating horizontal transfer of FimH-S66a alleles between phylogenetic groups A (ST10 and ST43) and B1 (ST410). The diversity of the C66/S66-bearing strains, genetic and epidemiologic, indicates relative population stability of FimH-C66 and -S66 variants compared to the FimH-R66 variant that disappears rapidly from circulation. Here, we define populational stability not as continuous, indefinite circulation of specific fimH alleles, but rather as the population's ability to circulate long enough to be detected in epidemiologically unlinked bacterial strains.
Dramatic functional trade-off effect of the R66 mutation.
To understand the differential populational stability of position 66 mutations, we utilized isogenic backgrounds to compare the functional properties of FimH variants to the respective parent (G66 bearing) FimH variants from which they derived. Similar to the R66 mutation, both C66 and S66 mutations conferred increases in 1M binding relative to the parent proteins, but the increases were significantly smaller than that mediated by R66 (Fig. 3B, panels ii and iii). The binding phenotype conferred by FimH-R66 was also significantly stronger than that of 10 other naturally occurring FimH alleles in our collection that (i) increase 1M binding, with the 1M/3M ratios falling in the 0.25 to 0.65 range (not shown); and (ii) can be considered relatively population stable, as they appear in multiple, genetically diverse E. coli strains (18). In fact, the only naturally occurring FimH mutations that have demonstrated binding comparable to that of FimH-R66 were FimH-A56 and FimH-R41, found only in single strains (18, 23). The latter variants were recovered during urinary tract infections and like FimH-R66 may have emerged in individual hosts under positive selection but then disappeared rapidly from circulation. Thus, the functional effect of R66 mutation is quite pronounced compared to those of other mutations that are relatively stable on the populational level.
It has been demonstrated that increased 1M binding due to FimH structural mutations is associated with increased sensitivity to inhibition by soluble mannosylated compounds (15, 16). This increased sensitivity could produce a significant trade-off for bacteria during binding to oropharyngeal or gut mucosa bathed with mannosylated glycoproteins. All mutations in position 66 resulted in an increase in sensitivity to mannose relative to parent proteins (Fig. 3E). However, the difference in sensitivities between the R66, C66, and S66 mutants was rather small, if any, indicating that other functional trade-offs may determine the differences in populational stability.
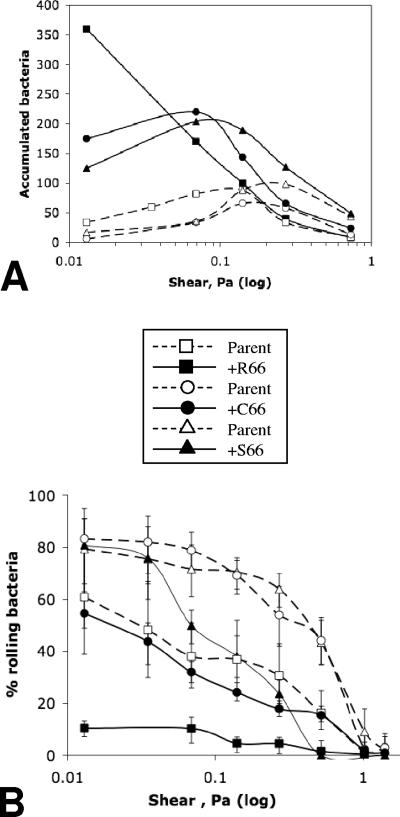
To identify possible additional ‘trade-off’ effects of the R66 mutation, we investigated the impact of position 66 mutations on bacterial binding under flow conditions. We tested the effect of flow conditions on E. coli surface accumulation mediated by the FimH-R66/S66/C66 variants and their respective parent proteins. As expected, bacterial binding under low shear (0.013 Pa) corresponded to the relative binding of the variants under static conditions, with FimH-R66 mediating 2- to 60-fold-higher binding than other FimH variants (Fig. 4A). With increased shear, all FimH variants except FimH-R66 exhibited increases in binding to some degree. In contrast, the R66 variant demonstrated a distinct, shear-inhibited binding pattern; FimH-R66 binding under the next shear level tested (0.069 Pa) dropped below that mediated by either FimH-C66 or FimH-S66 and, with further shear increase, the R66 variant bound as poorly as any other variant tested.
FIG. 4.
(A) Surface accumulation of isogenic strain pairs featuring FimH position 66 polymorphisms under a range of shear stresses. (B) Percentage of surface-rolling bacteria to total (rolling and firmly bound) bacteria for the isogenic strain pairs under a range of shear stresses. Results represent mean values with standard errors of the mean.
It has also been shown previously that under flow, type 1-fimbriated bacteria attach to the surface in either a loose-rolling mode or a firm, stationary-adhesion mode (20). Conversion of rolling into stationary adhesion is another manifestation of the shear-enhanced catch-bond properties of FimH (20). For the six FimH variants tested, the proportion of rolling bacteria was significantly higher under low shear than high shear (Fig. 4B). FimH-R66 bacteria, however, demonstrated the lowest proportion of rolling bacteria under all different shears, with only 10% rolling at even the lowest shear tested.
Although shear level (which depends directly on both fluid velocity and viscosity) is difficult to evaluate precisely in natural host compartments, the range of shears tested here are well within the physiological range. For example, shear level was measured at 0.017 Pa in the proximal renal tubule, 0.07 Pa at the tooth surface, 0.1 to 0.2 Pa at the wall of venous vessels, 1 to 2 Pa at arterial walls, and up to 10 Pa in the aorta (8). Based on the urethral lumen size and urine velocity, shear in the urethral passage could reach 0.3 to 0.5 Pa during voiding. Shear stress in the oropharynx or large intestine (habitats for intestinal type 1-fimbriated E. coli) has not been measured or estimated but, due to the high viscosity of mucosal fluids, could reach relatively high levels.
The ability to bind strongly under low-shear conditions may provide FimH-R66 an advantage in such compartments as the urinary bladder (between voids) or the renal tubules, especially at the early stages of infection, when the number of infecting bacteria is low. However, such strong binding might also promote apoptotic host cell responses and/or increased recruitment of host immune defense cells (10, 13) and, thus, represent a liability for the invading bacteria at later stages of the infection, when bacterial counts are high. The shear-inhibited phenotype of R66 clones may not allow free-floating bacteria to attach to urethral mucosa in the course of voiding and thus sustain themselves in the urinary tract. It also might prove to be a significant disadvantage in other compartments critical for populational stability of E. coli, where the shear level is relatively high. For example, recently it has been shown that strong and firm 1M binding inhibits the spread of developing E. coli biofilms under flow conditions (1).
While trade-off effects are also expected in FimH mutants with more modest phenotypes than FimH-R66, they are likely to be less dramatic and thereby allow the host strains to maintain a certain degree of populational stability. One should note, however, that although “moderate” FimH mutants are relatively stable in the E. coli population, they are not as persistent over evolutionary time scales as parental FimH variants (15, 18). This is evident from the lack of silent changes in fimH alleles after structural mutations have been acquired, as noted for the various C66/S66 alleles (Fig. 2B). Most silent polymorphisms are expected to be neutral and to accumulate at random, serving as a molecular evolutionary “clock.” The lack of silent mutations among the functional FimH mutants, therefore, indicates that the variant alleles have not persisted in the bacterial population long enough to accumulate such changes. This pattern contrasts with that seen in some of the parental FimH variants, in which the level of silent variability is comparable to that of housekeeping genes and reflects their long-term stability of fimH in the E. coli species (18). Evidently, moderate functional effects nonetheless produce a sufficient trade-off to result in the elimination of mutant FimH variants in the long term. Continuous emergence and elimination (whether slower or faster) of uropathogenicity-adaptive FimH mutations in E. coli populations are consistent with the “source-sink” model of adaptive evolution in bacterial pathogens (19). Under source-sink dynamics, bacteria continuously spread from stable reservoir habitats (sources) to transient virulence habitats (sinks), with mutations adaptive in the latter habitat being a liability in the former and leading to their rapid (at least on the evolutionary time scale) disappearance from circulation.
Taken together, our results indicate that pathoadaptive, 1M-enhancing FimH mutations can demonstrate distinctive populational dynamics. On the one hand, FimH mutations may emerge and disappear in a single host with urinary tract infection, under positive and negative selection, respectively. On the other hand, FimH mutations may circulate within the E. coli species for a sufficient length of time that the carrier clones accumulate a certain level of genetic diversity. It appears that the differential populational stability of the mutations is inversely proportional to the degree of functional effect induced. Mutations with very strong effects disappear quickly, while those with moderate effects are more stable. This pattern may result from functional trade-offs, wherein the mutations enhancing 1M binding also produce (i) increased sensitivity to mannosylated inhibitors, (ii) a lack of ability to increase (or even maintain) an essential level of surface accumulation under increasing flow, and (iii) inability to roll under low-flow conditions. Thus, FimH mutations may promote colonization of certain niches in the short term but represent a significant liability to carrier clones over the longer term.
Supplementary Material
Acknowledgments
We thank Steve Moseley for critical reading and thoughtful discussion of the manuscript; Mansour Samadpour for technical and material support of the PFGE analysis; and Marsha Cox, Cheryl Wobbe, and Ann Stapleton for providing clinical isolates from study archives.
This work was supported by Public Health Service grants DK64540, DK053369, AI057737 from the National Institutes of Health.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 14 May 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anderson, B. N., A. M. Ding, L. M. Nilsson, K. Kusuma, V. Tchesnokova, V. Vogel, E. V. Sokurenko, and W. E. Thomas. 2007. Weak rolling adhesion enhances bacterial surface colonization. J. Bacteriol. 189:1794-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeit, R. D. 1995. Laboratory procedures for the epidemiologic analysis of microorganisms, p. 190-208. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society of Microbiology, Washington, DC.
- 3.Bouckaert, J., J. Mackenzie, J. L. de Paz, B. Chipwaza, D. Choudhury, A. Zavialov, K. Mannerstedt, J. Anderson, D. Pierard, L. Wyns, P. H. Seeberger, S. Oscarson, H. De Greve, and S. D. Knight. 2006. The affinity of the FimH fimbrial adhesin is receptor-driven and quasi-independent of Escherichia coli pathotypes. Mol. Microbiol. 61:1556-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hommais, F., S. Gouriou, C. Amorin, H. Bui, M. C. Rahimy, B. Picard, and E. Denamur. 2003. The FimH A27V mutation is pathoadaptive for urovirulence in Escherichia coli B2 phylogenetic group isolates. Infect. Immun. 71:3619-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in The Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 6.Johnson, J. R., and T. A. Russo. 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli.” J. Lab. Clin. Med. 139:155-162. [DOI] [PubMed] [Google Scholar]
- 7.Krogfelt, K. A., B. A. McCormick, R. L. Burghoff, D. C. Laux, and P. S. Cohen. 1991. Expression of Escherichia coli F-18 type 1 fimbriae in the streptomycin-treated mouse large intestine. Infect. Immun. 59:1567-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malek, A. M., S. L. Alper, and S. Izumo. 1999. Hemodynamic shear stress and its role in atherosclerosis. JAMA 282:2035-2042. [DOI] [PubMed] [Google Scholar]
- 9.Mobley, H. L., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol. Microbiol. 10:143-155. [DOI] [PubMed] [Google Scholar]
- 10.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson, L. M., W. E. Thomas, E. V. Sokurenko, and V. Vogel. 2006. Elevated shear stress protects Escherichia coli cells adhering to surfaces via catch bonds from detachment by soluble inhibitors. Appl. Environ. Microbiol. 72:3005-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schilling, J. D., S. M. Martin, D. A. Hunstad, K. P. Patel, M. A. Mulvey, S. S. Justice, R. G. Lorenz, and S. J. Hultgren. 2003. CD14- and Toll-like receptor-dependent activation of bladder epithelial cells by lipopolysaccharide and type 1 piliated Escherichia coli. Infect. Immun. 71:1470-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokurenko, E. V., V. Chesnokova, R. J. Doyle, and D. L. Hasty. 1997. Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J. Biol. Chem. 272:17880-17886. [DOI] [PubMed] [Google Scholar]
- 15.Sokurenko, E. V., V. Chesnokova, D. E. Dykhuizen, I. Ofek, X. R. Wu, K. A. Krogfelt, C. Struve, M. A. Schembri, and D. L. Hasty. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. USA 95:8922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokurenko, E. V., H. S. Courtney, J. Maslow, A. Siitonen, and D. L. Hasty. 1995. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J. Bacteriol. 177:3680-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokurenko, E. V., H. S. Courtney, D. E. Ohman, P. Klemm, and D. L. Hasty. 1994. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J. Bacteriol. 176:748-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokurenko, E. V., M. Feldgarden, E. Trintchina, S. J. Weissman, S. Avagyan, S. Chattopadhyay, J. R. Johnson, and D. E. Dykhuizen. 2004. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol. Biol. Evol. 21:1373-1383. [DOI] [PubMed] [Google Scholar]
- 19.Sokurenko, E. V., R. Gomulkiewicz, and D. E. Dykhuizen. 2006. Source-sink dynamics of virulence evolution. Nat. Rev. Microbiol. 4:548-555. [DOI] [PubMed] [Google Scholar]
- 20.Thomas, W., M. Forero, O. Yakovenko, L. Nilsson, P. Vicini, E. Sokurenko, and V. Vogel. 2006. Catch-bond model derived from allostery explains force-activated bacterial adhesion. Biophys. J. 90:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas, W. E., L. M. Nilsson, M. Forero, E. V. Sokurenko, and V. Vogel. 2004. Shear-dependent ‘stick-and-roll’ adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol. 53:1545-1557. [DOI] [PubMed] [Google Scholar]
- 22.Thomas, W. E., E. Trintchina, M. Forero, V. Vogel, and E. V. Sokurenko. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell 109:913-923. [DOI] [PubMed] [Google Scholar]
- 23.Weissman, S. J., S. Chattopadhyay, P. Aprikian, M. Obata-Yasuoka, Y. Yarova-Yarovaya, A. Stapleton, W. Ba-Thein, D. Dykhuizen, J. R. Johnson, and E. V. Sokurenko. 2006. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 59:975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, G., W. J. Mo, P. Sebbel, G. Min, T. A. Neubert, R. Glockshuber, X. R. Wu, T. T. Sun, and X. P. Kong. 2001. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J. Cell Sci. 114:4095-4103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.