Abstract
The human malaria parasite, Plasmodium falciparum, utilizes multiple ligand-receptor interactions for the invasion of human erythrocytes. Members of the reticulocyte binding protein homolog (PfRh) family have been shown to be critical for directing parasites to alternative erythrocyte receptors that define invasion pathways. Recent studies have identified gene amplification, sequence polymorphism, and variant expression of PfRh paralogs as mechanisms underlying discrimination between pathways for invasion. In this study, we find considerable heterogeneity in the invasion profiles of clonal, uncultured P. falciparum parasite isolates from a low-transmission area in Senegal. Molecular analyses revealed minimal variation in protein expression levels of the PfRh ligands, PfRh1, PfRh2a, and PfRh2b, and an absence of gene amplification in these isolates. However, significant sequence polymorphism was found within repeat regions of PfRh1, PfRh2a, and PfRh2b. Furthermore, we identified a large sequence deletion (∼0.58 kb) in the C-terminal region of the PfRh2b gene at a high prevalence in this population. In contrast to findings of earlier studies, we found no associations between specific sequence variants and distinct invasion pathways. Overall these data highlight the importance of region-specific elaborations in PfRh sequence and expression polymorphisms, which has important implications in our understanding of how the malaria parasite responds to polymorphisms in erythrocyte receptors and/or evades the immune system.
Underlying the pathogenesis of malaria caused by Plasmodium falciparum infection is exponential growth of the parasite within erythrocytes of the bloodstream. Recent research has highlighted the importance of parasite factors contributing to the multiplication rate and of erythrocyte selectivity in severe malaria (5).
Both laboratory and field isolates of P. falciparum parasites demonstrate a remarkable plasticity in their ability to invade erythrocytes using alternative receptors (2, 7, 12, 15, 19, 21, 22). Genetic and antibody inhibition studies are beginning to elucidate the molecular basis for this phenomenon (7-9, 11, 12, 15, 18, 19, 22, 26, 27, 31). Several of these studies have established that alternative invasion pathway usage is specifically associated with variant expression of the P. falciparum reticulocyte binding protein homolog family (PfRh) (9, 18, 24, 26, 28, 29). These proteins belong to a superfamily of reticulocyte binding protein-like (RBL) proteins, which includes the Py235 family of Plasmodium yoelii and PvRBP proteins of Plasmodium vivax, which are postulated to be involved in the selection of specific host cells (10, 14). Members of the RBL family have been localized to the apical organelles of the invasive merozoite form and are believed to play a role in the apical recognition of the erythrocyte and tight junction formation.
Five PfRh genes have been identified in the 3D7 P. falciparum genome, PfRh1 (also previously published as PfNBP1), PfRh2a (also previously published as PfR2ha and PfNBP2a), PfRh2b (also previously published as PfR2hb and PfNBP2b), PfRh3 (also previously published as PfRH3), and PfRh4 (also previously published as PfRH4) (13, 24, 26, 30, 32); however, PfRh3 is not thought to be expressed as a protein in the asexual stage (30). PfRh1 interacts with an unidentified sialic acid-containing and trypsin-resistant receptor (Y) (26), and genetic evidence suggests that PfRh2b interacts with another undetermined chymotrypsin-sensitive and trypsin-resistant receptor (Z) in a sialic acid-independent manner (9). PfRh2a, which is identical to PfRh2b except for divergence in the small unique C-terminal region (24), has not been shown to define a dominant invasion phenotype. Expression of PfRh proteins in laboratory isolates of P. falciparum is variable, and the majority of parasite lines alternatively express either PfRh1 or PfRh2a and PfRh2b (9, 29, 31). Interestingly, sialic acid dependence of P. falciparum laboratory-cultured lines correlates with the level of PfRh1 expression, resulting from gene amplification, suggesting that the differential expression of PfRh1 or PfRh2a and PfRh2b specifies the use of alternative invasion pathways. Expression of another paralog, PfRh4, has also been associated with a switch to a sialic acid-independent pathway (28). Adding a further layer of complexity, recently Lobo and colleagues identified sequence polymorphisms in PfRh1, PfRh2a, and PfRh2b that were associated with specific invasion phenotypes in field samples from Brazil (17).
Here we report an investigation into the first-cycle invasion phenotypes of P. falciparum strains from Senegal, an area of low endemicity where most patients harbor clonal parasite lines. Although alternative erythrocyte invasion pathways were commonly used by these parasites, several features of the expression and sequence of the PfRh invasion ligands were unusual. Unlike most lab isolates, all the Senegal isolates studied expressed PfRh1, PfRh2a, and PfRh2b simultaneously, and although there was significant sequence polymorphism in these ligands, they did not associate with specific invasion pathways as has been previously reported (17). Furthermore, a C-terminal sequence deletion in PfRh2b previously identified only in a single lab isolate was present in the majority of these field isolates. The implications of these findings for our understanding of the role of variations in PfRh sequence and expression in defining invasion pathways are discussed.
MATERIALS AND METHODS
P. falciparum field strains.
This study was approved by the Institutional Review Board of the Harvard School of Public Health and by the Ethics Committee of the Ministry of Health in Senegal. Venous blood samples (∼5 ml) were collected from consenting patients during September and October 2004, the peak malaria transmission season. Two collection points were established, one in Pikine (Poste de Sante Touba Diacksao) and the other in Thies (Service de Lutte Anti-Parasitaire), approximately 15 and 70 km from Dakar (Senegal), respectively. A total of 54 samples were collected (27 from Thies and 27 from Pikine), placed on ice, and transported immediately to l'Hopital Aristide Le Dantec, Dakar, for analysis.
Erythrocyte invasion assays.
Invasion assays were carried out as previously described (9). Briefly, erythrocytes (O+, rhesus+) from a single donor were washed three times in phosphate-buffered saline (PBS) and treated with 2-3,6-8-neuraminidase (66 mU), trypsin (1 mg/ml), or chymotrypsin (1 mg/ml) or with a combination of 2-3,6-8-neuraminidase (66 mU) and trypsin (1 mg/ml; Sigma), which prevents invasion, as a negative control. Positive-control erythrocytes were washed in incomplete RPMI. Erythrocytes were placed on a rotating plate in a 37°C incubator for 1 h. Following treatment, erythrocytes were washed two times in PBS and resuspended in complete RPMI (incomplete RPMI supplemented with 5% [vol/vol] O+ and 0.25% [wt/vol] albumax [10 ml]) at a 2% hematocrit (calculated using a Hauser Scientific phase hemacytometer).
Infected samples were adjusted to 1% parasitemia with uninfected erythrocytes and then washed in PBS three times. Samples were then treated with 2-3,6-8-neuraminidase (66 mU; Calbiochem) and trypsin (1 mg/ml; Sigma for 1 h at 37′C) to prevent reinvasion of donor cells, washed two times, and resuspended in complete RPMI at 2% hematocrit.
Treated parasitized erythrocytes (50 μl) were added to treated nonparasitized erythrocytes (50 μl) in 96-well flat-bottom plates in triplicate. Assay plates were placed in a modular incubator chamber (Billups-Rothenberg) with 5% carbon dioxide-1% oxygen and incubated at 37°C until reinvasion occurred, routinely between 48 and 72 h. First-round parasitemias were determined by counting a minimum of 500 erythrocytes using a Miller graticule.
Western blots, PCR mapping, and Southern blots.
Parasite culture supernatants were separated on 5% or 10% Tris-glycine polyacrylamide gels and transferred to a nitrocellulose membrane (0.45 μM; Schleicher & Schuell). Membranes were blocked with 10% milk in PBS-Tween 20 before blotting with antisera specific for PfRh1, PfRh2a, and PfRh2b (used at a 1/300 dilution). The specificities of these antibodies have been described previously (26). Protein loading was controlled using mab2F3, specific for Sera5 (used at a 1/1,000 dilution), kindly provided by Brendan Crabb (WEHI). Parasite supernatants have been previously shown to correlate with protein levels in schizonts for PfRh1 in a semiquantitative fashion (31). Differences in PfRh2a and PfRh2b levels previously observed for laboratory isolates in supernatants (9) are also observed in schizonts (see Fig. S1 in the supplemental material).
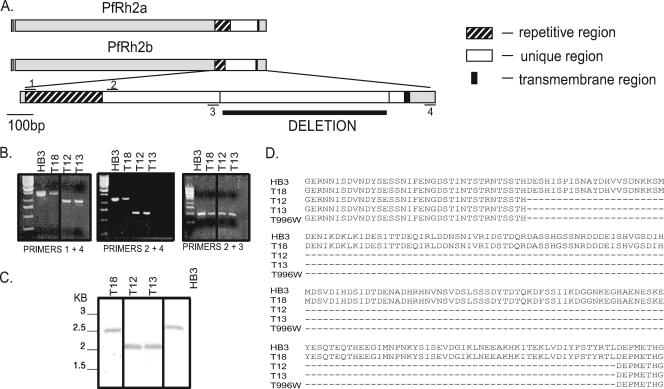
PCR mapping of the size polymorphism in PfRh2b was carried out using the following primers: P1-GCGGATCCGGTGATGAAAAATTAATATTAAAAG, P2-GCGGATCCCAACAACAAAGAAATATCCAAG, P3-GCGAATTCTTAATCATG TGTACTAGACGTGTTTC, and P4-GCGAATTCATCATCCATTTTGTTATGGTTTG (restriction sites indicated in bold) (see Fig. 4A). PCR typing of either the full-length PfRh2b or PfRh2bdel from the Senegalese isolates was carried out using the primers P2 and P4.
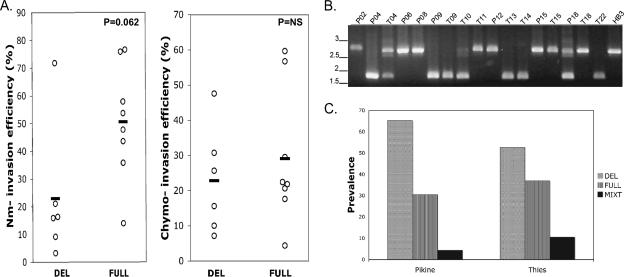
FIG. 4.
Identification of a large sequence polymorphism within the C terminus unique region of PfRh2b in Senegalese isolates. (A) Schematic of PfRh2a and PfRh2b showing the 3′ repetitive, unique, transmembrane, and cytoplasmic tail regions. Primers used for the PCR mapping of the deletion are indicated. (B) PCR mapping identifies a major sequence deletion in the unique region of PfRh2b in representative Senegalese isolates. (C) Southern blot of the C-terminal region of PfRh2b, confirming the presence of the deletion in the chromosomal DNA in a subset of Senegalese isolates. A fragment of the unique region upstream of the deletion, amplified with P1 and P3, was used as a probe. (D) The sequence of the PfRh2b deletion is identical in multiple Senegalese isolates and the Thai isolate T996w (T996 obtained from WEHI, Melbourne). Alignment of the unique region from Senegalese isolate with and without the deletion with those of HB3 and T996w.
Genomic blotting was carried out using standard methods. To confirm the presence of the deletion in the chromosome, genomic DNA was digested with AvaII and Nco1 and probed with the fragment produced by amplification with the primers P2 and P3 (see Fig. 4B), which corresponds to a PfRh2b sequence common to both the deleted and full-length forms of PfRh2b. The PfRh1 gene copy number was determined as previously described (31). Radioactive signals were quantitated using a phosphorimager.
Amplification and sequencing of the polymorphic regions of PfRh1, PfRh2a, and PfRh2b.
Gene fragments of PfRh2a and PfRh2b containing both the repeat region and the downstream unique region were amplified for sequencing using a single upstream primer, Rh2F (GTCGATGAAGCTACTTATTT), and separate downstream primers, Rh2aR (CCTGTGTGTATATAGCAAC) and Rh2bR (ATCATCCATTTTGTTATGGTTTG). A region surrounding the PfRh1 gene was amplified using the primers Rh1F (CATAATCATAATCATAATCATAATC) and Rh1R (CAAAAATACATTCTTTCATGGTTTCC). PCR primers were used for sequencing as well as internal primers Rh2a internal F (GAAAGAAAGAAAATCGAGTTAG), Rh2a internal R (CTAACTCGATTTTCTTTCTTTC), Rh2b internal F (TGCGTATGATCATGTTGTTT), and Rh2b internal R (AGCATCACGTTGGTCAGTA). Gene fragments were amplified with PCR using HotStarTaq DNA master mix (QIAGEN) on a DNA Engine Tetrad thermal cycler (MJ Research). PCR products were purified for direct sequencing using ExoSap-IT (US Biochemicals Corporation) and sequenced on an ABI3100 or ABI3730 capillary sequencer using BigDye Terminator v.3.0 (Applied Biosystems, CA) chemistry. Sequences were edited using Sequencher v.4.2.2 software (Gene Codes Corporation) and aligned manually using MacClade v.4.06.
RESULTS
Senegalese P. falciparum isolates utilize alternative pathways for first-round erythrocyte invasion in vitro.
Laboratory-adapted P. falciparum isolates display variable efficiencies of invasion of erythrocytes treated with either neuraminidase, which removes sialic acid, or the protease chymotrypsin or trypsin, which removes specific but nonoverlapping proteinaceous receptors, reflecting critical differences in dependence on alternate erythrocyte receptors for invasion (4, 9). We determined invasion efficiencies of receptor-restricted erythrocytes for a series of uncultured isolates from the same region of Senegal. Fifty-four P. falciparum isolates were collected from patients in the two field sites, Pikine and Thies. Twenty-eight had parasitemias greater than 1% and were used in invasion assays. Acceptable invasion data (which we defined as invasion of control erythrocytes at least onefold greater than the starting parasitemia) were obtained for 17 of the 28 parasite lines (Fig. 1A). The majority of these parasite lines (14/17) were clonal as measured by genotyping for size polymorphisms at the msp-1 and msp-2 loci (data not shown) (33).
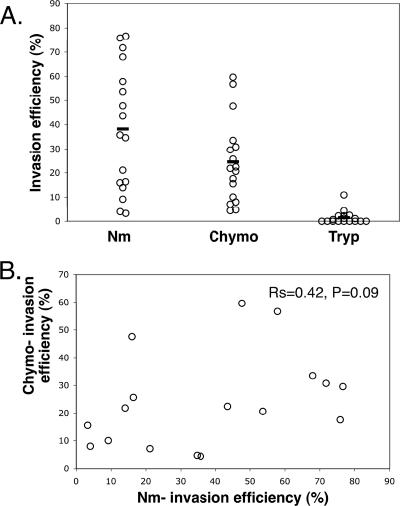
FIG. 1.
Plasmodium falciparum strains from Dakar, Senegal, routinely use alternate erythrocyte invasion pathways. (A) Efficiency of invasion by P. falciparum of neuraminidase (Nm)-treated erythrocytes, chymotrypsin (Chymo)-treated erythrocytes, and trypsin (Tryp)-treated erythrocytes. (B) Lack of significant correlation of efficiencies of invasion of enzyme-treated erythrocytes for the series of isolates. Rs and P values were derived by Spearman's rank correlation.
In the 17 parasite lines that invaded effectively, efficiency of invasion of sialic acid-depleted erythrocytes was very variable (30.8% ± 22.4% standard deviation [SD]). Similarly, invasion of chymotrypsin-treated erythrocytes also varied (25% ± 17.9% SD). No association was observed between the utilization of the sialic acid-dependent and chymotrypsin-sensitive invasion pathways, suggesting that they represent distinct invasion pathways (Fig. 1B). Despite the largely clonal nature of these isolates and the low transmission dynamics in the study population, there is clearly therefore wide variability in the abilities of P. falciparum isolates to use alternative invasion pathways. However, all isolates were largely unable to invade trypsin-treated erythrocytes (2% ± 3% SD).
Minimal variation in levels of expression of PfRh1, PfRh2a, and PfRh2b in Senegalese P. falciparum strains.
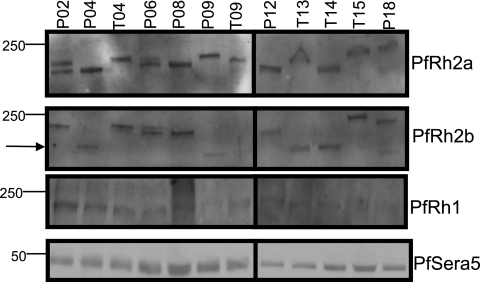
Protein expression of the PfRh ligands PfRh1, PfRh2a, and PfRh2b is variant in most laboratory-adapted P. falciparum strains—those that express PfRh1 often do not express detectable levels of PfRh2a and PfRh2b, while those that express PfRh2a and PfRh2b often express undetectable levels of PfRh1 (29, 31). We were interested in examining the extent of alternate expression of the different PfRh paralogs in the Senegalese isolates. To determine whether similar patterns of expression are observed with our field isolates, we probed parasite supernatants on Western blots with antibodies specific for PfRh1, PfRh2a, and PfRh2b (24, 26). Unlike the case with the majority of laboratory isolates, we found that all Senegalese P. falciparum field isolates tested expressed PfRh2a and PfRh2b, while almost all expressed PfRh1 (Fig. 2). We were not able to assess PfRh4 expression due to the unavailability of a specific antibody that recognizes PfRh4 in culture supernatants.
FIG. 2.
P. falciparum isolates from Senegal exhibit minimal variation in the expression of PfRh1, PfRh2a, and PfRh2b. Western blots of supernatant extracts probed with antisera specific for PfRh2a, PfRh2b, PfRh1, or PfSera5 as a loading control. The various sizes of PfRh2a and PfRh2b in the different isolates result from both sequence polymorphisms in the repetitive regions of these proteins and proteolytic breakdown products that are commonly observed even in laboratory isolates. Additionally, some isolates contain a large sequence deletion in PfRh2b (indicated by an arrow).
Absence of PfRh1 gene amplification in Senegalese parasites.
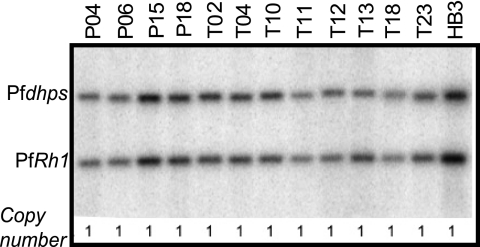
An increase in PfRh1 gene copy number has been associated with both the level of PfRh1 protein expression and sialic acid-dependent invasion in laboratory isolates (31). We were able to obtain genomic DNA in a large enough quantity for 12 Senegalese isolates, of which 9 were successful in invasion assays. We found that all of these isolates possessed a single copy of the PfRh1 gene (Fig. 3), consistent with the observed minimal variation in PfRh1 protein expression in this population.
FIG. 3.
Lack of PfRh1 amplification in Senegalese field isolates. Southern blot to determine PfRh1 copy number. Pfdhps is a single-copy gene used as a control. PfRh1 copy number was determined for each sample by comparing the Pfdhps/PfRh1 signal ratio for each Senegalese isolate to that for HB3, which is known to have a PfRh1 copy number of 1.
High-prevalence sequence polymorphism in the PfRh2b gene.
We noted significant polymorphism in the sizes of the PfRh2b proteins in a subset of the Senegalese isolates (Fig. 2). PCR mapping, using primers P1, P2, and P3, identified a significant deletion in the unique region found downstream of the repetitive region in a few representative isolates (Fig. 4A and B). To confirm that the PfRh2b deletion was not a PCR artifact and is encoded in the P. falciparum genome, Southern blotting was carried out with genomic DNA from these isolates (Fig. 4C). Bands corresponding to those expected for a genetic deletion of the size predicted from the PCR mapping were observed.
The sequence encoding the C-terminal region of PfRh2b was determined for 17 Senegalese isolates. Five of the 17 isolates contained the wild-type sequence for PfRh2b, while 12 isolates showed a single identical 582-bp deletion in the sequence, PfRh2bdel, corresponding to the amino acid residues 2867 (D) to 3061 (L), in the predicted protein sequence for full-length PfRh2b (MAL13P1.176) (Fig. 4D). No other polymorphisms were found within this region. The PfRh2bdel sequence is similar but not identical to one previously identified by Taylor and colleagues in the laboratory-adapted parasite line T996 from Thailand, differing only by a single point mutation at the boundary (29). However, resequencing of the T996 allele (provided by Tony Triglia, WEHI) revealed a sequence identical to that observed in the Senegalese samples.
We investigated the possibility that PfRh2bdel polymorphism is associated with the use of neuraminidase-sensitive invasion pathways. A positive association of borderline significance was found (P = 0.062, two-tailed Mann-Whitney U test) (Fig. 5A). No difference in the use of chymotrypsin-sensitive pathways was observed between the two groups.
FIG. 5.
The role of the PfRh2bdel polymorphism in the Senegalese isolates. (A) Association between invasion pathway utilization and the presence of the PfRh2bdel polymorphism. P value indicated was derived using a Mann-Whitney U test. DEL, deletion present; FULL, full-length sequence; NS, not significant. (B) PCR assay for the identification of polymorphisms in the downstream deletion in PfRh2b. (C) High prevalence of PfRh2b deletion in Senegalese population, determined using the PCR-based assay for the detection of the PfRh2bdel in isolates from Pikine (n = 23) or Thies (n = 19).
A larger series of field samples was typed by PCR for the presence of the PfRh2b sequence deletion (Fig. 5B). This analysis found PfRh2bdel to be prevalent in 64% of samples from Thies (n = 30) and Pikine (n = 30) (Fig. 5C).
Polymorphism in the repeat regions of PfRh1, PfRh2a, and PfRh2b.
Recently, Lobo and colleagues identified polymorphisms in repeat regions of PfRh1, PfRh2a, and PfRh2b that were associated with specific invasion pathways in Brazil (17). We sequenced the regions of the corresponding genes containing these polymorphisms to assess whether similar associations could be found in parasites from Senegal.
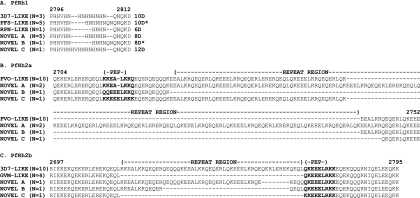
For PfRh1, we found that there is significant polymorphism in the repetitive region (Fig. 6). Three parasites of the 15 successfully sequenced had the FFS-like 10D variant (using the coding system of Lobo and colleagues [15]), previously reported to be associated with a sialic acid-dependent and trypsin-resistant invasion pathway in Brazil, while 5 isolates contained the 3D7-like 10D* variant and one the RPN-like 6D variant. Three novel variants, 8D (NOVEL A), 8D* (NOVEL B), and 12D (NOVEL C), were also found. Therefore, 6 different variants were identified from 16 determined sequences, demonstrating the complexity of this repeat region.
FIG. 6.
Extensive and novel polymorphisms in the repetitive regions of PfRh1, PfRh2a, and PfRh2b. (A) Alignment of PfRh1 repetitive sequence polymorphisms and their allelic designations according to the convention of Lobo et al. (17) (amino acids 2734 to 2741 according to the 3D7 sequence, accession no. AAQ10315). Isolates that contain polymorphisms previously described by Lobo et al. (17): 3D7-LIKE (P18, T14, and T15); FFS-LIKE (P04, P06, T04, T09, and T10); Dd2-LIKE (T22). Isolates that contain novel polymorphisms: NOVEL A (T11, T18, P02, P12, and T13); NOVEL B (P15), and NOVEL C (P09). (B) Alignment of sequence polymorphisms in the repetitive regions of PfRh2a in multiple Senegalese isolates. Polymorphism in the repetitive regions and the “pep” polymorphisms (amino acids 2734 to 2741 according to the FVO sequence, accession no. AAN39446) are indicated in bold. Isolates that contain polymorphisms previously described by Lobo et al. (17), FVO-LIKE (P04, P05, P06, P12, P41, P43, T09, T10, T11, and T13). Isolates that contain novel polymorphisms, NOVEL A (P15 and T15), NOVEL B (P18), and NOVEL C (T14). (C) Alignment of sequence polymorphisms in the repetitive regions of PfRh2b in multiple Senegalese isolates. The repetitive region and “pep” polymorphisms (amino acids 2769 to 2777 according to the 3D7 sequence, accession no. AAN39447) are indicated in bold. Isolates that contain polymorphisms previously described by Lobo et al. (17): 3D7-LIKE (P04,P05,P41,P43,T09,T10,T12,T13,T14,T18) and GVM-LIKE (P06,P15,P18,T22). Isolates that contain novel polymorphisms: NOVEL A (P09), NOVEL B (T11), and NOVEL C (P12). Isolates containing the downstream PfRh2bdel polymorphism are shown in bold.
Sequencing of PfRh2a revealed that the majority of isolates (12 of the 14) contained the FVO-like “pepA” sequence (KKEALKKQ) (24), while one contained the “pepC” sequence (KKEEELRKK). Only one isolate possessed a novel sequence, QQEEELKKQ, (NOVEL C) at this locus (Fig. 6B). Limited polymorphism was observed in the repetitive region of PfRh2a, with 12/14 isolates containing the same sequence as FVO. Only one isolate (P18) possessed the 50-amino-acid deletion downstream that occurs in all Brazilian isolates (NOVEL B) (17). Two other isolates contained a novel 58 amino acid insertion immediately downstream of the pepA sequence (NOVEL A). None of the isolates contained the upstream 15-amino-acid insertion polymorphism found in 7G8 (17).
The “pepC*” peptide sequence (QKEEELRKK) of PfRh2b was found in the majority of the Senegalese isolates (16/17) (Fig. 6C), while the remaining isolate (P12) contained a novel KKEEELRKK sequence (pepC**). The upstream repeat region was of two major types: the canonical sequence found in the 3D7 parasite line (10/17 isolates) and a 52-amino-acid sequence deletion found in Dd2 and FVO (5/17 isolates) (24) and the majority of Brazilian isolates, including GVM (17). Only two isolates (NOVEL A and NOVEL B) contained smaller deletions of 8 amino acids and 36 amino acids in the repeat regions that appear to be hybrids of the two forms.
No polymorphisms were observed in the unique regions downstream of the PfRh2a and PfRh2b repetitive regions, other than the PfRh2bdel sequence.
Associations between sequence polymorphisms of PfRh paralogs and invasion pathways.
The PfRh1 10D polymorphism has been associated with invasion of erythrocytes via neuraminidase-sensitive and trypsin-resistant receptors by Brazilian isolates (17). However, no association was found between this allele and erythrocyte invasion using either the neuraminidase-sensitive or chymotrypsin-sensitive pathway in these Senegalese isolates. The sensitivity of the isolates to trypsin precluded this analysis.
Similarly, the pepB sequences of PfRh2a and PfRh2b were found to associate with the neuraminidase-sensitive and trypsin-resistant pathway in the Brazilian isolates. None of the Senegalese isolates we studied contained the pepB sequences, consistent with the fact that they are largely dependent on trypsin-sensitive receptors for invasion but not consistent with their variable dependency on sialic acid receptors (Fig. 1A). No element in either the PfRh2a or the PfRh2b repetitive regions correlated with invasion pathway usage.
When we considered associations between the different sequence polymorphisms, a weak association was found between the presence of the canonical repeat region sequence in PfRh2b (found in 3D7) and the presence of the downstream PfRh2b deletion (P = 0.056, Fisher's exact; n = 20). This could be due either to a functional interaction between these regions in the PfRh2b protein or alternatively simply to genetic linkage.
DISCUSSION
Here we report an investigation into the first-round invasion phenotypes of P. falciparum strains from Senegal. Parasites displayed a variable dependence on sialic acid-containing and chymotrypsin-sensitive receptors, while most parasite lines were largely dependent on trypsin-sensitive receptors. Further, we have characterized the expression profiles of three key PfRh members, PfRh1, PfRh2a, and PfRh2b, and find that they are expressed in almost all of the clonal parasite lines. These paralogs contain numerous sequence polymorphisms, some of which have previously been associated with the use of different invasion pathways. Interestingly, we have identified a ∼194-amino-acid deletion in the C-terminal ectodomain region of Rh2b, previously identified only in a single laboratory isolate, which exists at a high prevalence in Senegalese isolates.
Our understanding of the extent of alternative invasion pathway usage in isolates of P. falciparum that actually infect people is still limited to a very few locations globally (2, 15, 17, 20, 21). We measured first-round invasion because of the possibility that invasion pathways may be altered in culture-adapted lines due to altered selection pressures in in vitro culture. Under these conditions and even though our study area is an area of low transmission with a large proportion of clonal isolates, we found the extensive utilization of alternative invasion pathways. In particular, there was significant variability in the abilities of these lines to use two independent alternative invasion pathways, sensitivity to neuraminidase or chymotrypsin, respectively. Taken together with the results of previous studies (2, 15, 21), our results emphasize that invasion pathway variation appears to be the norm for uncultured field isolates regardless of their origin. However, all isolates were highly dependent on trypsin-sensitive receptors, mirroring previous observations made with isolates from The Gambia (2). This may be a result of dependence of these isolates on glycophorin A for invasion, as previously suggested (2). However, several other trypsin-sensitive receptors, such as glycophorin C and receptor X, that may also play a role have been identified (7, 16, 18). Although invasion pathway variation is therefore widespread, some pathways are clearly more commonly used in specific regions than others, perhaps reflecting different distributions of erythrocyte surface polymorphisms in the exposed human population.
Alternative invasion pathways are known to be specified in part by the expression and sequence of the P. falciparum RBL invasion ligands PfRh1, PfRh2a, and PfRh2b. Previous studies with laboratory isolates from different parts of the world found that most isolates express either PfRh1 or PfRh2a and PfRh2b (9, 29, 31). Expression of PfRh1 has been associated with sialic acid-dependent invasion, while expression of PfRh2a and PfRh2b has been associated with sialic acid-independent invasion. Differential expression of either PfRh1 or PfRh2a and PfRh2b has also been seen in the only field studies to look at PfRh expression, in Kenya and Tanzania (3, 20). Surprisingly, the dramatic variation in expression of PfRh1 and PfRh2a/PfRh2b seen in laboratory isolates and previous field studies was not a feature of parasites obtained within this population, since almost all field isolates studied in Senegal expressed PfRh1, PfRh2a, and PfRh2b. This was reinforced by the striking absence of PfRh1 gene amplification in this population, which was previously associated with sialic-acid-dependent invasion in a series of laboratory isolates (31). Previous assumptions that differential expression of either PfRh1 or PRh2a and PfRh2b and/or gene amplification is a common feature of parasites using alternate invasion pathways clearly need to be qualified as a result of these studies. Further, a model has been put forth that suggests that expression alone does not dictate the dominant invasion pathway that is utilized by a parasite line for invasion (1). There is likely a hierarchy of molecular interactions, where even though some molecules are expressed, they may not be able to form definitive molecular interactions. This may partly explain the observed lack of association between the expression of the PfRh paralogs and invasion pathway usage.
Indeed, there are many members of the erythrocyte-binding antigen (EBA) and PfRh invasion ligand families that may be able to contribute to alternative invasion pathways in P. falciparum. The relationship between members of the EBA and Rh families is not clear. The targeted deletion of EBA-175 leads to an increase in PfRh4 expression (28). Similarly, in a parasite line with a targeted deletion of PfRh2b, there is an increased dependence on EBA-140 and EBA-175 for invasion (1). Therefore, the EBA and PfRh ligands exhibit a degree of functional equivalence. Expression levels of PfRh4 have been shown to be associated with alternative invasion pathways (28), although it was not possible to assess the expression of PfRh4 in these isolates, and this will be an important feature of future studies. It will also be of interest to determine whether different merozoites within schizonts express different PfRh paralogs, as observed for the rodent malaria parasite Plasmodium yoelii (23).
By contrast, the high prevalence of certain sequence polymorphisms observed in PfRh1, PfRh2a, and PfRh2b in the Senegalese parasites suggests that these polymorphisms may be of greater significance in defining alternative invasion pathways than the variant expression of the PfRh proteins, at least in this population. Many of the sequence polymorphisms have previously been found in globally diverse isolates (17). We found that the most prevalent polymorphisms in the repeat regions of PfRh1, PfRh2a, and PfRh2b in isolates from Brazil were also the most prevalent ones found in Senegal: the 10D/10D* polymorphisms in PfRh1, the pepC polymorphism in PfRh2a, and pepC* in PfRh2b. However, although specific polymorphisms in PfRh1, PfRh2a, and PfRh2b have previously been identified as associating with sialic acid-dependent and trypsin-resistant invasion pathways for Brazilian isolates (17), we were unable to find similar associations for our Senegalese isolates. This highlights the importance of studying these polymorphisms in different regions and suggests that the association of specific PfRh polymorphisms with invasion pathways may be more complex than previously thought. Nevertheless, nothing is known about the precise functional domains of the PfRh proteins. It is possible that these polymorphisms lie in the actual erythrocyte-binding region. Alternatively, they may result from selection for immune evasion.
Size polymorphisms in the repeat regions of PfRh2a and PfRh2b are likely to result from recombination between the repeats shared by both genes. Mitotic recombination between PfRh2a and PfRh2b has already been observed in a long-term-cultured isolate, highlighting the recombination potential of this region (6), and gene conversion between PfRh2a and PfRh2b has been suggested as a mechanism for maintaining the unusual shared structure of these two closely linked genes (25). Even though the prevalence of several PfRh2a and PfRh2b variants is high in Senegal, it is interesting to note that very few of the variants are unique to this region. The presence of the same repeat-region polymorphisms in isolates from Senegal and throughout the world suggests a degree of stability in these alleles through time and geography and that the recombination events that generate novel alleles are not frequent. Alternatively, it is possible that the limited number of novel variants reflects functional constraints on this region.
Downstream of the repeat region in PfRh2a and PfRh2b is a C-terminal ectodomain region that is divergent between them (Fig. 4A). Previously, we found through the use of genetic knockouts that PfRh2b but not PfRh2a is required for a discrete invasion pathway, providing genetic evidence that the difference in function of PfRh2a and PfRh2b maps to this unique region (9). A major finding of this study is the presence of a large deletion in the unique region of PfRh2b that is present at a high prevalence in Senegal. A similar deletion had previously been found in a single lab strain originally isolated from Thailand (T996 or Tak996 isolate) (29). We have also observed this deletion at a high prevalence in different populations throughout Senegal and Africa (Ahouidi et al., unpublished data), and the majority of Senegalese isolates had this deletion. Although the functional implications of this common mutation are currently unknown, here we provide some evidence that it is associated with a sialic acid-dependent invasion pathway (P = 0.062), further emphasizing that PfRh mutations can be associated with the use of alternative invasion pathways. Functional studies using reverse-genetics approaches are under way to confirm this association. If true, this is consistent with a model where the sialic acid-independent ligand PfRh2b harboring the deletion is nonfunctional, allowing the sialic acid-dependent ligand PfRh1 to play a more dominant role.
The PfRh proteins are exposed on the surface of the merozoite, where they can be inhibited by antibodies. In addition to different levels of expression of the PfRh proteins, sequence polymorphisms are likely to play a major role either in altering the specificity of parasite ligands for erythrocyte receptors and/or as a means for immune evasion. These studies emphasize the complexity of the association of PfRh expression and polymorphism profiles with alternative invasion pathways and suggest that we are still some way from being able to draw clear correlations. Further studies, both using field isolates from diverse locations and using reverse-genetics approaches in the laboratory, will make these correlations more clear and define their potential role in clinical severity.
Supplementary Material
Acknowledgments
We thank L. Ndiaye, D. Ndiaye, O. Ly, Y. Diedhiou, T. Sene, A. Mbaye, and D. Diop for collecting samples. We also thank N. C. Kane for her generous assistance.
This work was supported by a Fogarty International training grant, 5D43TW001503. M.T.D. was supported by NIH R01AI057919.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 30 April 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Baum, J., A. G. Maier, R. T. Good, K. M. Simpson, and A. F. Cowman. 2005. Invasion by P. falciparum merozoites suggests a hierarchy of molecular interactions. PLoS Pathogens 1:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum, J., M. Pinder, and D. J. Conway. 2003. Erythrocyte invasion phenotypes of Plasmodium falciparum in The Gambia. Infect. Immun. 71:1856-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bei, A. K., C. D. Membi, J. C. Rayner, M. Mubi, B. Ngasala, A. A. Sultan, Z. Premji, and M. T. Duraisingh. 2007. Variant merozoite protein expression is associated with erythrocyte invasion phenotypes in Plasmodium falciparum isolates from Tanzania. Mol. Biochem. Parasitol. 153:66-71. [DOI] [PubMed] [Google Scholar]
- 4.Camus, D., and T. J. Hadley. 1985. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science 230:553-556. [DOI] [PubMed] [Google Scholar]
- 5.Chotivanich, K., R. Udomsangpetch, J. A. Simpson, P. Newton, S. Pukrittayakamee, S. Looareesuwan, and N. J. White. 2000. Parasite multiplication potential and the severity of Falciparum malaria. J. Infect. Dis. 181:1206-1209. [DOI] [PubMed] [Google Scholar]
- 6.Cortes, A. 2005. A chimeric Plasmodium falciparum Pfnbp2b/Pfnbp2a gene originated during asexual growth. Int. J. Parasitol. 35:125-130. [DOI] [PubMed] [Google Scholar]
- 7.Dolan, S. A., J. L. Proctor, D. W. Alling, Y. Okubo, T. E. Wellems, and L. H. Miller. 1994. Glycophorin B as an EBA-175 independent Plasmodium falciparum receptor of human erythrocytes. Mol. Biochem. Parasitol. 64:55-63. [DOI] [PubMed] [Google Scholar]
- 8.Duraisingh, M. T., A. G. Maier, T. Triglia, and A. F. Cowman. 2003. Erythrocyte-binding antigen 175 mediates invasion in Plasmodium falciparum utilizing sialic acid-dependent and -independent pathways. Proc. Natl. Acad. Sci. USA 100:4796-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duraisingh, M. T., T. Triglia, S. A. Ralph, J. C. Rayner, J. W. Barnwell, G. I. McFadden, and A. F. Cowman. 2003. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 22:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galinski, M. R., C. C. Medina, P. Ingravallo, and J. W. Barnwell. 1992. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell 69:1213-1226. [DOI] [PubMed] [Google Scholar]
- 11.Gilberger, T. W., J. K. Thompson, T. Triglia, R. T. Good, M. T. Duraisingh, and A. F. Cowman. 2003. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J. Biol. Chem. 278:14480-14486. [DOI] [PubMed] [Google Scholar]
- 12.Hadley, T. J., F. W. Klotz, G. Pasvol, J. D. Haynes, and M. H. McGinniss. 1987. Falciparum malaria parasites invade erythrocytes that lack glycophorin A and B (MkMk). Strain differences indicate receptor heterogeneity and two pathways for invasion. J. Clin. Investig. 80:1190-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko, O., J. Mu, T. Tsuboi, X. Su, and M. Torii. 2002. Gene structure and expression of a Plasmodium falciparum 220-kDa protein homologous to the Plasmodium vivax reticulocyte binding proteins. Mol. Biochem. Parasitol. 121:275-278. [DOI] [PubMed] [Google Scholar]
- 14.Khan, S. M., W. Jarra, H. Bayele, and P. R. Preiser. 2001. Distribution and characterisation of the 235 kDa rhoptry multigene family within the genomes of virulent and avirulent lines of Plasmodium yoelii. Mol. Biochem. Parasitol. 114:197-208. [DOI] [PubMed] [Google Scholar]
- 15.Lobo, C. A., K. de Frazao, M. Rodriguez, M. Reid, M. Zalis, and S. Lustigman. 2004. Invasion profiles of Brazilian field isolates of Plasmodium falciparum: phenotypic and genotypic analyses. Infect. Immun. 72:5886-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobo, C. A., M. Rodriguez, M. Reid, and S. Lustigman. 2003. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl). Blood 101:4628-4631. [DOI] [PubMed] [Google Scholar]
- 17.Lobo, C. A., M. Rodriguez, C. J. Struchiner, M. G. Zalis, and S. Lustigman. 2006. Associations between defined polymorphic variants in the PfRH ligand family and the invasion pathways used by P. falciparum field isolates from Brazil. Mol. Biochem. Parasitol. 149:246-251. [DOI] [PubMed] [Google Scholar]
- 18.Maier, A. G., M. T. Duraisingh, J. C. Reeder, S. S. Patel, J. W. Kazura, P. A. Zimmerman, and A. F. Cowman. 2003. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 9:87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell, G. H., T. J. Hadley, M. H. McGinniss, F. W. Klotz, and L. H. Miller. 1986. Invasion of erythrocytes by Plasmodium falciparum malaria parasites: evidence for receptor heterogeneity and two receptors. Blood 67:1519-1521. [PubMed] [Google Scholar]
- 20.Nery, S., A. M. Deans, M. Mosobo, K. Marsh, J. A. Rowe, and D. J. Conway. 2006. Expression of Plasmodium falciparum genes involved in erythrocyte invasion varies among isolates cultured directly from patients. Mol. Biochem. Parasitol. 149:208-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okoyeh, J. N., C. R. Pillai, and C. E. Chitnis. 1999. Plasmodium falciparum field isolates commonly use erythrocyte invasion pathways that are independent of sialic acid residues of glycophorin A. Infect. Immun. 67:5784-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins, M. E., and E. H. Holt. 1988. Erythrocyte receptor recognition varies in Plasmodium falciparum isolates. Mol. Biochem. Parasitol. 27:23-34. [DOI] [PubMed] [Google Scholar]
- 23.Preiser, P. R., W. Jarra, T. Capiod, and G. Snounou. 1999. A rhoptry-protein-associated mechanism of clonal phenotypic variation in rodent malaria. Nature 398:618-622. [DOI] [PubMed] [Google Scholar]
- 24.Rayner, J. C., M. R. Galinski, P. Ingravallo, and J. W. Barnwell. 2000. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc. Natl. Acad. Sci. USA 97:9648-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rayner, J. C., T. M. Tran, V. Corredor, C. S. Huber, J. W. Barnwell, and M. R. Galinski. 2005. Dramatic difference in diversity between Plasmodium falciparum and Plasmodium vivax reticulocyte binding-like genes. Am. J. Trop. Med. Hyg. 72:666-674. [PubMed] [Google Scholar]
- 26.Rayner, J. C., E. Vargas-Serrato, C. S. Huber, M. R. Galinski, and J. W. Barnwell. 2001. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J. Exp. Med. 194:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sim, B. K. L., C. E. Chitnis, K. Wasniowska, T. J. Hadley, and L. H. Miller. 1994. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264:1941-1944. [DOI] [PubMed] [Google Scholar]
- 28.Stubbs, J., K. M. Simpson, T. Triglia, D. Plouffe, C. J. Tonkin, M. T. Duraisingh, A. G. Maier, E. A. Winzeler, and A. F. Cowman. 2005. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science 309:1384-1387. [DOI] [PubMed] [Google Scholar]
- 29.Taylor, H. M., M. Grainger, and A. A. Holder. 2002. Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect. Immun. 70:5779-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor, H. M., T. Triglia, J. Thompson, M. Sajid, R. Fowler, M. E. Wickham, A. F. Cowman, and A. A. Holder. 2001. Plasmodium falciparum homologue of the genes for Plasmodium vivax and Plasmodium yoelii adhesive proteins, which is transcribed but not translated. Infect. Immun. 69:3635-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Triglia, T., M. T. Duraisingh, R. T. Good, and A. F. Cowman. 2005. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol. Microbiol. 55:162-174. [DOI] [PubMed] [Google Scholar]
- 32.Triglia, T., J. Thompson, S. R. Caruana, M. Delorenzi, T. Speed, and A. F. Cowman. 2001. Identification of proteins from Plasmodium falciparum that are homologous to reticulocyte binding proteins in Plasmodium vivax. Infect. Immun. 69:1084-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viriyakosol, S., N. Siripoon, X. P. Zhu, W. Jarra, A. Seugorn, K. N. Brown, and G. Snounou. 1994. Plasmodium falciparum: selective growth of subpopulations from field samples following in vitro culture, as detected by the polymerase chain reaction. Exp. Parasitol. 79:517-525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.