The National Institute of Allergy and Infectious Diseases (NIAID) began a new bioinformatic venture in July 2004 intended to integrate the vast amount of genomic and other biological data that are both available and being produced by the rapid increase in biodefense research. Eight Bioinformatics Resource Centers for Biodefense and Emerging/Re-Emerging Infectious Disease (BRCs) were funded to provide the research community working on a variety of pathogens access to integrated genomic data to aid in the discovery and development of innovative therapeutics, vaccines, and diagnostics for these pathogenic organisms. The initial term of this effort is 5 years, and overall nearly $100 million dollars was awarded, facilitating the creation of what is likely to be the foremost bioinformatic effort to date dedicated to human pathogens.
Pathogenic species of biodefense and special public health interest have been classified by NIAID into three high-priority categories based on their relative capabilities for causing morbidity or mortality from disease in case of biowarfare (categories A, B, and C [http://www3.niaid.nih.gov/Biodefense/bandc_priority.htm]). The phylogenic domains contained in those categories not only encompass a large variety of microbial species and viral families but also embrace a small group of eukaryotic unicellular pathogens (Giardia and Entamoeba), protozoa such as Plasmodium and Toxoplasma, a group of fungal species (Microsporidia), and a plant (Ricinus communis, or castor bean). Invertebrate vectors of human pathogens are also included in the scope of the BRCs, such as Aedes aegypti, which is a vector for pathogen-caused diseases such as West Nile encephalitis, dengue hemorrhagic fever, and filarial diseases such as elephantiasis.
Among the chief goals of the BRCs is to offer users easy Web access and graphical user interfaces as well as other types of software interfaces (such as application programming interfaces or Web services) to pathogens' genomic and related data that are stored in a relational database management system, such as Oracle, Sybase, or MySQL. Such data are to include the genome sequences of multiple strains of these organisms and related plasmids, protein sequences, annotations, single nucleotide polymorphisms (SNPs), microarray data, epitope data, proteomic data, and epidemiological data, i.e., as much data as possible to facilitate comparative genomics. Eventually, the BRCs will evolve to contain data relevant to host-pathogen interactions. The variety of data types supported by the BRCs is dependent on several factors, including the abundance of data available either in public databases or in published literature, specific requests from the scientific community for the pathogens being supported, and ultimately the expertise of the BRCs' staff members. The eight awards are shown in Table 1.
TABLE 1.
NIAID BRCs
| BRC name (URL), principal investigator (institution) | Organisms assigned (no. of genomes in the BRC as of March 2007)a | Institutions |
|---|---|---|
| ApiDB (http://www.apidb.org), David Roos (University of Pennsylvania) | Apicomplexan species, including Toxoplasma gondii (1), Cryptosporidium spp. (2), Theileria spp. (2), and Plasmodium spp. (8) | University of Pennsylvania, University of Georgia |
| BioHealthBase (http://www.biohealthbase.org), Richard Scheuermann (University of Texas Southwestern Medical Center) | Giardia lamblia (*), Mycobacterium spp. (10), influenza virus (10,880), Francisella tularensis (5), Encephalitozoon cuniculi (1), and Ricinus communis (*) | Northrop Grumman Information Technology, University of Texas Southwestern Medical Center, Vecna Technologies, Amar International |
| ERIC (http://www.ericbrc.org), John Greene (SRA International, Inc.) | Diarrheagenic Escherichia coli (10), Shigella spp. (9), Salmonella spp. (6), Yersinia enterocolitica (0), and Yersinia pestis (7) | SRA International, Inc., University of Wisconsin—Madison |
| NMPDR (http://www.nmpdr.org), Rick Stevens (University of Chicago) | Staphylococcus aureus (11), pathogenic Vibrio spp. (11), Listeria monocytogenes (15), Campylobacter jejuni (10), Streptococcus pyogenes (12), and Streptococcus pneumoniae (2) | University of Chicago; FIG; University of Illinois, Urbana-Champaign |
| Pathema (http://pathema.tigr.org), Owen White (TIGR) | Bacillus anthracis (10), Clostridium botulinum (1), Burkholderia mallei (7), Burkholderia pseudomallei (7), Clostridium perfringens (3), and Entamoeba histolytica (1) | TIGR, University of Maryland |
| PATRIC (http://patric.vbi.vt.edu), Bruno Sobral (VBI) | Brucella sp. (4), Coxiella burnetii (4), Rickettsia sp. (10), caliciviruses (77), coronaviruses (217), hepatitis A virus (47), hepatitis E virus (65), and lyssaviruses (15) | VBI, Loyola University Medical Center, Social and Scientific Systems, University of Maryland, Swiss Institute of Bioinformatics |
| VBRC (http://www.vbrc.org), Elliott Lefkowitz (University of Alabama—Birmingham) | Viral pathogens belonging to the following families: Arenaviridae (106), Bunyaviridae (857), Filoviridae (27), Flaviviridae (498), Paramyxoviridae (169), Poxviridae (106), Togaviridae (83), and hepatitis C virus (*) | University of Alabama—Birmingham; University of Victoria, British Columbia, Canada; Columbia University |
| VectorBase (Invertebrate Vectors of Human Pathogens [http://www.vectorbase.org]), Frank Collins (University of Notre Dame) | Anopheles gambiae (1), Aedes aegypti (1), Culex pipiens (1), Pediculus humanus (1), and Ixodes scapularis (1) | University of Notre Dame; European Bioinformatics Institute; Imperial College of London; Institute of Molecular Biology and Biotechnology, Crete; Harvard University; Purdue University; University of California—Riverside |
*, will soon be available.
It is important that although the BRCs' focus is mostly on genomic information about organisms that could be manipulated and used as biological weapons, all data produced by the BRCs are freely and publicly available to the research community. In addition, the BRC program has a policy of publicly releasing any type of new data placed in the BRCs within 6 to 12 months from data deposition, and immediately upon publication. Any data supplied to the BRCs are to be credited to their source, and genome annotations should have evidence codes signifying whether the annotations were produced experimentally, computationally, or by other means. Standard operating procedures will soon be released to describe how annotations were generated and to share the process with the scientific community. The BRCs were required, in their proposals, to design robust, flexible, and scalable computational systems and infrastructure in support of the upcoming deluge of genomic data for the targeted species. All software developed from this program is to be made freely available as open source software.
A major focus is outreach to the research community both to establish and then serve its needs and to make its members aware of the significant resources provided by these new centers. Examples of outreach activities performed by the BRCs include training on the use of the bioinformatic tools and databases, booths and live demos at scientific meetings, and hands-on workshops or training sessions provided free of charge to groups of researchers. All of the BRCs have presented posters and talks at many conferences on infectious diseases, specific organisms, bioinformatics, and biodefense.
In addition to these direct interactions with the research community, each BRC has a scientific working group (SWG) of about 10 experts whose expertise ranges over the biology of that BRC's pathogens, bioinformatics, evolutionary genomics, biodefense, and infectious disease. SWGs meet one or two times a year to advise and provide feedback to each BRC on data sources, useful analysis tools, and user interfaces. Members of the SWGs are also involved in outreach activities. Altogether, the BRC program is advised by approximately 80 prominent investigators who help to provide vision for the BRCs, develop community ties, and embody the needs of the community of researchers working on each of these pathogens, ensuring that the BRCs are properly responding to those needs.
INTEROPERABILITY AND BRC CENTRAL
A great deal of emphasis is being placed on interoperability and establishing collaborations among the eight BRCs. A portal site, BRC Central (http://www.brc-central.org/), links to all eight websites for the awarded BRCs. BRC Central contains information required from each BRC about common data types, conferences and meetings to be attended by BRC representatives, notices of upcoming training on BRC tools, and publications by the BRCs and has a comprehensive list of released, downloadable software tools of various types, including tools for comparative genomics, computational annotation pipelines, annotation systems, sequence alignments and alignment editors, and genome viewers. BRC Central also provides search capabilities across all the BRCs, including searches for keywords, gene attributes, and organism names and sequence BLAST searches against a data set of protein sequences or contigs of the organisms supported by the BRCs. The website also permits downloads of flat files of sequences and annotation data from all the BRCs in the generic feature format, version 3 (GFF3) file format (http://www.sequenceontology.org/gff3.shtml).
The GFF3 file format was adopted to provide consistency in genome feature information. The full specifications for the BRC GFF3 file format are available at iowg.brcdevel.org/gff3.html, including the list of feature types and attribute tags. This document also describes a convention for storing genome, organism, and molecule descriptions in GFF3. The usage conventions are meant to be wholly compatible with the GFF3 specifications while preventing ambiguities between GFF3 codes from different centers. To ensure that the GFF3 conventions described in the document have been implemented properly, a GFF3 validator script has been written and provided to the BRCs. In general, Web services will evolve for each BRC to allow use of each other's analysis tools and to facilitate data exchange.
Among other program-wide activities, the BRCs recently met to discuss common quantitative measures of the quality and added value of the data provided by each BRC, focusing on annotation, literature curation, integration of a variety of data types, and service to the user community. Standard operating procedures for annotation are to be posted to the websites, as are metrics for comparing annotation consistency and quality.
GENOME ANNOTATION AND COMPARATIVE GENOMICS
In response to the growing need from the community to have well-curated gene annotation data with frequent updates, most of the BRCs have full-time curators, whose number ranges from two to six. In addition to gene annotation of relevant genomic regions, such as pathogenicity islands or genes involved in virus replication in the host, curators are responsible for following the literature on the pathogens, for development of an annotation standard operating procedure, and for collaboration with experts on these pathogenic organisms to solicit community annotation. The Enteropathogen Resource Integration Center (ERIC) is focusing its annotation on genes involved in pathogenicity and on genes determined by computational analysis to be unique to each of the individual enteropathogens assigned. The National Microbial Pathogen Data Resource Center (NMPDR) is focusing its annotation efforts on vertical integration across genomes that share proteins which perform the same biological functions. Pathema has released resource guide pages for the category A organisms Clostridium botulinum (neurotoxins) and Bacillus anthracis (anthrax). Each guide contains curated links to resource providers and primary data organized into topic-oriented Web pages, which include sequence, strain, structure, antibody, therapeutic, vaccine, inhibitor, protocol, reference, and related links.
Almost all of the species in the NIAID high-priority lists have at least one genome already sequenced and publicly available with annotation; however, tens of strains of these species are currently being sequenced by either NIAID-funded sequencing centers (see the microbial sequencing center effort described below) or other sequencing sources and will become available in the next 1 to 3 years. The amount of genome sequence data that will be generated and made publicly available by the largest sequencing centers is such that no large group of human curators will be able to perform a systematic annotation revision of each sequenced genome. Therefore, to ensure high-quality, efficient, and accurate annotation, especially across multiple strains of a single species, it is essential to rely on computational annotation pipelines that take full advantage of comparative genomic tools for ortholog identification and to identify genome rearrangements or lateral gene transfer that may be indicative of similar mechanisms of pathogenesis across species.
Each BRC is adopting its own approaches for dealing with the upcoming explosion in the number of sequenced microbial genomes. For example, NMPDR has established a system that allows comparative analysis of pathogen genomes in the context of more than 450 complete or draft genomes. Tools support the study of chromosomal clusters of synteny that are indicative of related function. NMPDR also presents subsystems for the comparative analysis of orthologous and analogous proteins. Subsystems are two-dimensional integrations of biological functions with genome sequences, which are represented in tables as columns of functional roles, rows of genomes, and cells populated by the genes responsible for each function.
Other BRCs are leveraging orthology information to create hidden Markov model (HMM) representations of gene families across a variety of species, in particular those families that are important for pathogenicity, as well as to identify genes that are unique to each pathogen. For example, ERIC has developed the EnteroFams, which are HMMs of essentially identical gene families across all enteropathogens available to date. Similarly, NMPDR is working with HMMs of the genomes in the related Fellowship for the Interpretation of Genomes (FIG) project to create FIGFams for the prediction of orthology for its assigned organisms. Both EnteroFams and FIGFams will be released publicly once they are fully validated. Other methods for determining orthology are also being used; the Apicomplexan Database (ApiDB) makes use of the OrthoMCL tool (1) to predict orthologues, using an all-against-all BLASTP approach followed by the identification of orthologous clusters of proteins by the Markov cluster algorithm.
RELATIONSHIPS WITH OTHER NIAID BIODEFENSE EFFORTS
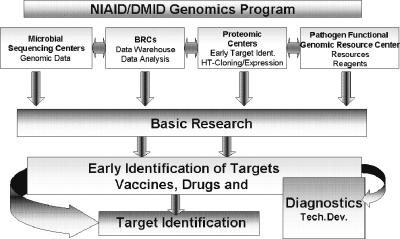
The BRC program is a component of a larger NIAID Genomics Program, which is responsible for the establishment of resources for the scientific community conducting basic and applied research to develop vaccines, diagnostics, and therapeutics (Fig. 1), using information derived from genome sequencing, proteomic, and functional genomic studies of the category A to C pathogens. Below are presented some of the genomic and other NIAID initiatives that are more closely relevant to the BRC program; more information about the NIAID Genomics Programs is available at http://www3.niaid.nih.gov/research/topics/pathogen/default.htm.
FIG. 1.
The Genomics Program of the Division of Microbiology and Infectious Diseases at NIAID, National Institutes of Health, has established a number of publicly accessible resources, which focus on microbial genome sequencing (two MSCs), functional genomics (a Pathogen Functional Genomics Resource Center), proteomics (seven PRCs), and bioinformatics (eight BRCs), to support researchers conducting biodefense basic and applied research to develop vaccines, diagnostics, and therapeutics.
The NIAID Microbial Sequencing Centers (MSCs; at The Institute for Genomic Research [TIGR] and the Broad Institute [http://www.niaid.nih.gov/dmid/genomes/mscs/default.htm]) provide rapid and cost-efficient resources for producing high-quality genome sequences of pathogens and invertebrate vectors of infectious diseases. The scientific community may request NIAID to sequence microorganisms by using a white-paper request process. Genomes that can be sequenced through these resources include microorganisms considered agents of bioterrorism, clinical isolates, closely related species, invertebrate vectors of disease, and microorganisms responsible for emerging and reemerging infectious diseases. It is a policy of the MSCs to deposit into GenBank the sequenced genomes and their first-pass computational annotations within 45 days of their availability. The BRCs then provide added value to the annotation generated by the MSCs, such as manual curation of the genomic features and of the relevant pathogen literature, comparative genomic analysis, sequence polymorphism identification, integration with other existing pathogen data sources, and other types of bioinformatic services. Hence, by serving as a companion initiative to the MSCs, the BRCs are intended to provide needed maintenance and further analysis of the genome sequence data and their annotation after the submission of that information by the MSCs to GenBank.
The seven Biodefense Proteomics Research Centers (PRCs) are another component of the NIAID Genomics Program, with the goal of discovering and validating potential candidate targets for the next generation of vaccines, therapeutics, and diagnostics by using proteomic technologies, such as yeast two-hybrid interactions, mass spectrometry, protein arrays, and other advanced proteomic technologies. In addition, this program has the goals of characterizing the proteomes of the NIAID category A to C high-priority pathogens and/or host cells to identify proteins associated with the biology of the microbes; elucidating mechanisms of microbial pathogenesis, and further understanding innate and adaptive immune responses to infectious agents and non-immunity-mediated host responses that contribute to microbial pathogenesis. In their effort to discover and validate targets, the PRCs will generate protein expression clones, protein-protein interaction data, protein expression data, and protein three-dimensional structure data, which will first be deposited into a database provided by the Administrative Resource Center for the PRCs (http://www.proteomicsresource.org). The BRCs will be able to use that data to improve the genome annotation quality and to integrate the proteomic and genomic data sets to better understand the biology of the pathogens. For example, in collaboration with the Pacific Northwest National Laboratory, ERIC will provide its users with links from ERIC's gene annotations to tables summarizing the state of the Salmonella enterica serovar Typhimurium proteome under different growth conditions. The presence of proteins in various subcellular fractions was assessed by mass spectrometry. Summaries will be presented within the ERIC site, with the ability to link to more detailed analysis at the Administrative Resource Center and the Pacific Northwest National Laboratory.
Several of the BRCs are collaborating with the NIAID Immune Epitope Database and Analysis Resource (IEDB; http://www.immuneepitope.org) (9) to link to their data on immunological epitopes important for virulence, pathogenesis, and host-pathogen interactions as well as to integrate their tools for predicting epitopes from protein sequences. The IEDB contains antibody and T-cell epitope data curated from scientific literature or submitted from the community, including data derived from experimentally validated epitopes generated by 14 NIAID Large-Scale Epitope Discovery efforts. The BioHealthBase BRC has used IEDB to integrate information from experimentally determined and computationally predicted influenza virus epitopes with genomic sequence and sequence polymorphism data. Pathema has contributed to the IEDB data by providing IEDB with epitope information extracted from the literature for the C. botulinum toxin.
It should be noted that the BRCs have not been funded to generate experimental data but instead to collect and integrate data produced by experimental laboratories external to the BRC network. Therefore, the success of the BRC program depends heavily on the public availability of such data, preferably in a computer parsable format, or in collaborations the BRCs may establish with such laboratories, for which they may provide bioinformatic and analysis services in exchange for data. In the future, the BRCs anticipate working with and supporting other NIAID efforts, including the Biodefense Regional Centers of Excellence (http://www.rcebiodefense.org/rce6/rce6pub.htm), whose areas of research and development include pathogenesis of microorganisms, immunology and vaccine development, drug development, and new methods for diagnosis and detection of infectious disease.
SURVEY OF BRCs
Although the eight BRCs have common contract terms and goals, they have taken varied approaches to reaching these goals—with different data, analysis tools, and focuses. For example, BioHealthBase has a high level of diversity in the type of organisms under study, whereas ERIC is focused on closely related enteropathogens. Pathema and NMPDR support specific data types for numerous microbial genomes. Below, we present a brief summary of each BRC, in alphabetical order. The list of coinvestigators on each BRC is also provided to acknowledge the valuable contributions of key individuals to the project.
ApiDB.
ApiDB (http://www.apidb.org) is an integrated BRC for protozoan pathogens (Table 1) in the phylum Apicomplexa, including the category B agents Cryptosporidium parvum and Toxoplasma gondii, and multiple (re)emerging infectious disease species within the phylum Plasmodium (the causative agents of malaria). A federation of three databases (PlasmoDB, CryptoDB, and ToxoDB) in ApiDB permits users to simultaneously query across all three.
The Genomics Unified Schema relational database system (http://www.gusdb.org) has enabled the integration of many differing data sets, including genomic sequences for multiple species (and SNPs identified through resequencing), synteny maps, curated annotations and comments from the user community, automated analysis of gene/protein attributes, crystal structures/models, interactome data, ortholog/paralog information, and functional genomic data sets (from both expression profiling and proteomic studies, on several platforms). Tools available through ApiDB include an integrated genome browser (GBrowse), BLAST queries, motif identification, ortholog identification, and metabolic pathway maps. Users may also exploit powerful tools to combine results by using Boolean operators to identify gene sets that exhibit very precise characteristics, and these results may be downloaded for further analysis and archiving.
The focus of the component databases is driven by community needs. Areas of particular interest currently include metabolic pathway modeling and reconstruction (CryptoDB), genetic diversity and identification of genes under positive or negative selection (PlasmoDB), and a new generation of photolithographic arrays suitable for both expression profiling and genotyping (ToxoDB).
The core client communities for these resources are relatively small and well integrated, and the active involvement of ApiDB staff with these communities (as independent researchers and through publications, meeting presentations, workshops, etc.) has resulted in daily use of ApiDB and its component databases by a majority of the ∼1,000 research labs studying these important organisms worldwide.
David Roos of the University of Pennsylvania is the principal investigator (PI) for ApiDB; the coinvestigators are Christian J. Stoeckert of the University of Pennsylvania and Jessica C. Kissinger of the University of Georgia.
BioHealthBase.
The Biodefense/Public Health Database (BioHealthBase; http://www.biohealthbase.org) is an integrated data repository and analysis portal supporting data on a broad range of pathogenic organisms, including Francisella tularensis, influenza virus, Mycobacterium tuberculosis, Microsporidia, Giardia lamblia, and Ricinus communis (Table 1).
BioHealthBase contains extensive genome sequence and protein function data for its designated pathogens. BioHealthBase aims to go beyond the traditional genomic and proteomic levels of experimental data to integrate data from a variety of different experimental technologies and to capture the interplay between pathogenic organisms and the human host. Building upon previous work of the biological and bioinformatic community has allowed BioHealthBase to quickly create a single cohesive resource for this wide variety of data to elucidate host-pathogen interactions.
One area of integration has been to combine information regarding the localization of immunological epitopes with sequence polymorphism analyses for influenza virus. Predicted major histocompatibility complex class I cytotoxic T-cell epitopes and validated immunological epitopes from the NIAID Immune Epitope Database for influenza virus are localized together with sequence conservation scores to indicate genomic regions under host selective pressure and protein motifs that can serve as prospective vaccine candidates. Future work will layer these data onto three-dimensional protein structure data to support an understanding of the relationships between protein structure, function, and host interactions.
Other collaborative efforts have been to initiate and expand annotations of metabolic and cellular signal transduction pathways for these pathogens and their human hosts. For example, the entire influenza virus life cycle framework and the host response pathways of Toll-like receptor 3 and RIG-I have been modeled and added to the Reactome database (6; http://www.reactome.org). Additionally, the pathways for transport reactions related to Mycobacterium tuberculosis H37Rv physiology have been added to the BioCyc pathway database (7; http://www.biocyc.org).
Richard Scheuermann of the University of Texas Southwestern Medical Center is the PI of BioHealthBase; the coinvestigators include Adolfo Garcia-Sastre of Mount Sinai School of Medicine, Stephen Johnston of Arizona State University, Barbara Mann of the University of Virginia, Hilary Morrison of Marine Biological Laboratory, Louis Weiss of Albert Einstein College of Medicine, and Ellen Vitetta of the University of Texas Southwestern Medical Center.
ERIC.
ERIC focuses on four enteropathogens and the closely related organism Yersinia pestis (Table 1), with a meticulous, disciplined focus on the annotation and curation of genes and gene families (http://www.ericbrc.org).
ERIC's Web portal currently centers around its pathogen annotation system, named ASAP (3), originally developed in the laboratory of coinvestigator Nicole Perna at the University of Wisconsin—Madison. This system's functions allow versioning of genomes, careful annotation of genes and other genome features (e.g., insertion elements) with evidence codes, using six annotators/curators, and the ability to allow direct community annotation.
In addition, the system makes integrated use of the MAUVE application for comparative genomics (2), which unlike most such applications, is not a pairwise genome aligner but is capable of aligning multiple genomes simultaneously and displaying chromosomal rearrangement, deletions, and insertions. One can zoom into the actual sequence, allowing MAUVE to also be of value in SNP location and prediction.
Other components to add value include GBrowse, for genome viewing with a direct link to ERIC's gene annotations, and a microarray database analysis system, originated for the National Cancer Institute's intramural program, to handle sophisticated analysis of microarray data. Work under development includes the integration of these resources to ask queries across these data types as well as advanced text mining of the literature, using SRA International, Inc.'s powerful NetOwl suite of tools, which promise to be able to extract heretofore difficult-to-find semantic relationships from the literature for the BRC Program.
ERIC is now providing training seminars on the use of the annotation system, which will be expanded over time to include the use of other tools, such as the mAdb microarray application. John Greene of SRA International, Inc., is the PI for ERIC; the coinvestigators are Nicole Perna and Frederick Blattner, both of the University of Wisconsin—Madison.
NMPDR.
NMPDR (http://www.nmpdr.org) contains the complete genomes of nearly 50 strains of pathogenic bacteria that are the focus of its annotators, as well as more than 400 (as of February 2007) other genomes that provide a broad context for comparative analysis. The NMPDR focus organisms are food-borne or nosocomial bacterial pathogens in NIAID category B (Table 1). NMPDR is both a central repository for a wide variety of scientific data on its core pathogens and a platform for software tools that support investigator-driven data analysis. Data resources include essential genes and candidate drug targets, which are provided for exploration and comparative analysis with tools such as Compare Regions and Functional Clusters.
The Signature Genes tool compares user-selected organisms to find the proteins in common to one group or those proteins that distinguish one group of organisms from another, e.g., virulent and avirulent strains. NMPDR integrates complete, public genomes with expertly curated biological subsystems to provide consistent functional annotations. Subsystems are defined here as sets of functional roles related by any biologically meaningful organizing principle which are vertically curated across microbial genomes. Investigators can browse subsystems and reactions to develop accurate, detailed reconstructions of networks involved in metabolism or pathogenesis.
The Drug Targets project represents the first step toward providing the user community with a comprehensive selection of potential targets for therapeutic intervention, including vaccines and antitoxins. The candidate targets have been determined experimentally to be either essential factors in virulence or involved in antibiotic resistance or sensitivity. Candidates have a close bacterial ortholog with an experimentally determined structure and no close ortholog in humans. Selected candidates will be used for in silico screening against libraries of small molecules, using computational docking methods. NMPDR will provide the results of in silico screening against both broad- and narrow-spectrum targets on its website as screening progresses. All candidates, not only those selected for in silico analysis, are presented at the NMPDR website, with links to physical and kinetic data useful for designing in vitro screening protocols.
NMPDR's training and outreach effort includes presentations at scientific meetings and training sessions both at the University of Illinois and at other institutions, upon request. NMPDR trainers also present lessons or lectures in undergraduate courses in microbiology at the University of Illinois at Urbana, and teaching materials are available online.
Rick Stevens of the University of Chicago is the PI of NMPDR; the coinvestigators include Ross Overbeek of FIG and Leslie McNeil of the National Center for Supercomputing Applications at the University of Illinois, Urbana-Champaign.
Pathema.
Pathema, developed at TIGR, is a website (http://pathema.tigr.org) composed of multiple databases and other computer resources that are meant to serve as an online focal point for the biodefense community (Table 1).
As mentioned earlier, Pathema has developed and released resource guide pages for Pathema's category A organisms, namely, the botulinum neurotoxin and Bacillus anthracis (anthrax) resource guides. Each guide contains 10 pages containing curated links to resource providers and primary data organized into topic-oriented Web pages, which include sequence, strain, structure, antibody, therapeutic, vaccine, inhibitor, protocol, reference, and related links. The C. botulinum guide has been reviewed by a number of botulinum researchers and will continue to be developed in close collaboration with the scientific community. In addition to updating existing pages and tables, Pathema investigators will continue to curate pathogenicity and virulence genes and proteins, identify toxigenic/nontoxigenic and capsulated/unencapsulated strains from the literature, provide reviews of diagnostic methods, and offer links to detection systems. Future work will include the development of similar resource guides for Pathema's category B organisms.
The Genome Properties system (4) is a comparative genomic system which incorporates both computer-generated and human-curated assertions of biological processes and properties of sequenced genomes. These genome properties are defined such that assertions or calculations made across many genomes are as standardized as possible, using controlled vocabularies or numerical values with controlled units. Many genome properties represent metabolic pathways and other biological systems; others define genome metadata, including the presence and type of flagella, pili, or capsule as well as the cell shape of the organism. The Pathema interface displays each property and enables one to find correlations between genomes based on manually annotated properties (e.g., optimal growth temperature, oxygen requirement, or human pathogen), taxonomic classifications, calculated values (e.g., GC content or average protein length), and the presence or absence of complete or partial metabolic pathways.
Pathema offers a service which will assemble the genomes of any category A to C organism and deposit a computer representation of that assembly into the GenBank Assembly Archive (10). Pathema provides this service for any genome for which sequence trace files and quality values are available; 24 genomes have been submitted to date. In all cases where SNPs or insertions-deletions were produced as a result of reassembly, these changes are faithfully reflected in the sequence found in the annotation division of GenBank.
Several SNP data sets were generated using a custom SNP clustering pipeline for the following groups of organisms: Burkholderia mallei, Burkholderia pseudomallei, Listeria monocytogenes, and B. anthracis. Users can view overall summaries, SNP positions for an individual organism, comparisons between strains, and phylogenetic trees based on SNP information.
TIGR offers a 3-day course in prokaryotic annotation and analysis to train investigators on how to evaluate the myriad microbial database resources and software tools that are available. The course familiarizes users with TIGR's prokaryotic annotation tools and the analysis of the prokaryotic data in Pathema. The Pathema website also displays extensive documentation describing manual annotation processes.
Owen White of TIGR is the PI for Pathema; the coinvestigator is Steven Salzberg of the University of Maryland, College Park.
PATRIC.
The PathoSystems Resource Integration Center (PATRIC; http://patric.vbi.vt.edu) integrates genomic and associated data types for three genera of proteobacteria and five single-stranded RNA viruses (Table 1).
PATRIC aims to provide a standardized curation for each pathosystem's genomes. Curation is based on existing annotations and the output of PATRIC's Genome Sequence Annotation Pipeline and Protein Annotation Pipeline (5). Importantly, PATRIC works with organism experts to ensure that curation efforts meet the expectations of the user base. These experts guide the selection of the reference genomes that receive thorough curation. Gene annotations are then propagated from reference genomes to orthologs in associated genomes to facilitate consistency of annotation between genomes.
Integration of gene expression and proteomic data types is another of PATRIC's fundamental goals that is currently in development. The data model being developed to link postsequence data generated by individual laboratories and NIAID programs, such as the PRCs, will be important for the BRC mission.
Close collaboration with organism communities helps the PATRIC project to understand how best to utilize PATRIC data. Bioinformatic research projects have been undertaken with members of the research community to identify gene sets of high importance for countermeasure development that may contain useful vaccine targets, to identify genes missed by previous annotation efforts, and to design pan-lyssavirus primer pools for virus identification and genotyping. Such gene sets are the focus of targeted curation efforts. Through this collaborative model, tools and workflows are being developed to help investigators put data to use.
Bruno Sobral of the Virginia Bioinformatics Institute (VBI) is the PI of PATRIC; the coinvestigators for PATRIC include Joao Setubal at VBI, Abdu Azad at University of Maryland, Baltimore County, and Susan Baker at the Loyola University Medical Center.
VBRC.
The Viral Bioinformatics Resource Center (VBRC) is focused on providing genomic data, analytical tools, and basic bioinformatic research focusing on pathogens that are members of viral taxonomic families (listed in Table 1). The VBRC (http://www.vbrc.org) is an extension of previous work to develop the Poxvirus Bioinformatics Resource Center (8; http://www.poxvirus.org).
The VBRC consists of a relational database, analytical tools, and Web interfaces that support the data storage, annotation, analysis, and information exchange goals of the BRC program. The database contains the complete genome sequences, along with comprehensive annotations of genes, for all of the human viral pathogens that are members of the VBRC taxonomic families. In addition, for comparative purposes, genomic information and annotations are included for all animal pathogens, as well as nonpathogenic viruses, that are also members of these viral families.
In addition to sequence data and computationally derived gene annotations, the VBRC provides literature-based manual curation for each of its viral genomes and gene records, resulting in a searchable, comprehensive minireview of gene function relating genotypes to biological phenotypes, with special emphasis on pathogenesis. The VBRC also includes a variety of analytical and visualization tools on its website to aid in the understanding of the available data, including tools for genome annotation, comparative analysis, whole-genome alignments, and phylogenetic analysis. Finally, an important aspect of the ongoing work is to solicit feedback from the scientific community, with the goals of enhancing and extending the VBRC, thereby making it both used and useful in support of basic and applied research on these viral pathogens.
Elliott Lefkowitz of the University of Alabama—Birmingham is the PI of VBRC; the coinvestigator for the project is Chris Upton at the University of Victoria, Canada.
VectorBase.
VectorBase (http://www.vectorbase.org) is responsible for annotating the genomes of a number of arthropod vectors of human pathogens. The provision of a predicted gene set with appropriate tools for interrogation and dissemination of these gene data are the primary goals. In terms of genome annotation, the VectorBase BRC has undertaken the reannotation of existing genomes for which it has assumed responsibility, as well as the annotation of new genomes in collaboration with the sequencing centers. Reannotation of Anopheles gambiae has improved the gene set by two approaches, namely, the manual annotation of chromosome 2L (one of the three chromosomes [about 40% of the genome]) and higher-quality automated prediction, with better discrimination of transposable elements, improved ab initio predictions, and noncoding RNA predictions.
The yellow fever vector mosquito Aedes aegypti genome annotation was recently incorporated into VectorBase. Aedes is estimated to have diverged from Anopheles approximately 150 million years ago, and comparative analysis between these genomes will improve predictions of further dipteran genomes, e.g., Culex pipiens quinquefasciatus.
The primary VectorBase Web presence consists of a genome browser powered by the Ensembl code base. This gives a rich, interlinked set of pages for genes and transcripts from which many annotations can be accessed. VectorBase provides orthologue assignments, Web links to the public sequence databases, microarray experiments, gene ontology terms, and protein domain features. The database can be interrogated by using a custom query tool or the BioMart system. Published microarray experiments are being incorporated into the database via the mapping of array probe sets onto the genomes of both mosquitoes. The array data are stored in a separate database (http://base.vectorbase.org) with Web links to the VectorBase gene pages and to features within the VectorBase genome browser.
Controlled vocabularies for mosquito anatomy have been developed (available through the Open Biomedical Ontologies website [http://obo.sourceforge.net]) and are used for annotating microarray experiments and for expanding the available gene ontology terms.
VectorBase has provided some training in the use of the Apollo annotation system to users who are actively involved in community-contributed manual annotation, and plans are under way to offer an expanded opportunity for both this training and more general training in the use of VectorBase.
Frank Collins of the University of Notre Dame is the PI of VectorBase; coinvestigators include William Gelbart of Harvard University; Ewan Birney of the European Bioinformatics Institute, United Kingdom; Kitsos Louis of the Institute of Molecular Biology and Biotechnology, Crete, Greece; and Fotis Kafatos of the Imperial College, United Kingdom.
Conclusions.
The NIAID-funded BRC program is tasked with supporting genomic and related data for a variety of different pathogenic organisms. The BRCs import data from existing repositories, generate related data types, store the data in databases, analyze them, and provide for investigator access via user interfaces.
As these BRCs mature, a significant challenge remaining will be to develop user interfaces not only for experts but also for relative novices. This will also impact training on these systems, as in addition to in-person workshops, manuals and online tutorials will need to be developed to reach the widest possible audience. For most of the BRCs, the basic infrastructure and analysis tools are in place, but there remains significant work to be done to enhance scientific usability and workflows through the systems.
Another challenge will be to develop easy-to-use, query-by-example methods to ask complex questions across genomic data types, integrating all this information. For example, one may ask the database to show all genes upregulated during infection as well as the annotations indicating which of these genes were already known to be involved in pathogenesis.
Perhaps most important for the success of the BRC program is the active involvement by the various components of the research community interested in these organisms, including pathogen specialists, biodefense researchers, evolutionary biologists, experts in one model gene family or organism, and genomic researchers. Each of these groups has different interests and needs, which the BRCs will endeavor to meet.
However, for the BRC program to succeed there is an absolute need for scientists from all of these communities to actively provide feedback, request refinements and enhancements, contribute data and annotations, and most importantly, use the valuable BRC resources relevant to their research. Such input and active use are the only way for these new, powerful bioinformatic resources for pathogens to develop to better serve the research communities, to ensure improved data curation in the future, and to fulfill their mission of facilitating the identification and refinement of molecular targets to develop vaccines, therapeutics, diagnostics, and countermeasures.
Acknowledgments
The Bioinformatics Resource Centers for Biodefense and Emerging/Re-emerging Infectious Disease are funded under R&D contracts from the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health. For financial disclosure purposes, J.M.G. is an employee and stockholder of SRA International, Inc.
We thank our coinvestigators and staffs for their hard work and express our gratitude to each other's teams for the selfless cooperation between the BRCs.
Editor: J. B. Kaper
Footnotes
Published ahead of print on 9 April 2007.
REFERENCES
- 1.Chen, F., A. J. Mackey, C. J. Stoeckert, Jr., and D. S. Roos. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34:D363-D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darling, A. C. E., R. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glasner, J. D., M. Rusch, P. Liss, G. Plunkett III, E. L. Cabot, A. Darling, B. D. Anderson, P. Infield-Harm, M. C. Gilson, and N. T. Perna. 2006. ASAP: a resource for annotating, curating, comparing, and disseminating genomic data. Nucleic Acids Res. 34:D41-D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haft, D. H., J. D. Selengut, L. M. Brinkac, N. Zafar, and O. White. 2005. Genome Properties: a system for the investigation of prokaryotic genetic content for microbiology, genome annotation, and comparative genomics. Bioinformatics 21:293-306. [DOI] [PubMed] [Google Scholar]
- 5.Hance, M. E., M. J. Czar, A. Azad, A. Purkayastha, E. E. Snyder, O. R. Crasta, J. C. Setubal, and B. W. Sobral. 2005. The pathogen resource integration center: implications for rickettsial research. Ann. N. Y. Acad. Sci. 1063:459-465. [DOI] [PubMed] [Google Scholar]
- 6.Joshi-Tope, G., M. Gillespie, I. Vastrik, P. D'Eustachio, E. Schmidt, B. de Bono, B. Jassal, G. R. Gopinath, G. R. Wu, L. Matthews, S. Lewis, E. Birney, and L. Stein. 2005. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 33:D428-D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karp, P. D., C. A. Ouzounis, C. Moore-Kochlacs, L. Goldovsky, P. Kaipa, D. Ahren, S. Tsoka, N. Darzentas, V. Kunin, and N. Lopez-Bigas. 2005. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 33:6083-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefkowitz, E. J., C. Upton, S. Changayil, C. Buck, P. Traktman, and R. M. Buller. 2005. Poxvirus Bioinformatics Resource Center: a comprehensive Poxviridae informational and analytical resource. Nucleic Acids Res. 33:D311-D316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters, B., J. Sidney, P. Bourne, H. H. Bui, S. Buus, G. Doh, W. Fleri, M. Kronenberg, R. Kubo, O. Lund, D. Nemazee, J. V. Ponomarenko, M. Sathiamurthy, S. Schoenberger, S. Stewart, P. Surko, S. Way, S. Wilson, and A. Sette. 2005. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 3:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzberg, S. L., D. Church, M. DiCuccio, E. Yaschenko, and J. Ostell. 2004. The genome Assembly Archive: a new public resource. PLoS Biol. 2:E285. [DOI] [PMC free article] [PubMed] [Google Scholar]