Abstract
Escherichia coli is the primary cause of urinary tract infection (UTI) in the developed world. The major factors associated with the virulence of uropathogenic E. coli (UPEC) are fimbrial adhesins, which mediate specific attachment to host receptors and trigger innate host responses. Another group of adhesins is represented by the autotransporter subgroup of proteins. The best characterized of these proteins, antigen 43 (Ag43), is a self-recognizing adhesin that is associated with cell aggregation and biofilm formation in E. coli K-12. The sequenced genome of prototype UPEC strain CFT073 contains two variant Ag43-encoding genes located on pathogenicity islands. The biological significance of both of these genes and their role in UPEC pathogenesis have not been investigated previously. Here we performed a detailed molecular characterization analysis of Ag43a (c3655) and Ag43b (c1273) from UPEC CFT073. Expression of Ag43a and Ag43b in a K-12 background revealed that they possess different functional properties. Ag43a produced a strong aggregation phenotype and promoted significant biofilm growth. Deletion mutants and strains constitutively expressing Ag43a and Ag43b were also constructed using CFT073. When these mutants were analyzed in a mouse model of UTI, Ag43a (but not Ag43b) promoted long-term persistence in the urinary bladder. Our findings demonstrate that Ag43a contributes to UPEC disease pathogenesis and reveal that there are pathogenicity-adapted variants of Ag43 with distinct virulence-related functions.
Urinary tract infections (UTI), which start as bladder infections and often evolve to encompass the kidneys, are among the most common infectious diseases of humans. It is estimated that 40 to 50% of adult healthy women have experienced at least one UTI episode in their lifetime, and there is a tendency for these infections to become chronic due to a high rate of recurrence. Escherichia coli is the cause of the majority (>80%) of UTI in humans and is one of the most common sources of gram-negative bacteremia in hospitalized patients. Almost all patients with an indwelling urinary catheter that is present for 30 days or longer develop catheter-associated UTI, which account for 40% of all nosocomial infections (11).
The ability of uropathogenic E. coli (UPEC) to colonize the urinary tract involves adhesins (e.g., type 1 and P fimbriae) and toxins (e.g., hemolysin) (27, 33). Adherence to the urinary tract epithelium enables the bacteria to resist the hydrodynamic forces of urine flow, to trigger host and bacterial cell signaling pathways, and to establish an infection. Among the adhesins, P fimbriae show the strongest disease association in clinical studies. P fimbriae contribute to the establishment of bacteriuria by binding to the α-d-galactopyranosyl-(1-4)-β-d-galactopyranoside receptor epitope in the globoseries of glycolipids (29) and activate innate immune responses in animal models and in the human urinary tract (3, 37, 38, 54-56). Type 1 fimbriae also enhance colonization and induce immune responses in the mouse UTI model (8). Type 1 fimbriae confer binding to α-d-mannosylated proteins, such as uroplakins, which are abundant in the bladder (53). Both P and type 1 fimbriae recognize their receptor targets by virtue of organelle tip-located adhesins (PapG and FimH, respectively) (27).
The phase-variable surface-located adhesin antigen 43 (Ag43) is also associated with urovirulence (1). Ag43 is a representative member of the autotransporter (AT) family. AT proteins are unique in that their primary sequence is sufficient to direct their transport across the bacterial membrane system and final routing of a variable passenger domain (alpha-domain) to the cell surface. AT proteins are generally very similar with respect to the structure of the transporter module (beta-domain) that assists the transport of the alpha-domain across the outer membrane, whereas they differ substantially in the alpha-domain, which determines the unique functional characteristics of AT proteins (17, 18). Once at the bacterial surface, the alpha-domain may be processed and released into the extracellular surroundings (e.g., Pet and EspP), or it may be cleaved but remain in contact with the cell surface via noncovalent interactions with the beta-domain (e.g., Ag43 and AIDA) (18). Thus, AT proteins have diverse functions and range from cell-associated adhesins to secreted toxins (17, 18).
Ag43 (encoded by the flu gene) is a self-recognizing AT adhesin that confers characteristic surface properties on host cells, such as autoaggregation and a frizzy colony morphology (14, 15). Large amounts of this protein are expressed (approximately 50,000 copies per cell), and the protein promotes bacterial biofilm formation (9, 24-26, 36, 42, 43, 44). The phase-variable expression of Ag43 occurs at switching rates of ∼10−3 per cell per generation due to the concerted actions of Dam methylase (positive regulation) and OxyR (negative regulation) (19, 43, 46, 52). Glycosylation of Ag43 occurs in some wild-type UPEC strains, and Ag43 glycosylation in recombinant E. coli enhances binding to HEp-2 cells (47).
A recent genome comparison analysis revealed that the sequenced genome of UPEC strain CFT073 encodes at least 10 putative AT proteins (35). CFT073 has two copies of the Ag43-encoding flu gene, both of which are located on pathogenicity islands. The two genes (fluA/upaF and fluB/upaD) are not identical; they share 91% conservation at the nucleotide sequence level and 90% identity at the amino acid level. The predicted alpha-domain of their protein products (Ag43a and Ag43b) shares 81.7 and 78.4% amino acid identity, respectively, with the K-12 Ag43 alpha-domain. The two predicted proteins themselves share 85% amino acid identity in the entire alpha-domain. Recently, the region of Ag43 responsible for autoaggregation was localized to the first 160 amino acids of the mature protein (26). The Ag43a and Ag43b proteins share only 76% amino acid identity in this region.
While the function of Ag43 is well documented, its precise role in UPEC disease pathogenesis remains to be elucidated. Ag43 is expressed on the surface of UPEC cells located within intracellular biofilm-like bacterial pods in the bladder epithelium, indicating that it may contribute to survival and persistence during prolonged infection (1). The in vivo expression of Ag43 by UPEC has also been demonstrated via direct examination of infected human urine using immunofluorescence microscopy (34). The biological significance of multiple copies of the flu gene is also unclear. Here, we characterized the function of both Ag43 variants of UPEC CFT073 and investigated their role in virulence. The fluA and fluB genes were cloned, their protein products were characterized, and their functions were studied both in the heterologous E. coli K-12 host and in UPEC CFT073. Deletion and expression-controlled mutants were compared to wild-type CFT073 in a mouse UTI model, and the results revealed an important contribution of Ag43a to long-term persistence in the bladder. The findings indicate a role for distinct Ag43 variants in UPEC disease pathogenesis and identify Ag43 as another class of pathogenicity-adapted cell surface proteins that possess amino acid alterations that are associated with distinct virulence-related functions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. The uropathogenic and commensal E. coli strains used to study the prevalence of the flu gene were obtained from the Princess Alexandra Hospital (Brisbane, Australia) or were part of the ECOR collection. Cells were routinely grown at 37°C on solid lysogeny broth (LB) medium or in liquid (4) supplemented with the appropriate antibiotics, including kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), ampicillin (100 μg/ml), apramycin (30 μg/ml), and zeocin (50 μg/ml). For growth in defined conditions, M9 and morpholinepropanesulfonic acid (MOPS) glucose media (41) were used as indicated below. Plasmid pCO2 contains the flu gene from MG1655 and was created by inserting a kanamycin resistance-encoding gene into the HindIII site of plasmid pKKJ143 (23). Plasmids pCO3 (with fluB from CFT073) and pCO4 (with fluA from CFT073) were constructed by insertion of a kanamycin resistance-encoding gene into the HindIII sites of plasmids pLH6 and pLH26, respectively (26). Resistance to kanamycin was required to facilitate transformation of these plasmids into the flu gfp+ K-12 strain OS56. In all constructs, expression of each flu gene is under control of the arabinose-inducible araBAD promoter (13). All plasmid constructs were verified by sequence analysis.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli K12 strains | ||
| MS427 | MG1655 Δflu | 36 |
| OS56 | MG1655 Δflu Gfp+, Ampr | 48 |
| MS1199 | pBAD/MycHisA in OS56 | This study |
| MS1230 | pCO2 in OS56, Ampr Kmr | This study |
| MS1231 | pCO3 in OS56, Ampr Kmr | This study |
| MS1232 | pCO4 in OS56, Ampr Kmr | This study |
| E. coli CFT073 strains | ||
| CFT073 | Wild-type UPEC isolate | 31 |
| CFTfluA | CFT073 ΔfluA::Apra, Aprar | This study |
| CFTfluB | CFT073 ΔfluB::cat, Cmr | This study |
| CFTfluA fluB | CFT073 ΔfluA::Apra ΔfluB::cat, double mutant, Aprar Cmr | This study |
| CFTfluB+ | CFT073 ampPcLfluB, constitutively expressed Ag43b | This study |
| CFTfluA fluB+ | CFT073 ΔfluA::Apra ampPcLfluB constitutively expressed Ag43b and Ag43a is absent, Aprar Ampr | This study |
| CFTfluB+ | CFT073 kmPcLfluA, constitutively expressed Ag43a | This study |
| CFTfluA+fluB | CFT073 kmPcLfluA ΔfluB::cat, constitutively expressed Ag43a and Ag43b is absent, Kmr Cmr | This study |
| CFTfluA+fluB+ | CFT073 kmPcLfluA ampPcLfluB, constitutively expressed Ag43a and Ag43b, Kmr Ampr | This study |
| CFToxyR | CFT073 ΔoxyR::Zeo, Zeor | This study |
| CFToxyR fluA | CFT073 ΔoxyR::Zeo ΔfluA::Apra, Zeor Aprar | This study |
| CFToxyR fluB | CFT073 ΔoxyR::Zeo ΔfluB::cat, Zeor Cmr | This study |
| CFToxyR fluA fluB | CFT073 ΔoxyR::Zeo ΔfluA::Apra ΔfluB::cat, Zeor Aprar Cmr | This study |
| Plasmids | ||
| pKKJ143 | flu gene from MG1655 in pBADMycHisA, Ampr | 23 |
| pLH6 | fluB gene (c1273) from CFT073 in pBADMycHisA, Ampr | 26 |
| pLH26 | fluA gene (c3655) from CFT073 in pBADMycHisA, Ampr | 26 |
| pCO2 | flu gene from MG1655 in pBADMycHisA-kan, Ampr Kmr | This study |
| pCO3 | fluB gene (c1273) from CFT073 in pBADMycHisA-kan, Ampr Kmr | This study |
| pCO4 | fluA gene (c3655) from CFT073 in pBADMycHisA-kan, Ampr Kmr | This study |
| pHHA13 | Type 1 fimbrial gene cluster from PC31 in pACYC184 | H. Hasman |
Km, kanamycin; Cm, chloramphenicol; Amp, ampicillin; Apra, apramycin; Zeo, zeocin.
DNA manipulations and genetic techniques.
DNA techniques were performed as described by Sambrook et al. (41). Isolation of plasmid DNA was carried out using a QIAprep Spin miniprep kit (QIAGEN). Restriction endonucleases were used according to the manufacturer's specifications (New England Biolabs). Chromosomal DNA purification was performed using a GenomicPrep cell and tissue DNA isolation kit (Amersham Pharmacia Biotech Inc.). Oligonucleotides were purchased from Sigma, Australia or Sigma, France. All PCRs were performed with the Expand High Fidelity polymerase system (Roche) used as described by the manufacturer. Amplified products were sequenced to ensure fidelity of the PCR. DNA sequencing was performed using an ABI Big Dye ver3.1 kit (ABI) by the Australian Genome Research Facility, Brisbane. Primers for flu gene prevalence studies were designed for conserved (5′-GGGTAAAGCTGATAATGTCG and 5′-GTTGCTGACAGTGAGTGTGC) and variant (5′-GCACTGGTTACCAGYAC and 5′-GGGACTTCWGCACGATAA) regions. The following program was used: 25 cycles of 94°C for 15 s, 50°C for 15 s, and 72°C for 1 min.
Construction of deletion and expression mutants.
In order to interrupt the fluA and fluB genes in CFT073, as well as to place chromosomal target genes under control of the PcL cassette (S. Da Re et al., submitted for publication), we used a three-step PCR procedure described previously (6, 30) and at http://www.pasteur.fr/recherche/unites/Ggb/matmet.html. The primers used to disrupt or insert the cassette upstream of the genes used in this study are described in Table 2.
TABLE 2.
Primers used in this study
| Target gene | Primer name | Primer sequence |
|---|---|---|
| fluA | c3655.500.5 | CACGCACGGTTCTGCGTGCCGTC |
| c3655.500-3 | GACTGAGCATATTTGAGTGCCC | |
| c3655.cat.L-3 | CTGCGAAGTGATCTTCCGTCACAGGGCTCTGCCCTCCGCCATCACGA | |
| c3655.cat.L-5 | GATGAGTGGCAGGGCGGGGCGTAACTGAATGTGACCTTCTGACAGAAC | |
| c3655.ext-5 | CAGCCTGCTGTTTAACAATAAC | |
| c3655.ext-3 | CGCATGTCAACAGGTTTAATTGC | |
| c3655.xBAD-500.5 | CACGCACGGTTCTGCGTGCCGTC | |
| c365.xBAD-500-3 | GCTCTGCCCTCCGCCATCACGA | |
| c3655.xBAD.L-3 | GAATCCCTGCTTCGTCCATTTGACAAGCTATTGCACGGACAAGAGAGACA | |
| c3655.xBAD.L-5 | GTTTCTCCATACCCGTTTTTTTGGTCCTGTTAGCAACGTGCGCAGATA | |
| c3655.PcL.L-3 | GTGAGAATTACTAACTTGAGCGAAAAGCTATTGCACGGACAAGAGAGACA | |
| c3655.PcL.L-5 | CGGTGATAATGGTTGCATGTATATCCTGTTAGCAACGTGCGCAGATA | |
| c3655.xBAD.ext-5 | CAGCCTGCTGTTTAACAATAAC | |
| c3655.xBAD.ext-3 | CCACTGCTCACCTCTGTTATCCAG | |
| 566 | CAACCAGAAAGGAAAGCTGCAGGTG | |
| 567 | TTCCCGGAAAGGGTAAGTATGGCGC | |
| fluB | c1273.500.5 | TCCGTACGGCCCTGCGCGCTGTTC |
| c1273.500-3 | TGATATCTTCAGGTGTGAGCCCG | |
| c1273.apra.L-3 | CACCCAGCCTGCGCGAGCAGGGGAGAAGCCTTCCCACGCAAATTCTG | |
| c1273.apra.L-5 | CGCCAGTCGATTGGCTGAGCTCATGAGAATGTGACCTTCTGATAATTCTG | |
| c1273.ext-5 | CGGTGGACTCCGGGTTATTC | |
| c1273.ext-3 | CAGCCAGCCAGTACAACAACCC | |
| c1273.xBAD-500.5 | CCGTACGGCCCTGCGCGCTGTTC | |
| c1273.xBAD-500-3 | GAAGCCTTCCCACGCAAATTCTG | |
| c1273.xBAD.L-3 | GAATCCCTGCTTCGTCCATTTGACTGATTGTTGCACGGACAAGAAAGGTG | |
| c1273.xBAD.L-5 | GTTTCTCCATACCCGTTTTTTTGGACCCGTTAGCAATGTATGCAGACA | |
| c1273.PcL.L-3 | GTGAGAATTACTAACTTGAGCGAATGATTGTTGCACGGACAAGAAAGGTG | |
| c1273.PcL.L-5 | CGGTGATAATGGTTGCATGTACTAACCCGTTAGCAATGTATGCAGACA | |
| c1273.xBAD.ext-5 | TCGGTGGACTCCGGGTTATTC | |
| c1273.xBAD.ext-3 | TTGATCACCGCCGTTAAGGACAG | |
| 568 | CAGAACGGCGAGCTGAGGGTTC | |
| 569 | CCGGCAAGGGTAAACGTGGCA |
Reverse transcription (RT)-PCR.
Cells were grown to the exponential phase in LB and resuspended directly in an equal volume of ice-cold RNAlater (Ambion). Total RNA was isolated and purified using an RNeasy mini kit (QIAGEN). After purification, RNA was treated with RNase-free DNase I to remove contaminating DNA and repurified using QIAGEN RNeasy columns. RNA samples were quantified spectrophotometrically at 260 nm and were additionally checked by gel electrophoresis. Purified total RNA was precipitated with ethanol and stored at −80°C until further use. RNA was converted to cDNA using SuperScript II as described by the manufacturer (Invitrogen Life Technologies). cDNA was used directly as a template for PCR using primers specific for fluA (primers 566 and 567) and fluB (primers 568 and 569). A negative control using the original RNA was always run in parallel to confirm the absence of contaminating DNA.
Autoaggregation assay.
In order to monitor the observed differences in autoaggregation, we devised an assay to monitor bacterial settling kinetics over time. Overnight LB cultures supplemented with the appropriate antibiotics and inducer were mixed well prior to the start of the assay. Four 50-μl samples were taken approximately 0.5 cm below the surface of the liquid cultures at 15-min intervals and transferred to a round-bottom microtiter plate (TPP, Europe), and the optical density at 600 nm (OD600) was determined with a microtiter plate reader. The data are shown below as the mean absorbance value ± standard deviation, where the degree of autoaggregation is inversely proportional to the turbidity.
Release of the alpha-domain of Ag43 and immunodetection.
Cells from overnight LB cultures were harvested by centrifugation, washed in 0.9% (wt/vol) NaCl, and resuspended in 1 ml of 75 mM NaCl-0.5 mM Tris (pH 7.4) to an OD600 of 15. The alpha-domain of Ag43 was released from the surface of the cells by heating at 60°C for 3 min. The cells were immediately removed by centrifugation, and the resultant protein in the supernatant was precipitated overnight at 4°C with 10% (vol/vol) trichloroacetic acid (TCA). The pellet was washed with 500 μl of 80% acetone and dried. Samples were boiled for 3 min prior to gel electrophoresis, and in each case an amount equivalent to 2.0 OD600 units was loaded onto the gel. Proteins were transferred to a polyvinylidene difluoride membrane, and immunodetection was performed using a 1:10,000 dilution of polyclonal rabbit antiserum raised against the alpha-domain of Ag43 (a kind gift from P. Owen).
Biofilm assays.
Biofilm formation on polystyrene surfaces was monitored by using 96-well microtiter plates (IWAKI) essentially as previously described (45). Briefly, cells were grown for 24 h in LB (containing 0.2% arabinose for induction of flu gene expression) at 37°C, washed to remove unbound cells, and stained with crystal violet. Quantification of bound cells was performed by addition of acetone-ethanol (20:80) and measurement of the dissolved crystal violet at OD600. Flow chamber experiments were performed as previously described (25, 44). Briefly, biofilms were allowed to form on glass surfaces in a multichannel flow system that permitted online monitoring of community structures. Flow cells were inoculated with OD600-standardized cultures pregrown overnight in MOPS medium containing chloramphenicol. Glucose was used as the sole carbon source at a concentration of 0.002%. Biofilm development was monitored by confocal scanning laser microscopy at 15 h after inoculation. Microfermentor experiments were performed as previously described (12). Briefly, overnight cultures were grown in LB and LB containing 0.2% arabinose at 37°C. Inoculation was performed by dipping the microfermentor removable spatula in a culture containing 108 bacteria/ml for 2 min and subsequently reintroducing the spatula into the microfermentor. The medium was pumped through the microfermentors at a constant rate (0.75 ml/min). The OD570 of each resulting suspension was then determined after 24 h. This optical density directly reflected the biomass on the spatula. All experiments were performed in triplicate.
Microscopy and image analysis.
All microscopic observation and image acquisition were performed with a scanning confocal laser microscope (TCS4D; Leica Lasertechnik, GmbH, Heidelberg, Germany) equipped with detectors and filters for monitoring green fluorescent protein. The images below (see Fig. 3) are representative of horizontal sections collected within each biofilm and vertical sections representing the yz plane and the xz plane. Vertical cross sections through the biofilms were generated by using the IMARIS software package (Bitplane AG, Zürich, Switzerland) running on a Silicon Graphics Indigo2 workstation (Silicon Graphics, Mountain View, CA). Images were further processed for display by using Photoshop software (Adobe, Mountain View, CA).
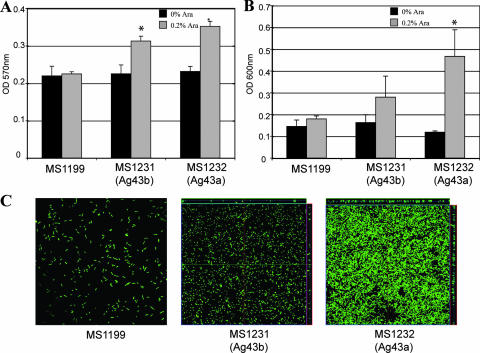
FIG. 3.
Biofilm formation by E. coli MG1655Δflu cells harboring plasmids expressing Ag43a and Ag43b. The effect of each Ag43 variant on biofilm formation was assessed in E. coli OS56 (MG1655Δflu, Gfp+) cells containing the following plasmids: (i) pBADMycHisA (MS1199), (ii) pCO3 (Ag43b, MS1231), and (iii) pCO4 (Ag43a, MS1232). All strains were grown in the presence of 0.2% arabinose to induce Ag43 expression. Three different assays were employed. (A) Static biofilm formation in polystyrene microtiter plates. Expression of Ag43 led to significant biofilm formation by MS1231 and MS1232 compared to the biofilm formation by control strain MS1199 (P < 0.05 for each strain, as determined by the independent sample t test). (B) Dynamic biofilm formation in a microfermentor system. Expression of Ag43a led to significant biofilm formation by MS1232 compared to the biofilm formation by control strain MS1199 (P < 0.05, as determined by the independent sample t test). MS1231 cells expressing Ag43b exhibited a small increase in biofilm formation compared to the biofilm formation by control strain MS1199, although the difference was not statistically significant. (C) Dynamic biofilm formation using a flow chamber model. Biofilm development was monitored by confocal scanning laser microscopy. The images are representative horizontal sections collected for each biofilm and vertical sections (to the right of and below each larger panel, representing the yz plane and the xz plane, respectively) at the positions indicated by the white lines.
Mouse model of UTI.
The mouse model of UTI described previously was used for this study (39). Female C57BL/6 mice (8 to 10 weeks old) were purchased from the Animal Resources Center, Western Australia, and were housed in sterile cages with ad libitum access to sterile water. Urine was collected from each mouse 24 h prior to challenge and examined microscopically with a hemocytometer and by culture. Mice with a combination of >5 × 102 CFU of bacteria per ml of urine and >2 × 105 white blood cells in urine were defined as having a preexisting condition and were excluded from the study. Mice were anesthetized by brief inhalation exposure to isoflurane, and the periurethral area was sterilized by swabbing it with a 10% povidone-iodine solution, which was removed with sterile phosphate-buffered saline (PBS). Mice were catheterized using a sterile Teflon catheter (inside diameter, 0.28 mm; outside diameter; 0.61 mm; length, 25 mm; Terumo) by inserting the device directly into the bladder through the urethra. A 25-μl inoculum containing 5 × 108 CFU of bacteria in PBS containing 0.1% India ink was instilled directly into the bladder using a 1-ml tuberculin syringe attached to the catheter. The catheter was removed immediately after the challenge, and mice were returned to their cages. Urine was collected from each mouse at 18 h after inoculation for quantitative colony counting. Groups of mice were euthanized at 18 h and 5 days after challenge by cervical dislocation; bladders were then excised aseptically, weighed, and homogenized in PBS. Bladder homogenates were serially diluted in PBS and plated onto LB agar for colony counting. Data are expressed below as the mean total number of CFU per 0.1 g of bladder tissue ± standard error of the mean. All animal experiments were repeated at least twice with a minimum group size of eight, and the data shown below are composites from all independent experiments.
Statistics.
Differences in aggregation and biofilm phenotypes (mean optical density values) between Ag43-expressing E. coli strains and mutants were compared using an independent sample t test in the SPSS v9.0.2 software package. Mean bacterial titers of UPEC strains and mutants recovered from bladders were compared using an independent sample t test with log-transformed data. Normal distribution parameters for the different data sets were estimated using P-P plots and histogram analysis. P values of <0.05 were considered significant.
RESULTS
Multiple copies of the flu gene are common in both uropathogenic and commensal E. coli.
CFT073 contains two copies of the flu gene (26). A comparison of the amino acid sequence encoded by these two genes with the K-12 Ag43 amino acid sequence is shown in Table 3. In order to remain consistent with our previous description of the two flu genes from CFT073 (26), we used the following terminology: fluA encoding Ag43a (also described as upaF encoding c3655) and fluB encoding Ag43b (also described as upaD encoding c1273). To assess the prevalence of the flu gene in UPEC and to compare UPEC to commensal E. coli, we devised a PCR using a single set of highly conserved primers designed from a multiple-sequence alignment of the flu genes available in the NCBI database. The primers were designed such that they could identify the flu gene irrespective of known nucleotide sequence variations in the region encoding the alpha-domain. PCR screening of the ECOR collection and our laboratory collection revealed that the flu gene was present in 83% (30/36) of UPEC strains, compared to 56% (35/62) of nonpathogenic E. coli strains. Next, we devised a second PCR that enabled rapid identification of one common class of flu gene variants, specifically those that contain a 216-bp deletion within the alpha-encoding domain (26). Using primers that flank this region, we screened our collections for the deletion and observed that 22% (8/36) of the uropathogenic and 18% (11/62) of the commensal E. coli strains tested contained two amplification products that were different sizes. Thus, multiple copies of the flu gene are common in both uropathogenic and commensal E. coli strains.
TABLE 3.
Properties of UPEC CFT073 Ag43 adhesins
| Gene | c No. | % Amino acid identity to K-12 Ag43 α-domain | % Amino acid identity to K-12 Ag43 β-domain | Overall % amino acid identity to K-12 Ag43 |
|---|---|---|---|---|
| fluA (upaF) | c3655 | 81.7 | 97.1 | 88.9 |
| fluB (upaD) | c1273 | 78.4 | 95.1 | 86.2 |
Cloning and expression of the flu genes from UPEC CFT073.
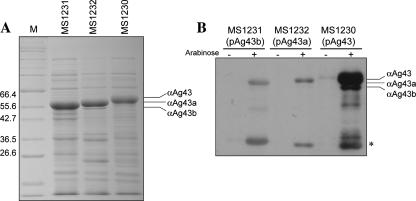
The fluA and fluB genes were amplified by PCR from UPEC CFT073 and cloned as transcriptional fusions behind the tightly regulated araBAD promoter in the pBAD/Myc-HisA expression vector (26). To characterize each of the encoded Ag43 proteins, the resultant plasmids (pCO3 [fluB] and pCO4 [fluA]), as well as pCO2 bearing the K-12 strain MG1655 flu gene, were all transformed into the previously described E. coli Δflu mutant strain OS56 (25). This strain is unable to mediate the classical cell aggregation and biofilm phenotypes associated with Ag43 expression (36). In the case of the E. coli K-12 Ag43 protein and other AT proteins associated with diarrheagenic E. coli (AIDA-I and TibA), the α-domain remains attached to the β-domain via noncovalent interactions. This results in a unique property in that the protein can be released and partially purified by brief heat treatment. E. coli K-12 OS56 cells transformed with either pCO4 (Ag43a; strain MS1232), pCO3 (Ag43b; strain MS1231), or pCO2 (K-12 Ag43; strain MS1230) were grown in LB in the presence of 0.2% arabinose, subjected to heat treatment, and purified by TCA precipitation. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis revealed that the Ag43 proteins could be expressed at equivalent levels after induction from each plasmid construct and released from the cell surface following a brief heat treatment (Fig. 1A). The identities of the Ag43 alpha-domain fragments were confirmed by immunodetection using a serum raised against the alpha-domain of Ag43 from E. coli K-12 (Fig. 1B). We noted that the antiserum reacted more strongly with the K-12 Ag43 alpha-domain subunit than with the CFT073 Ag43a and Ag43b alpha-domain subunits, which reacted similarly.
FIG. 1.
(A) Coomassie brilliant blue-stained SDS-PAGE gel demonstrating Ag43 expression in E. coli MG1655 Δflu strains, including MS1231 (pCO3, Ag43b), MS1232 (pCO4, Ag43a), and MS1230 (pCO2, Ag43 K-12). Production of each Ag43 variant was induced by 0.2% arabinose. Protein preparations were obtained by subjecting cells to a brief heat treatment, centrifugation, and purification of heat-released proteins by TCA precipitation. Lane M contained markers. (B) Western blot analysis using a serum directed against the alpha-domain of Ag43 from E. coli K-12. Expression of Ag43b (MS1231), Ag43a (MS1232), and Ag43 from K-12 (MS1230) was detected only following induction with 0.2% arabinose. The serum reacted most strongly against the K-12 Ag43 alpha-domain subunit; the Ag43a and Ag43b variants reacted more weakly, but the levels for these variants were comparable. The asterisk indicates specific degradation products of the different Ag43 variants.
Ag43a and Ag43b from CFT073 mediate different levels of bacterial aggregation.
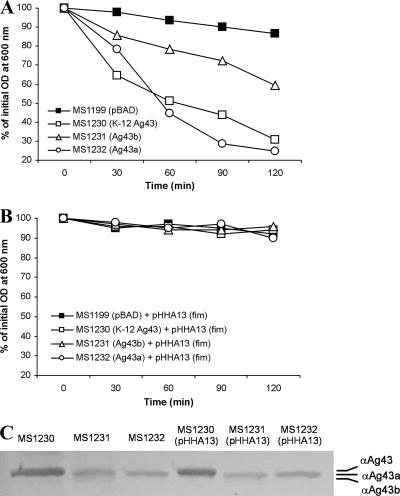
Cell-cell aggregation is a phenotype classically associated with Ag43 expression (16). To test the ability of the CFT073 Ag43a and Ag43b proteins to mediate this phenotype, E. coli K-12 OS56 cells containing pCO4 (Ag43a), pCO3 (Ag43b), or pCO2 (K-12 Ag43) were grown in LB with 0.2% arabinose and tested to determine their aggregation properties. Although produced in similar quantities, the Ag43a and Ag43b proteins promoted different degrees of cell aggregation. Aggregation mediated by Ag43a was the strongest and, analogous to the aggregation mediated by K-12 Ag43, resulted in rapid flocculation and settling of cells from standing overnight cultures (Fig. 2A). In contrast, aggregation mediated by Ag43b was less pronounced. In line with previous observations with other AT proteins that mediate cell aggregation, this property could be blocked by the introduction of a plasmid (pHHA13) conferring constitutive expression of type 1 fimbriae (Fig. 2B). Similar levels of the alpha-domain subunit were detected in the presence and in the absence of plasmid pHHA13, indicating that type 1 fimbria expression did not affect the surface presentation of Ag43a or Ag43b (Fig. 2C). Thus, Ag43a and Ag43b mediate phenotypes associated with different degrees of cell aggregation. Furthermore, aggregation can be physically blocked by fimbria expression, a property that may have in vivo consequences and affect CFT073 Ag43-mediated phenotypes during in vitro growth (see below).
FIG. 2.
(A) Autoaggregation assay demonstrating the settling profiles for liquid suspensions of E. coli OS56 (MG1655Δflu, Gfp+) containing the following plasmids: (i) pBADMycHisA (MS1199), (ii) pCO2 (K-12 Ag43, MS1230), (iii) pCO3 (Ag43b, MS1231), and (iv) pCO4 (Ag43a, MS1232). All strains were grown in the presence of 0.2% arabinose to induce Ag43 expression. Expression of Ag43 led to significant autoaggregation of MS1230, MS1231, and MS1232 compared to the autoaggregation of control strain MS1199 (at 60 to 120 min, P < 0.05 for each strain, as determined by the independent sample t test). Furthermore, MS1232 cells expressing Ag43a exhibited significantly stronger autoaggregation than their MS1231 Ag43b-expressing counterparts (at 60 to 120 min, P < 0.002, as determined by the independent sample t test). (B) Same assay in the presence of type 1 fimbria expression. MS1199, MS1230, MS1231, and MS1232 were transformed with pHHA13 (which encodes the ability to express type 1 fimbriae) and grown in the presence of 0.2% arabinose as described above to induce Ag43 expression. The presence of type 1 fimbriae on the cell surface resulted in prevention of Ag43-mediated aggregation. (C) Western blot of Ag43 alpha-domain subunit proteins liberated from E. coli MS1231 (Ag43b+), MS1232 (Ag43a+), MS1231(pHHA13) (Ag43b+ Fim+), and MS1232(pHHA13) (Ag43a+ Fim+). Overexpression of type 1 fimbriae did not affect the amount of Ag43 present at the cell surface of each strain.
Biofilm formation properties of the Ag43a and Ag43b adhesins.
To determine whether the CFT073 Ag43a and Ag43b proteins promote biofilm formation, gfp-tagged cells harboring each of the corresponding plasmids were tested to determine their abilities to form biofilms in static and dynamic biofilm assays. First, we employed a static nontreated polystyrene microtiter plate model and showed that both Ag43 variants promote biofilm formation after growth in LB (Fig. 3A). Next, we tested the abilities of the Ag43 proteins to promote biofilm formation in dynamic conditions using microfermentor and continuous-flow chamber models; the latter model allowed us to monitor bacterial distribution within an evolving biofilm at the single-cell level due to the combination of gfp-tagged cells and scanning confocal laser microscopy. Ag43a promoted the strongest biofilm growth under both experimental scenarios and in the flow chamber produced a structure with a depth of approximately 10 μm (Fig. 3B and C). Taken together, the data suggest that strong Ag43a-mediated aggregation enhances biofilm formation by E. coli K-12.
Both fluA and fluB are expressed in CFT073.
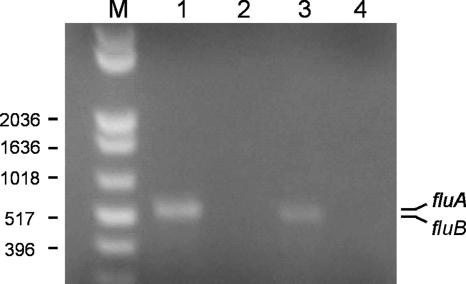
Our analysis of the function of the Ag43a and Ag43b proteins from CFT073 involved cloning of the genes and expression from an inducible promoter. To determine whether the fluA and fluB genes are expressed by wild-type strain CFT073, transcription of each gene was analyzed by RT-PCR. RNA was extracted from CFT073 cells grown in LB and converted to cDNA, and a specific PCR was performed for each of the genes (Fig. 4). In both cases, an amplification product was obtained and DNA sequencing confirmed the specificity of the reaction. No PCR products were obtained from the RNA sample prior to cDNA synthesis. These results demonstrate that the fluA and fluB genes located on the CFT073 chromosome are transcribed during growth in LB. However, the fluB gene appeared to be expressed at lower levels than the fluA gene.
FIG. 4.
RT-PCR analysis of fluA and fluB expression by CFT073. Total RNA was extracted from CFT073 during exponential growth in LB. Lane 1, fluA-specific PCR product (523 bp) obtained using cDNA as the template; lane 2, negative control using RNA prior to cDNA synthesis; lane 3, fluB-specific PCR product (518 bp) obtained using cDNA as the template; lane 4, negative control using RNA prior to cDNA synthesis; lane M, 1-kb Plus DNA ladder (Invitrogen).
Deletion of the flu genes in CFT073 and repression of flu expression by OxyR.
To further examine the role of Ag43a and Ag43b in UPEC virulence, specific deletion mutants were constructed in CFT073. Chromosomal construction was performed by λ-red recombination of linear DNA (6, 30). Mutants containing individual fluA and fluB deletions, as well as a fluA fluB double deletion, were created. These mutants did not display any differences in the ability to form a biofilm in the microtiter plate assay (Fig. 5). Western blot analysis using Ag43-specific antiserum failed to detect expression of Ag43a or Ag43b in CFT073 (Fig. 5). However, after prolonged exposure a faint band was observed in CFT073 that was absent in the fluA mutant, demonstrating that there was very low expression of Ag43a in CFT073 (data not shown). On the other hand, the same prolonged exposure did not result in detection of Ag43b.
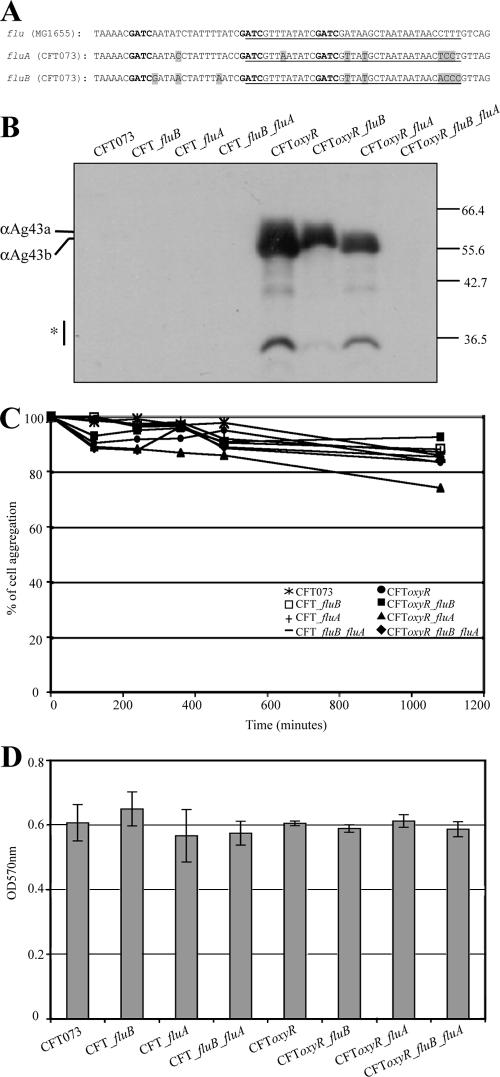
FIG. 5.
(A) Schematic representation of the OxyR binding sites and adjacent nucleotide sequences of the flu gene from E. coli K-12 and the fluA and fluB genes from CFT073. The putative OxyR binding sites are underlined, the GATC methylation sites are indicated by bold type, and the nucleotide sequence differences are indicated by shading. (B) Ag43 expression in CFT073 flu and oxyR mutants. Protein preparations were obtained by subjecting cells to a brief heat treatment, centrifugation, and subsequent SDS-PAGE and immunodetection analysis using a serum directed against the α-domain of Ag43 from E. coli K-12. Whereas no Ag43 variants could be detected in wild-type CFT073 in this image, longer exposure of the same membrane revealed the presence of Ag43a but not Ag43b in CFT073 (data not shown). Deletion of oxyR resulted in significant expression of both Ag43a and Ag43b, demonstrating that OxyR represses both the fluA and fluB genes in wild-type strain CFT073. (C) Autoaggregation assay demonstrating the settling profiles for liquid suspensions of CFT073, CFT073fluA, CFT073fluB, CFTfluA fluB, CFT073oxyR, CFT073oxyR fluB, CFT073oxyR fluA, and CFT073oxyR fluA fluB. Deletion of oxyR did not produce an aggregation phenotype. (D) Biofilm formation by CFT073, CFT073fluA, CFT073fluB, CFTfluA fluB, CFT073oxyR, CFT073oxyR fluB, CFT073oxyR fluA, and CFT073oxyR fluA fluB. Deletion of oxyR gave rise to a biofilm phenotype similar to that of the CFT073fluA+, CFT073fluB+, and CFT073fluA+ fluB+ strains.
In E. coli K-12, the expression of Ag43 is phase variable due to the concerted action of the Dam methyltransferase (positive regulation) and OxyR (negative regulation) (28). The CFT073 fluA and fluB promoter regions both contain three GATC sites which overlap a putative recognition sequence for OxyR. However, the putative OxyR binding sites are not the same as the site found in E. coli K-12 and contain 31/37 identical nucleotides (Fig. 5). In E. coli K-12, when the GATC sites are methylated, OxyR cannot bind; conversely, bound OxyR prevents methylation (29). The oxyR gene was deleted in wild-type strain CFT073, CFT073fluA, CFT073fluB, and CFT073fluA fluB by λ-red recombination of linear DNA. Each of these strains was then probed for Ag43 expression by immunodetection analysis using Ag43-specific antiserum. Ag43 expression was observed in all oxyR mutants except CFT073fluA fluB (Fig. 5), indicating that the expression of both Ag43a and Ag43b is repressed by OxyR. None of these strains displayed a positive aggregation or biofilm formation phenotype (Fig. 5C and D), further supporting the notion that the Ag43 function in CFT073 is masked by larger cell surface structures.
CFT073 fluA mutant displays reduced long-term persistence in the mouse bladder.
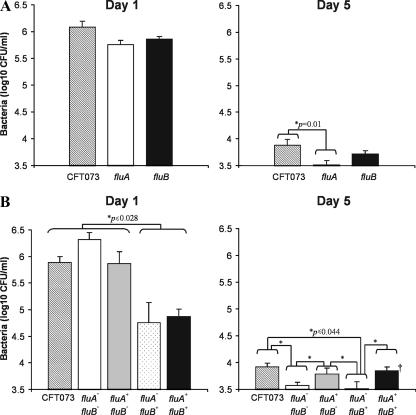
The CFT073 fluA and fluB mutants were tested to determine their abilities to survive in the mouse urinary tract. At day 1 following infection, we did not observe any significant difference in the abilities of the wild-type and mutant strains to colonize the mouse bladder. However, at day 5 following infection, we observed a significant reduction (P < 0.01) in the ability of the CFT073 fluA mutant to colonize the bladder (Fig. 6). No significant difference was observed when the fluB mutant was compared with the wild-type CFT073 strain. No colonization of the kidney was observed for any of the strains tested. Taken together, these data demonstrate that fluA (encoding Ag43a) is required for long-term colonization of the mouse bladder.
FIG. 6.
(A) Persistence of wild-type strain CFT073 and CFT073 fluA- and fluB-deficient mutants in the bladders of C57BL/6 mice following intraurethral challenge. At day 1 following infection, equivalent numbers of both flu deletion mutants and wild-type strain CFT073 were recovered. At day 5 following infection, significantly lower numbers of the fluA deletion mutant than of wild-type strain CFT073 were recovered (P = 0.01). The data are the mean total numbers of CFU per 0.1 g of bladder tissue ± standard errors of the means for 10 mice. (B) Persistence of E. coli CFT073 and CFTfluA fluB, CFTfluA+ fluB, CFTfluA fluB+, and CFTfluA+ fluB+ mutants in the bladders of C57BL/6 mice following intraurethral challenge. At day 1 following infection wild-type strain CFT073 and the CFTfluA fluB and CFTfluA+ fluB strains colonized the bladder epithelium significantly better than strains expressing Ag43b (P ≤ 0.028 for comparisons of all three of the strains with CFTfluA fluB+ and CFTfluA+ fluB+). At day 5 following infection constitutive expression of Ag43a in E. coli CFT073 promoted longer-term colonization in the bladder irrespective of the presence of Ag43b (P ≤ 0.044 for comparisons of CFTfluA fluB and CFTfluA fluB+ with CFT073, CFTfluA+ fluB, or CFTfluA+ fluB). The data are the mean total numbers of CFU per 0.1 g of bladder tissue ± standard errors of the means for at least two independent experiments. The following numbers of mice were used: on day 1, 29 mice for CFT073, 15 mice for CFTfluA fluB, 14 mice for CFTfluA fluB+, 15 mice for CFTfluA+ fluB, and 15 mice for CFTfluA+ fluB+; on day 5, 18 mice for CFT073, 15 mice for CFTfluA fluB, 14 mice for CFTfluA fluB+, 16 mice for CFTfluA+ fluB, and 15 mice for CFTfluA+ fluB+. An asterisk indicates that mean bacterial titers were compared using the independent sample t test after log transformation of data to achieve normal distributions according to P-P plots and histogram analysis (SPSS v9.0.2; SPSS, Chicago, IL).
Construction and analysis of CFT073 strains constitutively expressing Ag43a and Ag43b.
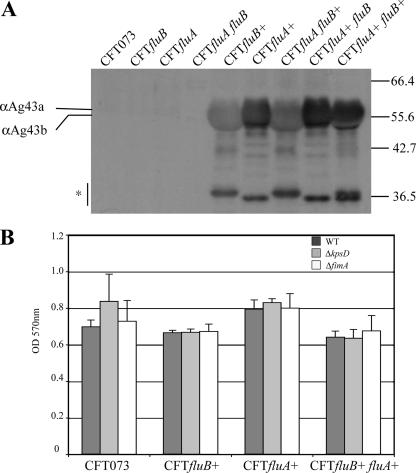
The phase-variable expression of Ag43 in E. coli K-12 prompted us to use a second approach to study the effect of Ag43a and Ag43b in CFT073. To this end, we constructed a unique set of Ag43 overexpression strains by inserting a chromosomally located constitutive promoter upstream of fluA or fluB. The strains constitutively expressing Ag43 were constructed using a modification of the RExBAD expression cassette system (40; S. Da Re et al., submitted for publication). This resulted in insertion of the λPR constitutive promoter in front of the fluA or fluB gene. We constructed the following Ag43a- and Ag43b-expressing strains: CFTfluA+ fluB (expressing only fluA), CFTfluA fluB+ (expressing only fluB), and CFTfluA+ fluB+ (expressing both fluA and fluB). Each strain produced the appropriate Ag43 variant(s), as shown by Western blot analysis (Fig. 7A). However, neither of the Ag43 proteins promoted aggregation or biofilm formation in this background (Fig. 7B; data not shown). Ag43 belongs to a class of short non-organelle-type adhesins that are predicted to protrude about 10 nm from the bacterial surface (22). Thus, Ag43-mediated cell aggregation requires close cell-cell contact. The inability of Ag43a and Ag43b to mediate aggregation and biofilm formation in CFT073 is probably due to the expression of other large cell surface structures, such as fimbriae, flagella, lipopolysaccharide, and the capsule, which have previously been shown to physically shield and block the close cell-cell contact required for Ag43-mediated cell aggregation (2, 14, 42, 48, 49, 51). We attempted to address this possibility by deletion of either the type 1 fimbria- or capsule-encoding genes in CFT073 strains expressing either fluA or fluB or both fluA or fluB; however, this did not increase the biofilm formation properties (Fig. 7B). It is therefore likely that several differentially expressed surface factors affect this phenotype.
FIG. 7.
(A) Immunodetection analysis of Ag43a and Ag43b proteins produced in CFT073 as a result of constitutive expression from PcL promoter insertions. Protein preparations were obtained by subjecting cells to a brief heat treatment, centrifugation, and subsequent SDS-PAGE and immunodetection analysis using a serum directed against the α-domain of Ag43 from E. coli K-12. Each strain produced the appropriate Ag43 variant(s), as demonstrated by the different sizes of the alpha-domain and the degradation products (indicated by an asterisk). (B) Biofilm formation in microtiter plates by CFT073 expressing Ag43 variants and capsule and type 1 fimbria mutant derivatives. WT, wild type.
Constitutive expression of Ag43a promotes long-term persistence in the mouse bladder.
Despite the in vitro phenotype of our CFT073 Ag43-expressing strains, the decreased bladder colonization observed with the CFT073 fluA mutant suggested that random in vivo phase variation of multiple cell surface structures may contribute to Ag43 function during disease. To assess the impact of the Ag43a and Ag43b proteins on the ability of CFT073 to survive in the mouse urinary tract, we tested the strains expressing either fluA or fluB or both fluA and fluB during early and long-term colonization of the mouse urinary tract (Fig. 6). At day 1 following infection, we observed significantly reduced colonization of the bladder by CFT073 strains expressing Ag43b. This effect occurred independent of Ag43a expression (compare the results for CFTfluA fluB+ and CFTfluA+ fluB+ to those for CFT073, CFTfluA fluB, or CFTfluA+ fluB). Thus, Ag43b overexpression appears to impair colonization of the bladder epithelium during the initial phase of infection. At day 5 following infection, however, we observed a distinct change in the survival pattern of the individual strains. The numbers of both CFT073 strains not expressing Ag43a recovered in the bladder were significantly lower than the numbers of CFT073 and the two Ag43a-expressing strains. This effect was observed independent of Ag43b expression (compare the results for CFTfluA fluB and CFTfluA fluB+ to those for CFT073, CFTfluA+ fluB, or CFTfluA+ fluB+). Taken together, these data demonstrate that while constitutive expression of Ag43b impairs the initial colonization, the presence of Ag43a is necessary for longer-term persistence in the mouse bladder. Moreover, at day 5, expression of Ag43a appears to compensate for the inhibitory effect of Ag43b expression observed at day 1. These findings are consistent with the reduced persistence of the CFT073 fluA mutant in the bladder.
DISCUSSION
Attachment to host tissues is the first critical step in colonization and is mediated by bacterial adhesins. UPEC produces a range of fimbrial and nonfimbrial adhesins that play a role in virulence and contribute to persistent infection of the urinary tract. Some UPEC fimbrial adhesins, including type 1 and P fimbriae, have been well characterized with respect to their expression, regulation, receptor-binding target, and role in virulence. Of the nonfimbrial adhesins, the AT family of proteins represent a novel group of virulence factors because of their role in adhesion, invasion, and biofilm formation in other organisms. Although UPEC possesses multiple AT-encoding genes, very little is known about their function and role in virulence. Here, we characterize two AT adhesins of UPEC CFT073 (Ag43a and Ag43b) and demonstrate that the Ag43a variant is able to promote biofilm formation and contributes to long-term persistence in the urinary bladder.
Our approach to studying the function of the UPEC CFT073 Ag43a and Ag43b proteins involved the use of two host strains, K-12 MG1655Δflu and CFT073. The K-12 MG1655Δflu strain is well characterized and has been shown to be an ideal background strain to assess the function of AT adhesins (24, 25). The Ag43a and Ag43b proteins had marked differences in the ability to mediate aggregation, and Ag43a-mediated aggregation was stronger. These different biological properties were not due to variations in the levels of Ag43a and Ag43b protein expression, as determined by SDS-PAGE analysis and Western blotting. The Ag43a- and Ag43b-mediated aggregation phenotype could be blocked by the concomitant expression of type 1 fimbriae, a finding that is consistent with other studies which demonstrated that the intimate cell-cell contact required for AT adhesin interaction can be physically blocked by the expression of larger surface structures, such as fimbriae, flagella, lipopolysaccharide, and the capsule (2, 14, 42, 48, 51). We also observed that Ag43-mediated aggregation was not manifested in CFT073 when the proteins were constitutively expressed or following deletion of the oxyR repressor. We attempted to address this issue by constructing a deletion in the type 1 fimbrial genes and the capsule-encoding kpsD gene; however, this did not have any effect on cell aggregation or biofilm formation, suggesting that additional factors may also mask the Ag43 function in CFT073.
One of the mechanisms by which UPEC promotes the formation of biofilms is via the expression of proteins that mediate cell-cell aggregation. Our previous work demonstrated that Ag43 (from E. coli K-12) is associated with the early stages of biofilm development (44). However, Ag43 has also been shown to be dispensable for biofilm formation as it can be replaced by alternative factors, such as conjugative pili (12, 36). Here we demonstrated that in a K-12 background only Ag43a is able to promote strong biofilm formation using dynamic flow model systems. The biofilm produced by the strain expressing Ag43a is similar to the biofilm reported for the K-12 Ag43 in terms of thickness and structure (25).
In UTI caused by UPEC, biofilm formation occurs within superficial umbrella cells of the bladders of mice after infection (1). The biofilms are pod-like structures that contain polysaccharide-encased cell clusters expressing Ag43, and their formation is postulated to be associated with long-term persistence in the urinary bladder (10). The strain used in these studies, UTI89, was recently sequenced and contains only one copy of the flu gene (c1139) (7). The corresponding gene product shares only 67% amino acid identity with the Ag43a and Ag43b adhesins of CFT073, and the vast majority of the differences occur in the passenger (exposed) domain of the protein. Therefore, it is not possible to associate the biofilm phenotype of UTI89 with that observed for Ag43a from CFT073. It is likely that sequence divergence within the passenger domain of Ag43 has resulted in the evolution of many different pathogenicity-adapted variants that possess enhanced biofilm formation properties. This may also be associated with the glycosylation of some Ag43 variants, as has been observed in UPEC strain 536 (47). Interestingly, UPEC CFT073 was a poor biofilm former in all of our assays. Although we were able to detect expression of the fluA and fluB genes by RT-PCR, we detected only a low level of Ag43a by immunodetection analysis using heat-extracted protein samples prepared from wild-type cultures. This type of differential expression of Ag43 variants has also been reported for UPEC strain 536, in which one of the two Ag43 variants, ORF52III, is expressed at higher levels than the other, ORF47V (2).
Two strategies were employed to determine the role of Ag43a and Ag43b in virulence in the mouse urinary tract. First, deletion mutants were constructed and revealed a novel role for Ag43a in long-term persistence in the bladder. The second strategy employed an approach analogous to the previously described RExBAD system (40), with the exception that we inserted a constitutive promoter (PcL) upstream of each target gene (S. Da Re et al., submitted). Since the two Ag43 proteins are phase variable, this avoids reliance on the natural promoter for flu expression and circumvents any pleiotropic effects that would have resulted from the use of our oxyR mutants. Furthermore, since the physiological conditions required for expression of Ag43a and Ag43b are not known, this enabled us to compare the survival of CFT073 mutants containing deletions in each gene. Expression of Ag43a in CFT073 was required for optimum colonization of the mouse bladder when it was assessed at 5 days as a prolonged infection model. This requirement for Ag43a for colonization was observed irrespective of Ag43b expression, although Ag43b appeared to have a detrimental effect on bacterial colonization after 1 day of infection. It is possible that Ag43b impedes the ability of CFT073 to establish early colonization in the bladder by virtue of its weaker aggregation- and biofilm-related properties. The differential roles of these two homologous proteins in survival in the urinary tract raise the possibility that they have evolved to possess different functional properties. Indeed, our in vitro aggregation and biofilm studies with these two proteins alluded to such differences. The sequence of the promoter region of the CFT073 fluA and fluB genes is not identical to the sequence of the corresponding promoter region of the K-12 flu gene. In the 300-bp region upstream of the protein-encoding sequence, fluA shares 89% (266/300) nucleotide sequence identity with the same region in K-12, while fluB shares 81% (243/300) nucleotide sequence identity. Despite these differences, derepression of both Ag43a expression and Ag43b expression was observed in a CFT073 oxyR background, demonstrating that OxyR represses the expression of both fluA and fluB in CFT073. The fact that the three GATC sites found in the K-12 flu promoter region are also conserved upstream of fluA and fluB in CFT073 suggests that Ag43a and Ag43b are both phase variable and that the mechanism by which this occurs is analogous to the mechanism for Ag43 in K-12 and is coordinated by the combined action of Dam and OxyR. The introduction of a constitutive promoter to control expression of each gene, therefore, provided the added advantage of allowing us to study the function of these adhesins in the context of a defined wild-type UPEC strain and bypassing of the phase variation mechanism for these two proteins. Our results suggest that the phase variation of Ag43a and Ag43b in the mouse urinary tract may be coordinated together with as-yet-undefined environmental or other factors that favor Ag43a expression and therefore persistence in the bladder. The ability of Ag43a to promote longer-term colonization in the bladder may be due to its capacity to enhance biofilm formation, which plays a role in resistance to host defenses and therefore the establishment of longer-term infection. Alternatively, Ag43 may exert its effect by influencing the susceptibility of UPEC to stress, a process that could be coordinately regulated via OxyR. In line with this, Ag43-mediated aggregation has been shown to enhance tolerance to bactericidal agents such as H2O2 (43).
Ag43a contributes to long-term persistence after establishment of an initial infection, suggesting that its function comes into play after fimbria-mediated specific adhesion. We detected a low level of fluA and fluB gene expression by RT-PCR, and Ag43a expression was observed by Western blot analysis. The failure to detect Ag43b may have been due to a combination of lower expression and weak cross-reactivity with our Ag43 antiserum. Although we did not observe a direct in vitro phenotype for CFT073 strains constitutively expressing Ag43a or Ag43b, our in vivo findings imply that phase variation of fimbriae may be an important aspect of long-term survival and adaptation in the mouse bladder. While cells expressing fimbriae would not be able to aggregate, cells devoid of fimbriae would be able to adapt to niche environments that favor aggregation and biofilm development. In our forced Ag43 expression strains this effect would be enhanced even further as a result of constitutive gene expression (i.e., the genes are already “on”). In support of this model others have demonstrated the existence of an inverse relationship between type 1 fimbrial gene expression and P fimbrial gene expression (20, 50, 57). It has been proposed that during the course of an infection type 1 fimbriae are important in the early stages; however, as the infection progresses, increased expression of motility and P fimbrial genes occurs at the expense of type 1 fimbrial expression (5). Such a model is also consistent with a previous report of fimbria-mediated coordination of Ag43 expression (46). The long-term survival of UPEC in the urinary tract has been linked to the formation of biofilm-like intracellular bacterial communities and the formation of quiescent intracellular reservoirs within the urinary bladder epithelium (21, 32). It would be now be interesting to determine the role of Ag43 in these survival mechanisms and to differentiate between the contributions of Ag43a and Ag43b to persistence in the bladder.
E. coli is a highly diverse species, and over 250 serotypes have been identified based on O, H, and K antigens. There is extensive variation in the DNA content in different strain types. The differences account for the versatility of E. coli with respect to disease since different strains have acquired different sets of virulence genes, which are typically located on pathogenicity islands or on plasmids. A PCR screen using primers designed for predicted conserved regions of the flu gene revealed a difference in the prevalence of this gene between uropathogenic and commensal E. coli strains. Three UPEC strains have now been entirely sequenced, and a different picture has emerged in each case with respect to the location and sequence of the flu gene. It therefore seems reasonable to conclude that the pathogenicity island localization of the flu gene contributes to its multiplicity in different E. coli strains. The variable sequence of the alpha-encoding domain of Ag43 is consistent with data for other AT adhesins and suggests that characterization of additional variants from other clinical isolates may reveal novel phenotypic properties of this versatile adhesin.
Acknowledgments
We thank Laura Zanichelli and Cheryl Ong for expert assistance. We are grateful to Peter Owen for his gift of Ag43 antiserum.
This work was supported by grants from the Australian National Health and Medical Research Council (grant 455914), the Australian Research Council (grant DP0557615), the University of Queensland, the Institut Pasteur, CNRS URA 2172, the Network of Excellence EuroPathoGenomics, the European community (grant LSHB-CT-2005-512061), and the Fondation BNP PARIBAS. J. Valle is a Marie-Curie Fellow.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 9 April 2007.
REFERENCES
- 1.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. [DOI] [PubMed] [Google Scholar]
- 2.Beloin, C., K. Michaelis, K. Lindner, P. Landini, J. Hacker, J. M. Ghigo, and U. Dobrindt. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 188:1316-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergsten, G., M. Samuelsson, B. Wullt, I. Leijonhufvud, H. Fischer, and C. Svanborg. 2004. PapG-dependent adherence breaks mucosal inertia and triggers the innate host response. J. Infect. Dis. 189:1734-1742. [DOI] [PubMed] [Google Scholar]
- 4.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan, A., P. Roesch, L. Davis, R. Moritz, S. Pellett, and R. A. Welch. 2006. Regulation of type 1 fimbriae by unlinked FimB- and FimE-like recombinases in uropathogenic Escherichia coli strain CFT073. Infect. Immun. 74:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 103:5977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connell, H., W. Agace, P. Klemm, M. Schembri, S. Marild, and C. Svanborg. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 93:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 10.Eto, D. S., J. L. Sundsbak, and M. A. Mulvey. 2006. Actin-gated intracellular growth and resurgence of uropathogenic Escherichia coli. Cell. Microbiol. 8:704-717. [DOI] [PubMed] [Google Scholar]
- 11.Foxman, B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S-13S. [DOI] [PubMed] [Google Scholar]
- 12.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 13.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasman, H., T. Chakraborty, and P. Klemm. 1999. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J. Bacteriol. 181:4834-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasman, H., M. A. Schembri, and P. Klemm. 2000. Antigen 43 and type 1 fimbriae determine colony morphology of Escherichia coli K-12. J. Bacteriol. 182:1089-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson, I. R., M. Meehan, and P. Owen. 1997. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol. Lett. 149:115-120. [DOI] [PubMed] [Google Scholar]
- 17.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, I. R., and P. Owen. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holden, N. J., M. Totsika, E. Mahler, A. J. Roe, K. Catherwood, K. Lindner, U. Dobrindt, and D. L. Gally. 2006. Demonstration of regulatory cross-talk between P fimbriae and type 1 fimbriae in uropathogenic Escherichia coli. Microbiology 152:1143-1153. [DOI] [PubMed] [Google Scholar]
- 21.Justice, S. S., C. Hung, J. A. Theriot, D. A. Fletcher, G. G. Anderson, M. J. Footer, and S. J. Hultgren. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 101:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajava, A. V., N. Cheng, R. Cleaver, M. Kessel, M. N. Simon, E. Willery, F. Jacob-Dubuisson, C. Locht, and A. C. Steven. 2001. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42:279-292. [DOI] [PubMed] [Google Scholar]
- 23.Kjaergaard, K., H. Hasman, M. A. Schembri, and P. Klemm. 2002. Antigen 43-mediated autotransporter display, a versatile bacterial cell surface presentation system. J. Bacteriol. 184:4197-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjaergaard, K., M. A. Schembri, H. Hasman, and P. Klemm. 2000. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J. Bacteriol. 182:4789-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 26.Klemm, P., L. Hjerrild, M. Gjermansen, and M. A. Schembri. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51:283-296. [DOI] [PubMed] [Google Scholar]
- 27.Klemm, P., and M. A. Schembri. 2000. Bacterial adhesins: function and structure. Int. J. Med. Microbiol. 290:27-35. [DOI] [PubMed] [Google Scholar]
- 28.Klemm, P., and M. A. Schembri. 15 November 2004, posting date. Chapter 8.3.2.6. Type 1 fimbriae, curli, and antigen 43: adhesion, colonization, and biofilm formation. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org. [DOI] [PubMed]
- 29.Leffler, H., and C. Svanborg-Eden. 1981. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect. Immun. 34:920-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesic, B., S. Bach, J. M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 52:1337-1348. [DOI] [PubMed] [Google Scholar]
- 31.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mysorekar, I. U., and S. J. Hultgren. 2006. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl. Acad. Sci. USA 103:14170-14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oelschlaeger, T. A., U. Dobrindt, and J. Hacker. 2002. Virulence factors of uropathogens. Curr. Opin. Urol. 12:33-38. [DOI] [PubMed] [Google Scholar]
- 34.Owen, P., M. Meehan, H. de Loughry-Doherty, and I. Henderson. 1996. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 16:63-76. [DOI] [PubMed] [Google Scholar]
- 35.Parham, N. J., U. Srinivasan, M. Desvaux, B. Foxman, C. F. Marrs, and I. R. Henderson. 2004. PicU, a second serine protease autotransporter of uropathogenic Escherichia coli. FEMS Microbiol. Lett. 230:73-83. [DOI] [PubMed] [Google Scholar]
- 36.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, J. A., M. B. Kaack, G. Baskin, M. R. Chapman, D. A. Hunstad, J. S. Pinkner, and S. J. Hultgren. 2004. Antibody responses and protection from pyelonephritis following vaccination with purified Escherichia coli PapDG protein. J. Urol. 171:1682-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, J. A., B. I. Marklund, D. Ilver, D. Haslam, M. B. Kaack, G. Baskin, M. Louis, R. Mollby, J. Winberg, and S. Normark. 1994. The Gal(alpha 1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl. Acad. Sci. USA 91:11889-11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roos, V., G. C. Ulett, M. A. Schembri, and P. Klemm. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect. Immun. 74:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roux, A., C. Beloin, and J. M. Ghigo. 2005. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J. Bacteriol. 187:1001-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and M. T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 186:1249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schembri, M. A., L. Hjerrild, M. Gjermansen, and P. Klemm. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J. Bacteriol. 185:2236-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 45.Schembri, M. A., and P. Klemm. 2001. Biofilm formation in a hydrodynamic environment by novel FimH variants and ramifications for virulence. Infect. Immun. 69:1322-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schembri, M. A., and P. Klemm. 2001. Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J. 20:3074-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherlock, O., U. Dobrindt, J. B. Jensen, R. Munk Vejborg, and P. Klemm. 2006. Glycosylation of the self-recognizing Escherichia coli Ag43 autotransporter protein. J. Bacteriol. 188:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherlock, O., R. M. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyder, J. A., B. J. Haugen, C. V. Lockatell, N. Maroncle, E. C. Hagan, D. E. Johnson, R. A. Welch, and H. L. Mobley. 2005. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect. Immun. 73:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulett, G. C., R. I. Webb, and M. A. Schembri. 2006. Antigen-43-mediated autoaggregation impairs motility in Escherichia coli. Microbiology 152:2101-2110. [DOI] [PubMed] [Google Scholar]
- 52.Waldron, D. E., P. Owen, and C. J. Dorman. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44:509-520. [DOI] [PubMed] [Google Scholar]
- 53.Wu, X. R., T. T. Sun, and J. J. Medina. 1996. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl. Acad. Sci. USA 93:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wullt, B., G. Bergsten, H. Connell, P. Rollano, N. Gebratsedik, L. Hang, and C. Svanborg. 2001. P-fimbriae trigger mucosal responses to Escherichia coli in the human urinary tract. Cell. Microbiol. 3:255-264. [DOI] [PubMed] [Google Scholar]
- 55.Wullt, B., G. Bergsten, H. Connell, P. Rollano, N. Gebretsadik, R. Hull, and C. Svanborg. 2000. P fimbriae enhance the early establishment of Escherichia coli in the human urinary tract. Mol. Microbiol. 38:456-464. [DOI] [PubMed] [Google Scholar]
- 56.Wullt, B., G. Bergsten, M. Samuelsson, and C. Svanborg. 2002. The role of P fimbriae for Escherichia coli establishment and mucosal inflammation in the human urinary tract. Int. J. Antimicrob. Agents 19:522-538. [DOI] [PubMed] [Google Scholar]
- 57.Xia, Y., D. Gally, K. Forsman-Semb, and B. E. Uhlin. 2000. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the PapB protein. EMBO J. 19:1450-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]