Abstract
Invasive group A streptococcal (GAS) disease reemerged in The Netherlands in the late 1980s. To seek an explanation for this resurgence, the genetic compositions of 22 M1 and 19 M28 GAS strains isolated in The Netherlands between 1960s and the mid-1990s were analyzed by using a mixed-genome DNA microarray. During this four-decade period, M1 and especially M28 strains acquired prophages on at least eight occasions. All prophages carried a superantigen (speA2, speC, speK) or a streptodornase (sdaD2, sdn), both associated with invasive GAS disease. Invasive and noninvasive GAS strains did not differ in prophage acquisition, suggesting that there was an overall increase in the pathogenicity of M1 and M28 strains over the last four decades rather than emergence of hypervirulent subclones. The increased overall pathogenic potential may have contributed to the reemergence of invasive GAS disease in The Netherlands.
Group A streptococcus (GAS) or Streptococcus pyogenes is associated with a wide range of different clinical manifestations (7). A remarkable resurgence of invasive GAS disease, often complicated by the development of toxic shock-like syndrome, was noted throughout the Western world in the late 1980s (4, 6, 10, 11). GAS can be categorized on the basis of antigenic differences in the M protein, an important virulence factor that confers antiphagocytic properties. Several studies have suggested that there is a direct relationship between the genetic content of GAS M types and their clinical importance in terms of frequency and severity of disease (9, 13, 15, 17). Thus, alterations in the bacterial genome composition over time may have contributed to the resurgence of GAS disease in the Western world. However, whether the reemergence of invasive GAS disease is related to a population-wide increase in virulence or to the emergence of certain hypervirulent clones is largely unknown.
We compared the genetic compositions of M1 and M28 GAS strains from invasive and noninvasive diseases that were isolated long before and after the mid-1980s using a mixed-whole genome microarray to study whether genomic changes underlie the resurgence of invasive GAS disease in The Netherlands (5, 21). M1 and M28 are both predominant M types in The Netherlands. M1 is highly virulent and represents the predominant M type in The Netherlands (20); serotype M28 is particularly associated with puerperal sepsis. This is the first microarray study that compared genetic alterations in invasive and noninvasive GAS strains of different M types, spanning a period of four decades.
All M1 and M28 strains isolated after the 1980s were enriched in prophages encoding superantigens or streptodornases, irrespective of their source of isolation. As these phage-encoded virulence factors are associated with invasive disease (12, 18), these findings suggest that there was an overall increase in virulence among these strains. Thus, population-wide alterations in virulence rather than emergence of virulent subclones within given M types may have contributed to the resurgence of invasive GAS disease in The Netherlands.
MATERIALS AND METHODS
Bacterial strains.
GAS M1 and M28 isolates were obtained from across the country by the Dutch National Institute of Public Health (RIVM) in the periods from 1959 to 1983 and from 1992 to 1996. The estimated annual incidence of invasive GAS disease in The Netherlands varied between 2.0 and 4.0 per 100,000 person years between 1994 and 2003 (20). For each M type and decade, one half of the isolates were obtained from normally sterile body sites, representing invasive GAS disease, and the other half were obtained from the skin and pharynx, representing noninvasive GAS disease. M1 isolates (n = 22) were isolated in 1959, 1960, 1966, 1968, 1971, 1982 (n = 3), 1983, and 1992 to 1996 (n = 13); M28 isolates (n = 19) were isolated in 1962, 1966, 1982, 1983 (n = 2), and 1992 to 1996 (n = 14). Historical isolates in our National Institute of Public Health are mainly M types 1 and 28. Therefore, it is likely that these M types were predominant in GAS disease before the 1980s. However, the possibility of a collection bias cannot be excluded as these isolates were not obtained as part of a nationwide prospective study, as was the case in the 1990s. Due to the lack of ancient isolates of other M types in The Netherlands, this study was confined to M1 and M28. The emm genotype was determined by sequencing the emm amplicon as described on the Centers for Disease Control website (www.cdc.gov/ncidod/biotech/strep/emmtypes.htm). Strains were grown overnight on blood agar plates with 5% CO2 at 37°C.
Microarray construction.
Random DNA fragments obtained from eight historical and more recent GAS strains were used to produce a mixed-genome DNA microarray (21). In short, random DNA fragments (1 to 1.9 kb) from these strains were cloned into Escherichia coli, PCR amplified, and spotted on the microarray. The probability P [0;1] to cover the entire GAS genome with an average length G (1.9 × 106 bp) and an average open reading frame length t (1,500 bp) can be estimated using Poisson distribution statistics with the following formula: P = 1 − [1 − (t + I − 2r)G]N, where N is the number of significant biomarkers (2,704) (see Results) with average insert size I (1,450 bp). With an estimated minimal overlap size for successful hybridization between spotted biomarkers and labeled DNA of about 80 nucleotides (r), the probability for complete genome coverage was estimated to be 98%.
PCR-amplified probes specific for 34 known virulence genes (superantigens, adhesins, hemolysins/proteolytic enzymes, immunoreactive antigens, and regulatory elements) were also spotted on the microarray. Main genomic differences between the 41 GAS strains were identified by differentially hybridizing fragments on the microarray. DNA of differentiating biomarkers, defined as spots that had a positive Cy3 signal (reference DNA) for all isolates tested and no Cy5 signal (tester DNA) for at least one of the analyzed strains, was sequenced.
This microarray approach has several advantages. As the microarray is spotted with random DNA fragments obtained from relevant GAS strains, it does not require prior genome sequence information and allows identification of new genes. Furthermore, most GAS genomes published thus far are from North America, whereas the current approach provides a dedicated microarray to study well-documented GAS strains from The Netherlands.
Data analysis.
Hierarchical clustering of differentiating biomarkers from all strains was done with TIGR software (14) (available at http://www.tigr.org/software/tm4). Clusters of biomarkers that emerged or disappeared in one of the M types over time were sequenced. To determine the possible function of a given sequence, ERGO bioinformatics was used (http://ergo.integratedgenomics.com/ERGO/), as were BLAST searches in the GenBank database. For the sake of simplicity we use the terms “acquisition” and “loss” to describe genetic differences between historical and recent strains.
Bacteriophage induction and lysis assay.
GAS strains were grown overnight at 37°C in Todd-Hewitt medium plus yeast extract (THY). Each overnight GAS culture was diluted 1:100 with prewarmed THY and grown to an optical density at 660 nm of 0.2. Mitomycin C was added to the cultures to a final concentration of 0.2 μg/ml. Cultures were incubated for an additional 3 h at 37°C (2).
Mitomycin-treated bacteria were centrifuged at 4,000 × g for 15 min, and the supernatant was sterilized with a 0.22-μm-pore-size filter (Millipore). GAS target strains were grown to an optical density at 660 nm of 1.0, taken up in THY soft agar, and plated on THY agar plates. A phage mixture (10 μl) was spotted onto air-dried lawns. Plates were incubated overnight at 37°C, and lysis was defined as a visible clear area under the point where phage was applied. Experiments were performed three times.
Phage restriction analysis.
Mitomycin-treated bacteria (see above) were centrifuged at 4,000 × g for 15 min. The supernatant was centrifuged at 141,000 × g for 4 h at 10°C, and the pellet was suspended in 400 μl of phage suspension buffer (0.15 M NaCl, 10 mM Tris HCl [pH 7.5], 5 mM MgCl2, 1 mM CaCl2). The phage particles were lysed with 0.5% sodium dodecyl sulfate, 10 mM EDTA, and 400 μg of proteinase K/ml for 1 h at 37°C. Phage DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), followed by chloroform-isoamyl alcohol (24:1), and precipitated with 0.2 volume of 3 M sodium acetate (pH 4.2) and 2 volumes of ethanol at −70°C overnight. Finally, DNA was washed with 70% ethanol and suspended in 40 μl distilled water (2). Digestion of the phage DNA was performed with restriction endonucleases EcoRI (Roche) and PinAI (Gibco BRL) according to the guidelines of the manufacturer. Restriction endonucleases were selected on the basis of their ability to distinguish between the different GAS prophages (8) using Webcutter 2.0 software (available at http://rna.lundberg.gu.se/cutter2/). Phage restriction fragments were analyzed on 1.0% agarose gels.
RESULTS
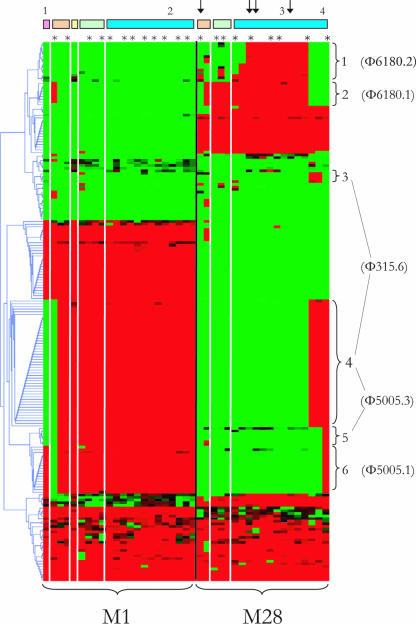
Hybridization of 41 GAS isolates belonging to M types M1 and M28 isolated in The Netherlands from the 1960s to the 1990s to a mixed-genome DNA microarray revealed that 203 of 2,704 biomarkers (8%) were differentially present (Fig. 1). These biomarkers exhibited 100% reproducible patterns in all duplicate experiments, and the distribution of 13 randomly chosen biomarkers and 22 virulence factors was confirmed by PCRs and sequencing for all isolates tested (data not shown). Biomarkers (n = 114) that differentiated between old and more recent isolates grouped in six clusters (Fig. 1) and were sequenced and analyzed using ERGO bioinformatics and BLAST searches in the GenBank database. All biomarkers showed >95% homology to genes present in at least one of the completed GAS genome sequences and represented 38 phage-related open reading frames. Using the BLAST algorithm, five of the six clusters each corresponded to a particular prophage in the GenBank database, either Φ6180.1 (8), Φ6180.2 (8), Φ315.6 (3), Φ5005.1 (19), or Φ5005.3 (19) (Table 1). One cluster (cluster 4 in Fig. 1) matched both Φ315.6 and Φ5005.3 and consisted of biomarkers that span a 33-kb genetic fragment which is identical in these two prophages (1). All these phages carry at least one superantigen or streptodornase (Table 2).
FIG. 1.
Two-dimensional hierarchical clustering, illustrating differential biomarkers in recent (isolated after 1980) and old (isolated before 1980) M1 and M28 strains. The GAS strains are arranged along the x axis by M type and year of isolation (except isolates from the 1990s, which are [given the short time span] clustered in the biomarker profile). Isolates associated with invasive GAS disease are indicated by asterisks. Isolates associated with noninvasive GAS disease are indicated by dashes. The dendrogram on the y axis shows the clustering of 203 differentiating biomarkers. Red represents the presence of a biomarker, and green represents the absence of a biomarker. The bars at the top show the decade of isolation for each M type, as follows: pink indicates isolates obtained in the 1950s, brown indicates isolates obtained in the 1960s, yellow indicates isolates obtained in the 1970s, green indicates isolates obtained in the 1980s and blue indicates isolates obtained in the 1990s. The arrows indicate strains with the emm28.0′′ allele. Braces indicate clusters of biomarkers, representing prophages. The numbering of the clusters corresponds to the numbering in Table 1.
TABLE 1.
Differential biomarkers for old (isolated before 1980) and new (isolated after 1980) M1 and M28 strains and their putative functions
| Clustera | Phageb | Accession no. | Biomarker | Coordinates (beginning … end) | Name | Function |
|---|---|---|---|---|---|---|
| 1 | 6180.2 (SpeK and Sla) | NC_007296 | 1 | 1266943 … 1267323 | M28_Spy1283 | Phage protein |
| 1266570 … 1266929 | M28_Spy1282 | Phage transcriptional regulator, Cro/CI family | ||||
| 2 | 1265583 … 1265774 | M28_Spy1281 | Phage protein | |||
| 3 | 1264844 … 1265572 | M28_Spy1280 | Phage antirepressor protein | |||
| 4 | 1256286 … 1256507 | M28_Spy1257 | Phage protein | |||
| 5 | 1255757 … 1256014 | M28_Spy1256 | Phage protein | |||
| 6 | 1251854 … 1253065 | M28_Spy1250 | Terminase large subunit | |||
| 7 | 1246284 … 1246700 | M28_Spy1243 | Phage protein | |||
| 8 | 1239705 … 1240421 | M28_Spy1235 | Phage protein | |||
| 9 | 1240418 … 1243699 | M28_Spy1235 | Phage protein | |||
| 10 | 1237561 … 1239708 | M28_Spy1234 | Phage endopeptidase | |||
| 11 | 1234137 … 1236023 | M28_Spy1231 | Phage protein | |||
| 1232591 … 1232776 | M28_Spy1227 | Phage protein | ||||
| 1231278 … 1232480 | M28_Spy1226 | Phage-associated cell wall hydrolase | ||||
| 12 | 1228677 … 1229471 | M28_Spy1223 | SpeK | |||
| 2 | 6180.1 (SpeC) | NC_007296 | 13 | 1001137 … 1001460 | M28_Spy0981 | Phage protein |
| 14 | 997487 … 1001122 | M28_Spy0980 | Phage protein | |||
| 15 | 994625 … 996679 | M28_Spy0977 | Phage protein | |||
| 3 | 315.6 (Sdn) | NC_004589 | 16 | 32577 … 34589 | SpyM3_1417 | Phage infection protein |
| 17 | 36745 … 37476 | SpyM3_1411 | Phage-associated cell wall hydrolase | |||
| 4 | 315.6 (Sdn) | NC_004589 | 18 | 2307 … 1927 | SpyM3_1456 | Phage protein |
| 2695 … 2324 | SpyM3_1455 | Phage transcriptional regulator, Cro/CI family | ||||
| 19 | 5022 … 5222 | SpyM3_1447 | Phage protein | |||
| 5313 … 5609 | SpyM3_1446 | Phage protein | ||||
| 20 | 7389 … 9308 | SpyM3_1443 | Phage-encoded DNA polymerase | |||
| 21 | 9316 … 11697 | SpyM3_1442 | DNA primase | |||
| 22 | 20647 … 21531 | SpyM3_1429 | Phage protein | |||
| 23 | 24338 … 28717 | SpyM3_1421 | Phage protein | |||
| 24 | 28732 … 29571 | SpyM3_1420 | Phage protein | |||
| 25 | 29584 … 31560 | SpyM3_1419 | Phage endopeptidase | |||
| 5005.3 (SdaD2) | NC_007297 | 18 | 1423403 … 1423804 | M5005_Spy_1465 | Phage protein | |
| M5005_Spy_1464 | Phage transcriptional regulator, Cro/CI family | |||||
| 19 | 1420497 … 1420796 | M5005_Spy_1454 | Phage protein | |||
| M5005_Spy_1453 | Phage protein | |||||
| 20 | 1416408 … 1418330 | M5005_Spy_1450 | Phage-encoded DNA polymerase | |||
| 21 | 1414019 … 1416403 | M5005_Spy_1449 | DNA primase | |||
| 22 | 1404178 … 1404747 | M5005_Spy_1435 | Phage scaffold protein | |||
| 1403278 … 1404165 | M5005_Spy_1434 | Phage protein | ||||
| 23 | 1396092 … 1400474 | M5005_Spy_1426 | Phage protein | |||
| 24 | 1395238 … 1396080 | M5005_Spy_1425 | Phage protein | |||
| 25 | 1393246 … 1395228 | M5005_Spy_1424 | Phage endopeptidase | |||
| M5005_Spy_1423 | Hyaluronoglucosaminidase | |||||
| 5 | 5005.3 (SdaD2) | 26 | 1389641 … 1391548 | M5005_Spy_1422 | Phage protein | |
| 27 | 1389641 … 1391548 | M5005_Spy_1421 | Phage infection protein | |||
| 28 | 1389471 … 1389632 | M5005_Spy_1420 | Phage protein | |||
| 1388848 … 1389468 | M5005_Spy_1419 | Phage protein | ||||
| 29 | 1387049 … 1388245 | M5005_Spy_1416 | Phage-associated cell wall hydrolase | |||
| 30 | 1385761 … 1386933 | M5005_Spy_1415 | SdaD2 | |||
| 6 | 5005.1 (SpeA2) | NC_007297 | 31 | 989426 … 991177 | M5005_Spy_1006 | Phage structural protein |
| 32 | 991293 … 994733 | M5005_Spy_1007 | Phage protein | |||
| 33 | 996219 … 998024 | M5005_Spy_1009 | Phage protein | |||
| 34 | 996219 … 998024 | M5005_Spy_1010 | Phage protein | |||
| 35 | 1001072 … 1001962 | M5005_Spy_1018 | Phage protein | |||
| M5005_Spy_1019 | Phage scaffold protein | |||||
| 36 | 1003948 … 1005273 | M5005_Spy_1021 | Phage protein | |||
| M5005_Spy_1022 | Portal protein | |||||
| 37 | 1006537 … 1006917 | M5005_Spy_1023 | Terminase large subunit | |||
| 38 | 1006537 … 1006917 | M5005_Spy_1024 | phage protein | |||
| M5005_Spy_1025 | Phage-encoded transcriptional regulator, ArpU |
Clusters of biomarkers shown in Fig. 1.
Superantigens, streptodornases, and a phospholipase, encoded on the different prophages, are indicated in parentheses.
TABLE 2.
Oligonucleotide primers used for detection of phage-specific virulence factors
| Virulence gene | Phage (reference) | Annealing temp (°C) | Amplicon size (bp) | Primer sequence (5′-3′) |
|---|---|---|---|---|
| speC | Φ6180.1 (8) | 42 | 584 | GATTTCTACTTATTTCACC |
| AAATATCTGATCTAGTCCC | ||||
| speK | Φ6180.2 (8) | 42 | 782 | TACTTGGATCAAGACG |
| GTAATTAATGGTGTAGCC | ||||
| sla | Φ6180.2 (8) | 52 | 442 | CTCTAATAGCATCGGCTACGC |
| AATGGAAAATGGCACTGAAAG | ||||
| sdn | Φ315.6 (3) | 52 | 489 | AACGTTCAACAGGCGCTTAC |
| ACCCCATCGGAAGATAAAGC | ||||
| speA2 | Φ5005.1 (19) | 50 | 754 | ATGGAAAACAATAAAAAAGTATTG |
| TACTTGGTTGTTAGGTAGACTTC | ||||
| sdaD2 | Φ5005.3 (19) | 50 | 687 | TTCCCGAACTTTATCGTACAA |
| CAGTAGAAGATAAGAGTCCACCG |
All M1 strains had the emm1.0 genotype. Recent M1 GAS strains contained prophage clusters Φ5005.1 and Φ5005.3. In contrast, two strains from 1959 and 1960 lacked cluster Φ5005.3, whereas one of these strains also lacked cluster Φ5005.1. M28 strains showed a higher degree of variability. The 19 M28 isolates all belonged to emm28.0. However, within this cluster two different emm alleles were found. Fifteen of 19 isolates had sequences that were identical to the emm28.0 reference sequence (http://www.cdc.gov/ncidod/biotech/strep/emmdata.htm#emm28). The remaining four isolates showed an identical pattern consisting of seven base pair mutations in the emm sequence outside the emm typing region. These seven base pair mutations corresponded to three altered amino acids. These four M28 isolates were therefore designated emm28.0′′ and are indicated in Fig. 1.
M28 strains from the 1990s possessed the Φ6180.2 cluster, the Φ315.6 cluster, or the Φ5005.3 and Φ5005.1 clusters. These clusters were all absent in strains isolated between the 1960s and the 1980s. All three GAS emm28.0′′ isolates from the 1990s contained the Φ6180.1 cluster, in contrast to a GAS emm28.0′′ isolate from the 1950s. Interestingly, the Φ6180.2 cluster was present in recent emm28.0 and emm28.0′′ strains, whereas both of these emm variants lacked this cluster in the 1960s (Fig. 1).
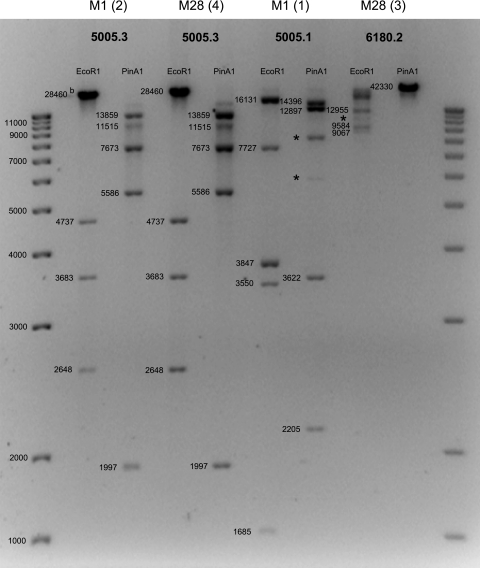
To confirm that the different clusters containing prophage genes correspond to the prophages described in the GenBank database, additional prophage-specific PCRs, phage lytic assays, and phage restriction analyses were performed. For all 41 strains, the results of PCRs and sequencing of prophage-specific virulence factors (Table 2) were completely in line with the microarray-predicted prophage content (data not shown). All strains were challenged with mitomycin C to examine the phage release and phage sensitivity within the M1 and M28 strain collection using a bacterial lysis assay. Only from induced M1 and M28 strains that possessed the Φ5005.3 cluster were phages that demonstrated lytic activity collected. These phages lysed only M1 and M28 strains that did not contain either the Φ5005.3 or Φ315.6 cluster (data not shown). Restriction profiling of DNA isolated from mitomycin-induced phages essentially confirmed their identification based on microarray, PCR, and lysis experiments (Fig. 2).
FIG. 2.
EcoRI and PinAI DNA restriction profiles of prophages isolated from M1 and M28 strains. For each strain-prophage combination a representative restriction profile is shown. The observed restriction patterns corresponded to the predicted restriction patterns, except for a few fragments indicated with an asterisk. These aberrant fragments may be explained by mutations in the corresponding restriction sites. The numbers in parentheses identify isolates used for phage DNA restriction profiling in Fig. 1. The numbers next to the bands indicate the predicted DNA fragment sizes for the restriction profiles of prophage 5005.1, 5005.3, and 6108.2 sequences deposited in the GenBank database, as determined using Webcutter 2.0.
DISCUSSION
This study determined genetic alterations in invasive and noninvasive M1 and M28 strains in The Netherlands over a period of more than four decades. All recent M1 and M28 strains were enriched in prophages carrying superantigens and streptodornases, irrespective of their clinical profile (Fig. 1). Prophages were identified as phage 6180.1, 6180.2, 315.6, 5005.1, or 5005.3 based on microarray analysis, PCR, lysis profiles, and phage restriction patterns.
Previous findings in the United States showed that M1 isolates have acquired prophages 5005.1 and 5005.3 over time (19). We show that in The Netherlands, acquisition of phages 5005.1 and 5005.3 may already have occurred in the 1950s. The oldest M1 strain in our collection, isolated in 1959, appears to represent an “intermediate” M1 isolate that had already obtained φ5005.1 but was still lacking φ5005.3. Since all subsequent M1 isolates possessed both φ5005.1 and φ5005.3, these phages, or the M1 clone harboring them, may have rapidly gained predominance among all M1 isolates. The phage induction and lysis experiments suggest that φ5005.3 remained highly mobile in all strains harboring this phage. The predominance of this phage among M1 strains, despite its mobility, suggests that it confers a selective advantage to the bacterium. Indeed, φ5005.3 is likely to enhance the virulence of its host; Aziz and colleagues showed that phages 5005.1 (sphinx) and 5005.3 (phyramid) constituted the main difference between SF370, a sequenced strain that is infrequently associated with invasive infections, and a epidemic virulent M1 strain (1).
Prophages 5005.1 and 5005.3 were also present in an emm28.0 isolate from the 1990s. To our knowledge, this is the first report of the acquisition of M1 phages by another M type.
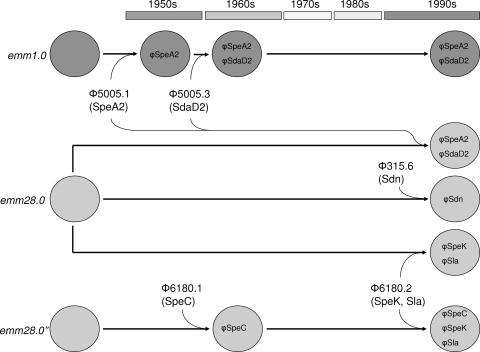
Analogous to M1, acquisition of these phages may have increased the virulence of M28 strains in The Netherlands. Given the early acquisition and predominance of prophages 5005.1 and 5005.3 in M1 isolates, these phages may have first been introduced into M1 and subsequently been transferred to M28. Indeed, lysis experiments indicated that phage 5005.3 could be induced from M1 strains and was able to infect and lyse M28 isolates that lacked this phage. Other emm28.0 isolates are likely to have obtained an M3 prophage (φ315.6) containing the streptodornase-encoding sdn gene (2). Both microarray hybridization patterns and lysis assays suggest that prophages 315.6 and 5005.3 are mutually exclusive, probably since these phages are highly homologous and have the same phage repressor module. Phage 6180.2 may have been acquired by strains with both allelic variants of emm28.0 (emm28.0 and emm28.0′′) on separate occasions (Fig. 3). Alternatively, phage 6180.2 may have been obtained by a single emm28.0 strain, which diverged into two emm28.0 variants after acquisition of the prophages. Since both emm28.0 subtypes (without this phage) were already present in 1960s isolates and all emm28.0′′ strains had identical emm gene sequences, the latter explanation is unlikely. Phages 6180.1 and 6180.2 have recently been described as part of a completed M28 genome sequence (8). However, this is the first report describing the acquisition of these phages in M28 over time, based on the notion that the oldest M28 isolate in this study did not contain either one of these phages and φ6180.1 was identified only in M28 strains isolated in the 1990s.
FIG. 3.
Hypothetical model of evolutionary events accommodating the data presented in this paper. M1 isolates may have acquired prophage 5005.1, encoding SpeA2, and φ5005.3, encoding SdaD2, on separate occasions around 1960. A similar event may have taken place in emm28.0 in the 1990s. In these years, other emm28.0 isolates may have obtained an M3 prophage (φ315.6) containing the sdn gene. M28 isolates that did not acquire these M1 and M3 prophages may have obtained the M28 phages 6180.1 and 6180.2 with corresponding virulence factors. These events involved both types of the allelic variants of emm28.0. The presence of φ6180.1 in historical emm28 isolates and its absence in recent emm28 isolates harboring prophage 315.6 or prophages 5005.1 and 5005.3 suggest that acquisition of the latter phages could be associated with loss of φ6180.1 despite the fact that they do not have the same chromosomal insertion site (8) or repressor module (GenBank).
Although the prophages themselves have been described previously, the apparent acquisition of M1 and M3 phages by M28 and the early presence of the 5005.1 and 5005.3 phages in M1 are novel. In contrast to the genome composition of M28 GAS strains, the genome composition of M1 strains remained relatively stable after the mid-1960s. The clonality of the latter M type over time may reflect its continued success as an established virulent M type.
Taken together, our results suggest that over a period of four decades, M1 and especially M28 strains have acquired prophages on at least eight different occasions (Fig. 3). All the prophages carried superantigens (speA2, speC, speK), a phospholipase (sla), or streptodornases (sdaD2, sdn). These superantigens, streptodornases, and possibly the phospholipase are all associated with invasive GAS disease (12, 16, 18). Since prophage enrichment was similar in GAS strains associated with invasive disease and GAS strains associated with noninvasive disease, our results suggest that there was an overall increase in the virulence of M1 and M28 strains over the last four decades rather than emergence of hypervirulent subclones. This increased overall virulence potential may have enhanced the frequency of invasive GAS and hence contributed to the reemergence of invasive GAS disease in The Netherlands.
Editor: A. Camilli
Footnotes
Published ahead of print on 23 April 2007.
REFERENCES
- 1.Aziz, R. K., R. A. Edwards, W. W. Taylor, D. E. Low, A. McGeer, and M. Kotb. 2005. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 187:3311-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks, D. J., B. Lei, and J. M. Musser. 2003. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 71:7079-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisno, A. L. 1991. Group A streptococcal infections and acute rheumatic fever. N. Engl. J. Med. 325:783-793. [DOI] [PubMed] [Google Scholar]
- 5.Borucki, M. K., S. H. Kim, D. R. Call, S. C. Smole, and F. Pagotto. 2004. Selective discrimination of Listeria monocytogenes epidemic strains by a mixed-genome DNA microarray compared to discrimination by pulsed-field gel electrophoresis, ribotyping, and multilocus sequence typing. J. Clin. Microbiol. 42:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carapetis, J., R. Robins-Browne, D. Martin, T. Shelby-James, and G. Hogg. 1995. Increasing severity of invasive group A streptococcal disease in Australia: clinical and molecular epidemiological features and identification of a new virulent M-nontypeable clone. Clin. Infect. Dis. 21:1220-1227. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, N. M., S. Zhang, S. F. Porcella, M. J. Nagiec, K. D. Barbian, S. B. Beres, R. B. Lefebvre, and J. M. Musser. 2005. Genome sequence of a serotype M28 strain of group A streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192:760-770. [DOI] [PubMed] [Google Scholar]
- 9.Hoe, N. P., R. M. Ireland, F. R. DeLeo, B. B. Gowen, D. W. Dorward, J. M. Voyich, M. Liu, E. H. Burns, Jr., D. M. Culnan, A. Bretscher, and J. M. Musser. 2002. Insight into the molecular basis of pathogen abundance: group A Streptococcus inhibitor of complement inhibits bacterial adherence and internalization into human cells. Proc. Natl. Acad. Sci. USA 99:7646-7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaul, R., A. McGeer, D. E. Low, K. Green, and B. Schwartz. 1997. Population-based surveillance for group A streptococcal necrotizing fasciitis: clinical features, prognostic indicators, and microbiologic analysis of seventy-seven cases. Ontario Group A Streptococcal Study. Am. J. Med. 103:18-24. [DOI] [PubMed] [Google Scholar]
- 11.Kiska, D. L., B. Thiede, J. Caracciolo, M. Jordan, D. Johnson, E. L. Kaplan, R. P. Gruninger, J. A. Lohr, P. H. Gilligan, and F. W. Denny, Jr. 1997. Invasive group A streptococcal infections in North Carolina: epidemiology, clinical features, and genetic and serotype analysis of causative organisms. J. Infect. Dis. 176:992-1000. [DOI] [PubMed] [Google Scholar]
- 12.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55:77-104. [DOI] [PubMed] [Google Scholar]
- 13.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88:2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz, B., R. R. Facklam, and R. F. Breiman. 1990. Changing epidemiology of group A streptococcal infection in the USA. Lancet 336:1167-1171. [DOI] [PubMed] [Google Scholar]
- 16.Sitkiewicz, I., M. J. Nagiec, P. Sumby, S. D. Butler, C. Cywes-Bentley, and J. M. Musser. 2006. Emergence of a bacterial clone with enhanced virulence by acquisition of a phage encoding a secreted phospholipase A2. Proc. Natl. Acad. Sci. USA 103:16009-16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens, D. L., M. H. Tanner, J. Winship, R. Swarts, K. M. Ries, P. M. Schlievert, and E. Kaplan. 1989. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N. Engl. J. Med. 321:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Sumby, P., K. D. Barbian, D. J. Gardner, A. R. Whitney, D. M. Welty, R. D. Long, J. R. Bailey, M. J. Parnell, N. P. Hoe, G. G. Adams, F. R. DeLeo, and J. M. Musser. 2005. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl. Acad. Sci. USA 102:1679-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sumby, P., S. F. Porcella, A. G. Madrigal, K. D. Barbian, K. Virtaneva, S. M. Ricklefs, D. E. Sturdevant, M. R. Graham, J. Vuopio-Varkila, N. P. Hoe, and J. M. Musser. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group a Streptococcus involved in multiple horizontal gene transfer events. J. Infect. Dis. 192:771-782. [DOI] [PubMed] [Google Scholar]
- 20.Vlaminckx, B. J. M., W. van Pelt, L. M. Schouls, A. van Silfhout, C. P. Elzenaar, E. M. Mascini, J. Verhoef, and J. Schellekens. 2004. Epidemiological features of invasive and noninvasive group A streptococcal disease in the Netherlands, 1992-1996. Eur. J. Clin. Microbiol. Infect. Dis. 23:434-444. [DOI] [PubMed] [Google Scholar]
- 21.Vlaminckx, B. J. M., F. H. J. Schuren, R. C. Montijn, M. P. M. Caspers, A. C. Fluit, W. J. B. Wannet, L. M. Schouls, J. Verhoef, and W. T. M. Jansen. 2007. Determination of the relationship between group A streptococcal genome content, M type, and toxic shock syndrome by a mixed genome microarray. Infect. Immun. 75:2603-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]