Abstract
Previously, we reported that the mouse intestine selected mutants of Escherichia coli MG1655 that have improved colonizing ability (M. P. Leatham et al., Infect. Immun. 73:8039-8049, 2005). These mutants grew 10 to 20% faster than their parent in mouse cecal mucus in vitro and 15 to 30% faster on several sugars found in the mouse intestine. The mutants were nonmotile and had deletions of various lengths beginning immediately downstream of an IS1 element located within the regulatory region of the flhDC operon, which encodes the master regulator of flagellum biosynthesis, FlhD4C2. Here we show that during intestinal colonization by wild-type E. coli strain MG1655, 45 to 50% of the cells became nonmotile by day 3 after feeding of the strain to mice and between 80 and 90% of the cells were nonmotile by day 15 after feeding. Ten nonmotile mutants isolated from mice were sequenced, and all were found to have flhDC deletions of various lengths. Despite this strong selection, 10 to 20% of the E. coli MG1655 cells remained motile over a 15-day period, suggesting that there is an as-yet-undefined intestinal niche in which motility is an advantage. The deletions appear to be selected in the intestine for two reasons. First, genes unrelated to motility that are normally either directly or indirectly repressed by FlhD4C2 but can contribute to maximum colonizing ability are released from repression. Second, energy normally used to synthesize flagella and turn the flagellar motor is redirected to growth.
Intestinal colonization is defined as the indefinite persistence of a bacterial population in the intestine of an animal in stable numbers without repeated introduction of the bacterium into the animal. Commensal Escherichia coli strains colonize the mouse intestine by growing in the intestinal mucus layer which covers the epithelium (18, 24, 25, 28, 38, 39, 43). The intestinal mucus layer is constantly being synthesized, degraded by indigenous bacteria, and sloughed into feces (15, 27). Colonization requires bacteria to penetrate the mucus layer, compete for nutrients with the indigenous flora, and divide at a rate that is at least equal to the washout rate caused by sloughing of the mucus layer into feces (7, 16); however, the role of motility in mucosal colonization is complex and appears to differ in different microorganisms (12).
Motility and chemotaxis in E. coli are controlled by a master regulator encoded by flhD and flhC, which comprise the flhDC operon. Together, FlhD and FlhC form the FlhD4C2 complex (20, 44), which activates transcription of class II flagellar genes that encode components of the flagellar basal body and export machinery (30). One of the class II flagellar genes, fliA, encodes an RNA polymerase sigma factor, σ28, which switches on expression of the class III genes coding for the cell-distal structural components of the flagellum (30). The sequenced E. coli MG1655 strain has an IS1 element in the regulatory region of the flhDC promoter (3), which increases the expression of the flhDC operon and makes this strain hypermotile compared to the isogenic strain in which this element is missing (2).
Recently, we showed that the mouse intestine selects nonmotile ΔflhDC mutants of E. coli MG1655 that originate immediately downstream of the IS1 element (17). These mutants have improved colonizing ability and grow 10 to 20% faster than their parent in mouse cecal mucus in vitro and 15 to 30% faster on a variety of sugars as sole carbon and energy sources in vitro (17). E. coli MG1655 flhDC operon deletion mutants might utilize carbon sources better than their parent as a result of release of repression of genes normally regulated by the FlhD4C2 regulatory complex (e.g., the complex is known to repress gltA [citrate synthase], sdhCDAB [succinate dehydrogenase], mdh [malate dehydrogenase], and mglBAC [galactose transport] [31, 32]) or as a result of increased energy that is available for other cellular processes in the absence of hyper-flagellum synthesis and rotation. In the present study, these possibilities were examined.
MATERIALS AND METHODS
Bacterial strains.
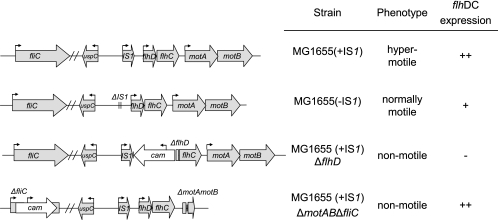
E. coli MG1655 Strr is a spontaneous streptomycin-resistant mutant of the sequenced wild-type E. coli MG1655 strain (CGSC 7740) (24). It has an IS1 element in the flhDC promoter (3). E. coli MG1655 Strr Nalr is a spontaneous nalidixic acid-resistant mutant of E. coli MG1655 Strr (24). It is referred to below as E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression). Its flhDC locus is illustrated in Fig. 1. E. coli MG1655 (CGSC 6300) was obtained from the E. coli Genetic Stock Center and is a wild-type strain that lacks the IS1 element in the flhDC promoter. E. coli MG1655 (−IS1) Strr Nalr, constructed as described below, is isogenic with MG1655 Strr except that it is nalidixic acid resistant and contains the flhDC promoter region of MG1655 (CGSC 6300). Because this strain is missing the IS1 element in the flhDC promoter, it is less motile than E. coli MG1655 (+IS1) (Fig. 2); it is referred to below as E. coli MG1655 (−IS1) (normally motile, normal flhDC expression). Its flhDC locus is illustrated in Fig. 1. E. coli MG1655 Strr ΔflhD::cam has a 546-bp deletion beginning immediately downstream of ISI in the regulatory region of flhD and continuing into flhD. The deletion inactivates the entire flhDC operon (17). E. coli MG1655 Strr ΔflhD::cam is streptomycin and chloramphenicol resistant and is referred to below as E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression). Its flhDC locus is illustrated in Fig. 1. E. coli MG1655 Strr ΔmotAB ΔfliC::cam was constructed from E. coli MG1655 Strr and has the IS1 element in the flhDC promoter. It is both streptomycin and chloramphenicol resistant, has a 1,711-bp deletion encompassing the motA and motB genes which inactivates the flagellar motor, and has a 583-bp deletion in fliC, preventing flagellin synthesis. It is referred to below as E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression) and was constructed as described below. Its flhDC, motAB, and fliC loci are illustrated in Fig. 1. E. coli MG1655 Strr Δedd::kan, referred to below as E. coli MG1655 Δedd, was constructed from E. coli MG1655 Strr (17). It grows poorly using gluconate as a sole carbon and energy source compared to the wild type (17), lacks 6-phosphogluconate dehydratase, and is both streptomycin and kanamycin resistant. E. coli MG1655 Strr Δedd*::kan, referred to below as E. coli MG1655 Δedd*, was isolated from the feces of a mouse 20 days after feeding of MG1655 Strr Δedd::kan (17). It has a 2,384-bp deletion beginning immediately downstream of IS1 in the regulatory region of flhD and continuing through flhD, flhC, and motA and into motB (17). E. coli MG1655 Δedd* grows better than E. coli MG1655 Δedd on gluconate and on a number of other sugars and is a better mouse intestine colonizer than E. coli MG1655 Δedd (17).
FIG. 1.
flhDC, motAB, and fliC loci of E. coli MG1655 (+IS1), E. coli MG1655 (−IS1), E. coli MG1655 (+IS1) ΔflhD, and E. coli MG1655 (+IS1) ΔmotAB ΔfliC. The arrows above genes indicate promoters and directions of transcription.
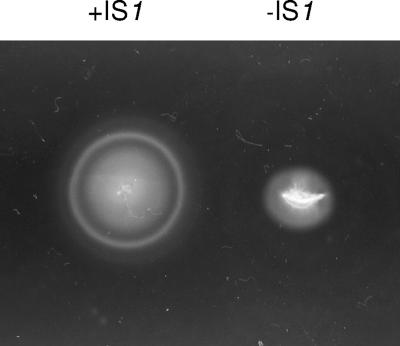
FIG. 2.
Spread of E. coli MG1655 (+IS1) and E. coli MG1655 (−IS1) on Luria motility agar after 8 h at 37°C.
Media and growth conditions.
Luria broth (LB) was made as described by Revel (34). Luria agar is LB containing 12 g of Bacto agar (Difco) per liter. MacConkey agar (Difco) was prepared according to the manufacturer's instructions. M9 minimal medium (23) was supplemented with NaCl to bring the total NaCl concentration to 200 mM and with either reagent grade glycerol (0.4%, wt/wt) or a mixture of l-arabinose, l-fucose, d-galactose, d-gluconate, N-acetyl-d-glucosamine, d-glucuronate, d-maltose, d-mannose, d-melibiose, and d-ribose, each at a concentration of 0.005% (wt/wt). Inocula were prepared as follows. Overnight cultures grown in LB were started from single colonies picked from Luria agar plates. The LB cultures were washed twice in M9 minimal medium (containing 200 mM NaCl and no carbon source), and 10-μl portions of the washed cultures were transferred to M9 minimal glycerol medium (containing 200 mM NaCl) and incubated overnight. The next morning, the cultures were washed twice as described above and were diluted to an A600 of 0.05 into fresh 20-ml aliquots of M9 minimal medium (containing 200 mM NaCl) containing the 10-sugar mixture. Cultures were incubated at 37°C with shaking in 125-ml tissue culture bottles. Growth was monitored spectrophotometrically at A600 using a Pharmacia Biotech Ultrospec 2000 UV/visible spectrophotometer.
Construction of E. coli MG1655 (−IS1).
Primers used to construct deletion mutants were designed based on the MG1655 genome sequence (3). The DNA procedures used have been described previously (24). E. coli MG1655 (CGSC 7740) contains an IS1 element in the promoter of the flhDC operon (3). E. coli MG1655 Strr ΔflhD::cam has a 546-bp deletion originating immediately downstream of IS1 in the flhD promoter and extending into flhD (Fig. 1). E. coli MG1655 (CGSC 6300) is missing the IS1 element in the flhDC promoter (2). Using the system of Datsenko and Wanner (8), the region 121 bp upstream of the IS1 element in CGSC 7740 and the 546-bp deletion in flhD were replaced with the wild-type sequence of E. coli MG1655 (CGSC 6300), which lacks the IS1 element, as follows. The primers used were: forward primer, 5′-CCTGTTTCATTTTTGCTTGCTAGC-3′; and reverse primer, 5′-GGAATGTTGCGCCTCACCG-3′. For isolation of the desired transformant, since MG1655 (CGSG 6300) is motile and E. coli MG1655 Strr ΔflhD::cam is nonmotile, 2-μl aliquots of the transformation mixture were spotted onto Luria motility agar plates (see below), which were then incubated at 37°C overnight. The next day, motile colonies were observed emanating from the site of each spot. Several colonies were picked and tested for sensitivity to chloramphenicol and ampicillin. One isolate, which was sensitive to both chloramphenicol (replacement of the deleted flhD gene with wild-type flhD) and ampicillin (loss of pKD46 [8]), was sequenced (see below) and thereby shown to be missing the IS1 element in the flhDC promoter, to have the same flhDC promoter sequence as E. coli MG1655 (CGSC 6300), and to have the wild-type flhD gene. This strain was named E. coli MG1655 (−IS1) Strr. A spontaneous nalidixic acid-resistant mutant was isolated from E. coli MG1655 (−IS1) Strr after 108 CFU was plated on Luria agar plates containing nalidixic acid (50 μg/ml). This strain was named E. coli MG1655 (−IS1) Strr Nalr and is referred to below as E. coli MG1655 (−IS1) (Fig. 1).
Construction of E. coli MG1655 (+IS1) ΔmotAB ΔfliC.
The E. coli MG1655 (+IS1) ΔmotAB deletion mutant was constructed by allelic exchange as described by Datsenko and Warner (8). A total of 1,711 bp was deleted from motA (which encodes the proton conductor component of the flagellum motor) and motB (which encodes the protein that enables flagellar motor rotation) beginning immediately downstream of the motA GTG start codon and ending 96 bp upstream of the motB TGA stop codon. The motAB deletion primers (bold type indicates MG1655 DNA, and underlining indicates chloramphenicol resistance cassette DNA) were: primer 1, 5′-TTGTTCTCGGTACAGTTTTCGGCGGTTAGTGTAGGCTGGAGCTGCTTCG-3′; and primer 2, 5′-CCAGGGCGCTTACTGGCTCATTCTGGCTTTCGGCTATGAATATCCTCCTTAGT-3′. The chloramphenicol resistance cassette was then removed (8). The E. coli MG1655 (+IS1) ΔmotAB mutant was then used to construct the E. coli MG1655 (+IS1) ΔmotAB ΔfliC deletion mutant (Fig. 1). A total of 583 bp was deleted from fliC (which encodes flagellin) beginning at the ATG start codon and ending 910 bp upstream of the TAA stop codon. The fliC deletion primers (bold type indicates MG1655 DNA, and underlining indicates chloramphenicol resistance cassette DNA) were: primer 1, 5′-AAACCCAATACGTAATCAACGACTTGCAATATAGGATAACGAATCGTGTAGGCTGGAGCTGCTTCG-3′, and primer 2, 5′-GGCTGCTTCCGTAGAAAGGGTAATTCCAGTAAGTTTAATATTGTTTCATATGA ATATCCTCCTTAGT-3′.
Sequencing.
DNA sequencing was done at the URI Genomics and Sequencing Center, University of Rhode Island, Kingston, using the CEQ8000 genetic analysis system (Beckman Coulter, Fullerton, CA). A Dye Terminator cycle sequencing Quick Start kit (Beckman Coulter) was used in the sequencing reactions. The primers used to amplify PCR products for sequencing to determine the precise location of the deletion in motAB were: forward primer (upstream of motA), 5′-CATCCTGTCATGGTCAACAGTGG-3′; and reverse primer (downstream of motB), 5′-CGCTGAAGCCAAAAGTTCCTGC-3′. The same primers were used in the sequencing reactions. The primers used to amplify PCR products for sequencing to determine the precise location of the deletion in fliC were: forward primer (upstream of fliC), 5′-GGGGTTATCGGTCTGAATTGC-3′; and reverse primer (extension begins 759 bp downstream of the fliC coding sequence), 5′-AGTCGCCATTGTCACTGTACC-3′. These primers were also used in the fliC sequencing reactions. The primers used to PCR amplify the flhDC promoter region in MG1655 (−IS1) were the primers used above for making the strain. The primer used for sequencing was the forward primer used for PCR amplification (i.e., 5′-CCTGTTTCATTTTTGCTTGCTAGC-3′), which is upstream of IS1.
Motility.
Normally, Luria motility agar is LB containing 3.5 g of Bacto agar per liter. However, for measurement of motility in the presence of various concentrations of NaCl, three colonies of each strain were transferred with toothpicks to motility agar plates containing LB diluted 10-fold with distilled water and each of the NaCl concentrations. The diluted LB was used to better control the NaCl concentration. Motility agar plates were incubated for 5.5 h at 37°C, and then spreading was measured. For testing the motility of fecal isolates from mice, 50 colonies from each mouse on each day of sampling were transferred with toothpicks to motility agar made from LB that had been diluted 10-fold with distilled water but contained no added NaCl. The preparations were incubated overnight at room temperature. Under these conditions, it was possible to distinguish motile colonies from nonmotile colonies on a single plate inoculated with 50 colonies.
Mouse colonization experiments.
The method used to compare the large intestine-colonizing abilities of E. coli strains in mice has been described previously (38, 39, 43). Briefly, three male CD-1 mice (5 to 8 weeks old) were given drinking water containing streptomycin sulfate (5 g/liter) for 24 h to eliminate the resident facultative bacteria (22). Following 18 h of starvation without food and water, the mice were fed 1 ml of 20% (wt/vol) sucrose containing LB-grown E. coli MG1655 strains, as described in the Results. After ingestion of the bacterial suspension, both the food (Harlan Teklad mouse and rat diet; Harlan Teklad, Madison, WI) and streptomycin-containing water were returned to the mice, and 1 g of feces was collected after 5 and 24 h and on odd-numbered days at the times indicated below. Mice were housed individually in cages without bedding and were placed in clean cages daily. Fecal samples (no older than 24 h) were homogenized in 1% Bacto tryptone, diluted in the same medium, and plated on MacConkey agar plates with appropriate antibiotics. Plates contained streptomycin sulfate (100 μg/ml) and nalidixic acid (50 μg/ml), streptomycin sulfate (100 μg/ml) and kanamycin sulfate (40 μg/ml), or streptomycin sulfate (100 μg/ml) and chloramphenicol (30 μg/ml). Antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). All plates were incubated for 18 to 24 h at 37°C prior to counting. Each colonization experiment was performed at least twice, and essentially identical results were obtained. Pooled data from at least two independent experiments are presented below.
Enumeration of E. coli MG1655 (−IS1) and E. coli MG1655 (+IS1) ΔflhD in mouse small intestine mucus and cecal mucus.
On day 16 after three mice were fed E. coli MG1655 Δedd*, E. coli MG1655 (−IS1), and E. coli MG1655 (+IS1) ΔflhD, the mice were sacrificed by CO2 asphyxiation, and their ceca and small intestines, exclusive of the ileum, were removed. Cecal mucus and small intestine mucus from each mouse were scraped into 5 ml of HEPES-Hanks' buffer (pH 7.4) as previously described (6). The numbers of E. coli MG1655 (−IS1) and E. coli MG1655 (+IS1) ΔflhD cells in each sample were determined by dilution and plating on MacConkey agar plates containing streptomycin sulfate (100 μg/ml) and either nalidixic acid (50 μg/ml) to enumerate E. coli MG1655 (−IS1) or chloramphenicol (30 μg/ml) to enumerate E. coli MG1655 (+IS1) ΔflhD. The plates were incubated for 18 to 24 h at 37°C prior to counting. Counts were corrected for the total volume of each sample.
Statistics.
Where indicated below, log10 means of the numbers of CFU/gram of feces of two strains from six mice were compared by Student's t test (P values). A P value of >0.1 was interpreted as indicating no significant difference, and a P value of <0.05 was interpreted as indicating a significant difference.
RESULTS
E. coli MG1655 (+IS1) and E. coli MG1655 (−IS1).
Two different strains of wild-type E. coli MG1655 exist. The first strain, CGSC 7440, which has been sequenced (3), has an IS1 element in the regulatory region of the flhDC operon, hyperexpresses the flhDC operon, and is hypermotile compared to the second strain, CGSC 6300, which is missing the element (2). Previously, a streptomycin-resistant, nalidixic acid-resistant mutant of CGSC 7440, which is referred to in this paper as E. coli MG1655 (+IS1), was used to study colonization of the mouse intestine (17). For the purposes of the present investigation, a strain was constructed that is isogenic to E. coli MG1655 (+IS1) except that it is missing the IS1 element in the regulatory region of the flhDC operon (see Materials and Methods). This strain is referred to in this paper as E. coli MG1655 (−IS1). The flhDC locus for each of the strains is illustrated in Fig. 1. As expected, E. coli MG1655 (+IS1) is hypermotile compared to E. coli MG1655 (−IS1) (Fig. 2).
Kinetics of appearance of nonmotile E. coli MG1655 (+IS1) ΔflhDC mutants in the mouse intestine.
Previously, we reported that the mouse intestine selected nonmotile mutants of E. coli MG1655 (+IS1) with deletions beginning immediately downstream of the IS1 element within the regulatory region of the flhDC operon that have improved colonizing ability (17). These mutants grew 10 to 20% faster than their parent in mouse cecal mucus in vitro and 15 to 30% faster on several sugars found in the mouse intestine (17). In the present investigation, experiments were performed to determine the kinetics of appearance of these mutants in vivo.
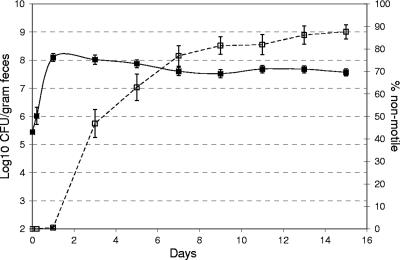
A total of 6,300 E. coli MG1655 (+IS1) colonies, plated directly from 42 overnight LB cultures that had each been started from a separate single colony, were tested for motility (150 colonies per culture). All of the colonies were motile. However, when fecal samples from 42 mice fed E. coli MG1655 (+IS1) (105 CFU/mouse) from these LB cultures were examined for motility, by day 3 after feeding 45 to 50% of the colonies were nonmotile and by day 15 after feeding between 80 and 90% were nonmotile (Fig. 3). Between 1 and 15 days after feeding the total numbers of E. coli MG1655 (+IS1) in feces remained relatively constant (Fig. 3). Ten nonmotile colonies (three from one mouse on day 3 after feeding and one each from seven individual mice on various days after feeding) were examined for deletions in the flhDC operon. DNA sequencing revealed that all 10 nonmotile isolates had deletions in the flhDC operon beginning at the same base pair immediately downstream of the IS1 target sequence and ranging in size from 102 to 6,166 bp. However, despite this strong selection for ΔflhDC mutants, 10 to 20% of the E. coli MG1655 colonies remained motile over a 15-day period (Fig. 3), suggesting that there is an as-yet-undefined intestinal niche in which motility is an advantage.
FIG. 3.
Kinetics of appearance of nonmotile E. coli MG1655 (+IS1) in the mouse intestine. Forty-two mice were fed 105 CFU of wild-type E. coli MG1655 (+IS1). At the indicated times, 50 colonies isolated from the feces of each mouse were tested for motility (□) as described in Materials and Methods; the error bars represent the standard deviations of the means for the percentage of nonmotile E. coli MG1655 (+IS1). ▪, log10 mean E. coli MG1655 (+IS1) CFU per gram of feces for 42 mice (the error bars represent the standard errors of the log10 means).
High osmolarity reduces E. coli MG1655 (−IS1) motility but not E. coli MG1655 (+IS1) motility.
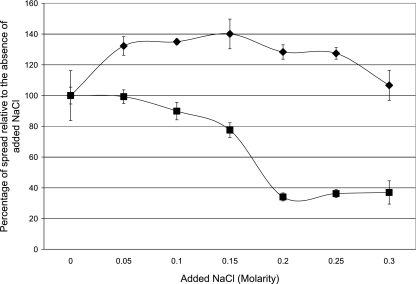
In E. coli K-12, expression of the flhDC operon lacking the IS1 element decreases drastically with increasing osmolarity due to the repressing effect of phosphorylated OmpR binding to the flhDC promoter (37). Phosphorylated OmpR is generated under high-osmolarity conditions (9). The NaCl concentration in the human intestine has been reported to be 140 mM (1), which is considerably higher than that in most laboratory media. To determine whether the hypermotile phenotype of E. coli MG1655 (+IS1) is affected by NaCl concentrations found in the intestine, the motilities of E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) and E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) were tested with a wide range of NaCl concentrations. E. coli MG1655 (+IS1) was twofold more motile than E. coli MG1655 (−IS1) at the lowest NaCl concentration and about fourfold more motile at the higher concentrations (Fig. 4). Furthermore, the motility of E. coli MG1655 (+IS1) did not decrease to a significant extent with increasing NaCl concentrations, whereas the motility of E. coli MG1655 (−IS1) decreased more than twofold between 100 mM NaCl and 200 mM NaCl such that it was nearly nonmotile. It therefore appears that the presence of the IS1 element in the regulatory region of the flhDC operon not only results in hypermotility but also interferes with the negative regulation of the flhDC operon that occurs at higher NaCl concentrations, such as those found in the intestine.
FIG. 4.
Spread of E. coli MG1655 (+IS1) (⧫) and E. coli MG1655 (−IS1) (▪) on motility agar with various concentrations of NaCl. Sets of three colonies of each strain were transferred with toothpicks to motility agar containing the indicated concentrations of NaCl as described in Materials and Methods. Incubation was for 5.5 h at 37°C. The data for each strain are expressed as the percentage of the spread observed in the absence of added NaCl. Each data point was corrected for the diameter of growth of the nonmotile E. coli MG1655 (+IS1) ΔflhD mutant on motility agar (1.6 ± 0.3 mm). The corrected spread for E. coli MG1655 (+IS1) without added NaCl was 9.4 ± 2 mm, and that for E. coli MG1655 (−IS1) was 4.7 ± 0.3 mm. The error bars represent standard deviations.
Growth on a mixture of 10 sugars.
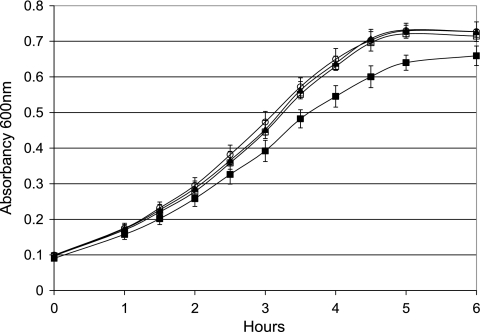
E. coli MG1655 (+IS1) ΔflhDC mutants (nonmotile, no flhDC expression), selected in the intestine from the parent E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression), grow 15 to 30% faster than their parent on a number of sugars present in mouse cecal mucus and are better colonizers of the mouse intestine than their parent (17). The same is true of an E. coli MG1655 (+IS1) ΔflhD mutant (nonmotile, no flhDC expression) constructed in the laboratory that contains a 546-bp deletion beginning immediately downstream of ISI in the regulatory region of flhD and continuing into the flhD gene (17). Because the promoter is deleted, the entire flhDC operon is inactivated, but since the deletion ends in flhD, the strain is referred to as E. coli MG1655 (+IS1) ΔflhD. To determine how well E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) grows relative to E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) and E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression), the strains were grown in M9 minimal medium containing 200 mM NaCl to mimic the salt concentration in the intestine and a mixture of 10 sugars (l-arabinose, l-fucose, d-galactose, d-gluconate, N-acetyl-d-glucosamine, d-glucuronate, maltose, d-mannose, d-melibiose, and d-ribose), each at a concentration of 0.005%. As shown in Fig. 5, E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) and E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) grew at the same rate, and both grew faster than E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression). E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) and E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) both reached a higher final absorbance than E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) (Fig. 4). It might be argued that hyperflagellation causes E. coli MG1655 (+IS1) to clump more than the other strains, which would lower its absorbance readings; however, when the organisms were viewed microscopically, none of the strains clumped at any time during growth. Therefore, it appeared that either the reduced expression of the flhDC operon in the normally motile strain E. coli MG1655 (−IS1) compared to the expression in the hypermotile strain E. coli MG1655 (+IS1) or the reduced amount of energy required for flagellum synthesis and rotation in the normally motile strain E. coli MG1655 (−IS1) compared to the amount required in the hypermotile strain E. coli MG1655 (+IS1) was responsible for the higher growth rate and higher growth yield of strain E. coli MG1655 (+IS1). Indeed, the energy required to synthesize flagella and rotate the motor has been estimated to be about 2% of the total energy that is normally consumed during growth (21). To distinguish between the two possibilities, the growth rate on the mixture of 10 sugars of E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression), which is unable to synthesize flagellin or rotate the flagellar motor, was also determined in the experiment depicted in Fig. 5. E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression) grew as rapidly as E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) and E. coli (−IS1) (normally motile, normal flhDC expression) and faster and with a higher final yield than E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression). Therefore, the growth data suggest that it is the energy required to hypersynthesize and rotate its flagella rather than hyper-flhDC expression that slows the growth of E. coli MG1655 (+IS1) compared to the growth of the other three strains in minimal medium containing the 10-sugar mixture.
FIG. 5.
Growth of E. coli MG1655 (+IS1) (▪), E. coli MG1655 (−IS1) (▴), E. coli MG1655 (+IS1) ΔflhD (○), and E. coli MG1655 (+IS1) ΔmotAB ΔfliC (□) on a mixture of 10 sugars. Incubation was at 37°C with aeration. The means and standard deviations of the A600 values at the indicated times are presented for three independent cultures of each strain.
Mouse-colonizing abilities of E. coli MG1655 (+IS1), E. coli MG1655 (+IS1) ΔflhD, E. coli MG1655 (−IS1), and E. coli MG1655 (+IS1) ΔmotAB ΔfliC.
E. coli MG1655 Δedd is defective in the Entner-Doudoroff pathway and is motile. E. coli MG1655 Δedd* is also defective in the Entner-Doudoroff pathway, was selected by the mouse intestine as being a better colonizer than its E. coli MG1655 Δedd parent, and was found to be nonmotile due to deletion of the entire flhDC operon (17). It also grew faster than both its parent and E. coli MG1655 (+IS1) on a number of sugars found in the mouse intestine (5, 17). When low numbers (105 CFU/mouse) were fed to mice, E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) was eliminated from the mouse intestine by high numbers (1010 CFU/mouse) of E. coli MG1655 Δedd* (17), but low numbers (105 CFU/mouse) of E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) grew to high numbers in mice fed high numbers (1010 CFU/mouse) of E. coli MG1655 Δedd* (17). These results showed that E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) was a better colonizer than E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) (17). In the present study, using the ability to grow from low numbers to high numbers in the presence of high numbers of E. coli MG1655 Δedd* in the mouse intestine as a measure of colonizing ability, a series of experiments was conducted to determine whether the increased intestine-colonizing ability of E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) relative to that of E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) was due solely to the increase in energy available for growth when hypermotility was prevented by the deletion.
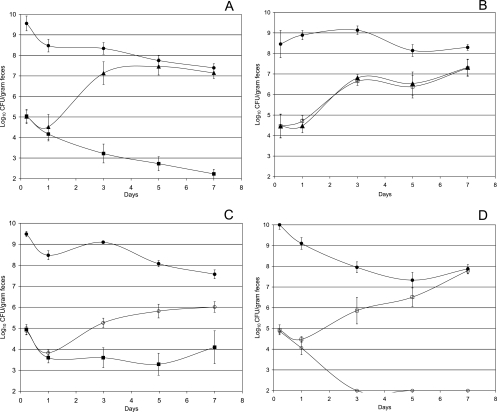
Mice were fed high numbers (1010 CFU/mouse) of E. coli MG1655 Δedd* along with low numbers (105 CFU/mouse) of the strains to be tested. As observed previously (17), E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) was a better colonizer than E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) (P > 0.1 at 5 h after feeding, but P < 0.001 at both 5 and 7 days after feeding); i.e., E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) was eliminated from the intestine in the presence of high numbers of E. coli MG1655 Δedd*, whereas E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) grew to high numbers (Fig. 6A). Compared to nonmotility and no flhDC expression, normal motility and normal flhDC expression did not detract from colonizing ability; that is, E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) and E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) both grew from low to high numbers at the same rate in the presence of high numbers of E. coli MG1655 Δedd* (Fig. 6B) (P > 0.1 at 5 h and 5 and 7 days after feeding). As suggested by the growth studies (Fig. 5), energy directed to hyper-flagellum synthesis and flagellar motor operation did indeed appear to decrease colonizing ability; i.e., E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression) was a better colonizer than E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) (P > 0.1 at 5 h after feeding, but P < 0.02 at 5 days after feeding and P < 0.05 at 7 days after feeding) (Fig. 6C). However, it appears that hyperexpression of flhDC, independent of the energy expended for hypermotility, also decreases colonizing ability; that is, E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression) was not nearly as good a colonizer as E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) (P > 0.1 at 5 h after feeding, but P < 0.001 at 5 and 7 days after feeding) (Fig. 6D). Since normal motility requires more energy expenditure than nonmotility, these data suggest that E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression) is a worse colonizer than E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) because of regulatory effects of hyperexpression of flhDC unrelated to its effects on hyper-flagellum synthesis and rotation. Table 1 summarizes these results.
FIG. 6.
Colonization of the mouse intestine. (A) Sets of three mice were fed 105 CFU of E. coli MG1655 (+IS1) (▪), 105 CFU of MG1655 (+IS1) ΔflhD (▴), and 1010 CFU of E. coli MG1655 Δedd* (•). (B) Sets of three mice were fed 105 CFU of E. coli MG1655 (−IS1) (□), 105 CFU of MG1655 (+IS1) ΔflhD (▴), and 1010 CFU of E. coli MG1655 Δedd* (•). (C) Sets of three mice were fed 105 CFU of E. coli MG1655 (+IS1) (▪), 105 CFU of MG1655 (+IS1) ΔmotAB ΔfliC (○), and 1010 CFU of E. coli MG1655 (+IS1) Δedd* (•). (D) Sets of three mice were fed 105 CFU of E. coli MG1655 (−IS1) (□), 105 CFU of MG1655 (+IS1) ΔmotAB ΔfliC (○), and 1010 CFU of E. coli MG1655 (+IS1) Δedd* (•). At the indicated times, fecal samples were homogenized, diluted, and plated as described in Materials and Methods. Error bars representing standard errors of the log10 mean CFU per gram of feces for six mice are presented for each time point.
TABLE 1.
Summary of strain genotypes, phenotypes, and colonization abilities
| E. coli strain | flhDC expressiona | Motilityb | Intestine-colonizing abilityc |
|---|---|---|---|
| MG1655 (+IS1) ΔflhD | − | − | Best |
| MG1655 (−IS1) | + | + | Best |
| MG1655 (+IS1) ΔmotAB ΔfliC | ++ | − | Next best |
| MG1655 (+IS1) | ++ | ++ | Worst |
−, no flhDC expression; +, normal flhDC expression; ++, hyper-flhDC expression.
−, nonmotile; +, normally motile; ++, hypermotile.
See Fig. 6.
Growth of E. coli MG1655 in mouse small intestine mucus in vivo does not require motility.
Although E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) and E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) appeared to be equally good colonizers (Fig. 6B), it was still possible that growth in a specific segment of the mouse intestine might require that E. coli MG1655 strains be motile. If so, that segment should be available to E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) but not to E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression). E. coli MG1655 colonizes the mouse intestine by growing in intestinal mucus (24). Since both E. coli MG1655 (−IS1) and E. coli MG1655 (+IS1) ΔflhD appear in equal numbers in feces, it would be expected that they would appear in equal numbers in cecal mucus. The question was whether E. coli MG1655 (−IS1) and E. coli MG1655 (+IS1) ΔflhD appear in equal numbers in small intestine mucus, which harbors far fewer total E. coli MG1655 cells than cecal mucus. Therefore, at day 16 after feeding of high numbers of E. coli MG1655 edd* cells and low numbers of E. coli MG1655 (−IS1) and E. coli MG1655 (+IS1) ΔflhD cells, cecal mucus and small intestine mucus, exclusive of the ileum, which mimics cecal mucus, were isolated from the mice and plated to determine the amounts of E. coli MG1655 (−IS1) and E. coli MG1655 (+IS1) ΔflhD. As expected, the amounts of the two strains in cecal mucus were 20- to 30-fold higher than the amounts in the mucus isolated from the entire small intestine (Table 2); however, E. coli MG1655 (−IS1) and E. coli MG1655 (+IS1) ΔflhD were found in equal numbers in cecal mucus and in equal numbers in small intestine mucus (Table 2). It should be noted that all 50 E. coli MG1655 (−IS1) colonies isolated from the cecal mucus of each mouse and tested for motility and all 50 E. coli MG1655 (−IS1) colonies isolated from the small intestine mucus of each mouse and tested for motility were motile. These data suggest that although E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) and E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) grow to higher numbers in cecal mucus, they grow equally well relative to one another both in small intestine mucus and in cecal mucus in vivo. It therefore does not appear that growth in small intestinal mucus in vivo requires motility.
TABLE 2.
E. coli MG1655 (−IS1) and E. coli MG1655 ΔflhDC in mouse cecal mucus and small intestine mucus
| Location | Log10 CFUa
|
|
|---|---|---|
| E. coli MG1655 (−IS1) | E. coli MG1655 ΔflhDC | |
| Small intestine mucus | 3.93 ± 0.26 | 3.60 ± 0.59 |
| Cecal mucus | 5.24 ± 0.39 | 5.16 ± 0.24 |
| Feces | 6.97 ± 0.27 | 7.18 ± 0.18 |
The values are log10 means ± standard errors of the means for three mice. Mucus preparations were isolated on day 16 after feeding. The CFU values for the small intestine and cecal mucus are corrected for the entire volume of each mucus preparation. The fecal values are the CFU/gram of feces at 15 days after feeding.
DISCUSSION
A large and growing body of evidence indicates that commensal E. coli strains grow in the intestine largely on nutrients acquired from cecal mucus and that growth is inhibited in cecal contents (18, 24, 28, 38, 39, 43). The cecal mucus layer of the mouse intestine appears to turn over about every 2 h (33) and is continuously being degraded by the indigenous microflora. The degraded cecal mucus is shed into the cecal contents and ultimately is compacted in feces. If commensal E. coli strains do not attach to underlying intestinal epithelial cells (24, 29) and growth takes place predominantly in the mucus layer, it seems reasonable that to maintain a stable population in the intestine, the bacterial growth rate must keep pace with the turnover rate of the mucus layer. In addition, it is becoming increasingly clear that in the face of competition from a complex microflora, E. coli MG1655 simultaneously uses several, presumably limiting, sugars for growth in mouse cecal mucus, including l-fucose, d-gluconate, N-acetylglucosamine, and sialic acid (5). As a consequence, any slight advantage that an E. coli MG1655 mutant might have in utilizing these nutrients to attain even a small increase in the growth rate would be magnified into predominance of the mutant over the wild type in the intestine over time.
In a previous study, it was shown that E. coli MG1655 (+IS1) ΔflhDC mutants were selected by the mouse intestine (17). These mutants were better intestinal colonizers than their parent and grew better in vitro in cecal mucus and in vitro on a variety of sugars normally found in cecal mucus (17). The E. coli MG1655 (+IS1) ΔflhDC mutants were generated with deletions immediately downstream of the IS1 element that is present in the regulatory region of the flhDC promoter of E. coli MG1655 (+IS1). The data presented here show that although less than 1 in 6,300 of the cells in LB are E. coli MG1655 ΔflhDC mutants (i.e., < 0.016%), almost 50% of the colonies present in feces at 3 days after feeding and about 90% of the colonies by day 15 days after feeding are ΔflhDC mutants (Fig. 3) There appear to be two components that contribute to this strong selection. First, E. coli MG1655 (+IS1) is hypermotile, and the energy involved in making and rotating its flagella appears to decrease both its growth rate and its colonizing ability; i.e., E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression) grows faster than E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) on the mixture of 10 sugars in vitro (Fig. 5) and is the better colonizer (Fig. 6C), yet the only difference between the two strains is that E. coli MG1655 (+IS1) ΔmotAB ΔfliC is unable to make flagella and rotate the flagellar motor. Second, it appears that hyperexpression of the FlhD4C2 complex in E. coli MG1655 (+IS1) may result in negative regulation of one or more as-yet-unidentified genes that contribute to maximum colonizing ability. This is suggested by the fact that although E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression), E. coli MG1655 (−IS1) (normally motile, normal flhDC expression), and E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) grow at the same rate on the 10-sugar mixture in vitro (Fig. 5), E. coli MG1655 (−IS1) (normally motile, normal flhDC expression) and E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) are better colonizers than E. coli MG1655 (+IS1) ΔmotAB ΔfliC (nonmotile, hyper-flhDC expression) (Table 1 and Fig. 6).
E. coli MG1655 (−IS1) is as good an intestinal colonizer as E. coli MG1655 (+IS1) ΔflhD, yet it makes the FlhD4C2 complex and is motile; however, it is about fourfold less motile than E. coli MG1655 (+IS1) because it presumably makes less FlhD4C2 complex. The fact that it makes less FlhD4C2 complex may account for its better colonizing ability relative to that of E. coli MG1655 (+IS1); i.e., the concentration of the FlhD4C2 complex in E. coli MG1655 (−IS1) may not be high enough to either repress expression of genes unrelated to motility that are important for maximum colonization or to hyperexpress motility genes. If this is true, then normal motility would not be detrimental to colonization and E. coli MG1655 (−IS1) might be motile in the intestine. Alternatively, it is possible that the high osmolarity in the intestine shuts off expression of the E. coli MG1655 (−IS1) flhDC operon, as it appears to do on motility agar plates containing high concentrations of NaCl (Fig. 4). If this is true, E. coli MG1655 (+IS1) ΔflhD and E. coli MG1655 (−IS1) would be functionally equivalent in the intestine (i.e., no flhDC expression and no motility).
At the present time, we do not know the identity of the gene(s) that appears to be repressed by the FlhD4C2 regulatory complex in E. coli MG1655 (+IS1) in the intestine. However, in preliminary experiments, when E. coli MG1655 (+IS1) (hypermotile, hyper-flhDC expression) and E. coli MG1655 (+IS1) ΔflhD (nonmotile, no flhDC expression) were grown in cecal mucus in vitro and the gene expression patterns were compared, it was found that several genes encoding catabolic enzymes for sugars not present in the 10-sugar mixture were upregulated at least twofold in E. coli MG1655 (+IS1) ΔflhD. These genes were lacZ (lactose), garD (galactarate), gudX (glucarate [putative]), and garR (glucarate and galactarate). Indeed, it will be of great interest to examine the roles of lactose, glucarate, and galactarate catabolism in colonization of the mouse intestine by E. coli MG1655 (−IS1) and E. coli MG1655 (+IS1) ΔflhDC mutants.
While we have shown that the mouse small intestine does not appear to be a distinct intestinal segment in which motility is required, in mice fed E. coli MG1655 (+IS1) 10 to 20% of the cells remained motile between days 9 and 15 after feeding (Fig. 3). It therefore appears that motility may be favored in a small niche that is present in the mouse intestine. One possibility for the site of this niche is the part of the intestinal mucus layer that is closest to the epithelium; i.e., it may be that motility is required for strains of E. coli MG1655 to reach and remain at that site. In addition, the mucosal tissues are oxygen rich, and it is well known that oxygen diffuses into the intestine at appreciable levels (13, 19). It is therefore likely that E. coli MG1655 (+IS1) encounters a higher oxygen tension close to the epithelium than at the luminal surface. Since a major source of nutrients in the intestine could be derived from exfoliated lysed epithelial cells, it is also possible that E. coli MG1655 (+IS1) encounters a higher concentration of nutrients closer to the epithelium than at the luminal surface. Nutrient stress has been reported to increase the frequency of transposition mediated by insertion elements (42). Therefore, since the environment close to the epithelium may more closely approximate the favorable growth conditions in LB, it may be that flhDC deletions do not occur at an appreciable rate close to the surface of the epithelium or that flhDC deletion mutants are not successful in that niche because they are not motile.
In contrast to the situation in E. coli MG1655, it appears that motility and chemotaxis do play a major role in salmonellae penetrating the mucus layer in the ileum (35, 45). Moreover, it appears that motility and/or chemotaxis is required for Campylobacter jejuni colonization of the suckling mouse intestine (40), for Helicobacter pylori colonization of both the mouse and gnotobiotic piglet stomachs (10), and for rapid penetration of rabbit ileal mucus by Vibrio cholerae (11) However, it is becoming clear that in some instances, motility is important at some but not all stages of the life cycle of a pathogen and that flagella can sometimes be important for adhesion to mucosal surfaces rather than for motility (12). Nevertheless, it should be noted that in the strain collection of the Statenserum Institut in Copenhagen, Denmark, 370 of 1,000 enteropathogenic E. coli strains are nonmotile (37%), 125 of 520 enterotoxigenic E. coli strains are nonmotile (24%), 45 of 135 enteroinvasive E. coli strains are nonmotile (33%), and 548 of 1,890 enterohemorrhagic strains are nonmotile (29%) (Karen A. Krogfelt, personal communication).
It should also be noted that selection of E. coli MG1655 flhDC deletion mutants in the mouse intestine is not the only instance in which mutation within the flhDC operon has been reported to occur in nature or to be advantageous. For example, E. coli O157:H− nonmotile strains, found in up to 40% of human hemolytic-uremic syndrome cases in Germany, have recently been shown to contain a 12-bp deletion in flhC (26). In addition, an flhD mutant of Salmonella enterica serovar Typhimurium was more virulent than its parent in C57BL/6J mice and appeared to grow more rapidly than its wild-type parent in the spleen and in mouse macrophages in tissue culture (36). Moreover, deletion of the Yersinia enterocolitica flhDC operon has been reported to upregulate the yop (Yersinia outer protein) regulon (4, 14). Lastly, it appears that a major reason that Shigella strains are nonmotile is damage in the flhDC operon caused by either insertions or deletions associated with insertion elements that have occurred during niche adaptation (41).
In summary, the data presented here underscore the complexity of the role of the flhDC operon and motility in E. coli MG1655 colonization of the mouse intestine. It appears that while motility is not necessary for colonization, there is an as-yet-undefined intestinal niche in which 10 to 20% of E. coli MG1655 (+IS1) cells reside and in which motility is an advantage. Identifying where that niche is may shed light on why motility is important for its occupation by E. coli MG1655 (+IS1) and whether occupation of the niche might also be important for pathogens. The E. coli MG1655 (+IS1) flhDC deletion mutants that are selected by the intestine appear to be selected for two reasons. First, genes unrelated to motility that are either directly or indirectly repressed by FlhD4C2 but can contribute to maximum colonizing ability are released from repression. Second, energy normally used to hyperexpress and rotate flagella is redirected to growth.
Acknowledgments
This research was supported by Public Health Service grant AI 48945 to T.C. and P.S.C. E.J.G. and R.M.-L. were supported by a USDA Strengthening Research grant (Environmental Biotechnology at URI) to P.S.C.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Banwell, J. G., S. L. Gorbach, N. F. Pierce, R. Mitra, and A. Mondal. 1971. Acute undifferentiated human diarrhea in the tropics. II. Alterations in intestinal fluid and electrolyte movements. J. Clin. Investig. 50:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, C. S., B. M. Prüß, and P. Matsumura. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 186:7529-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Bleves, S., M. Marie-Noëlle, G. Detry, and G. R. Cornelis. 2002. Up-regulation of the Yersinia enterocolitica yop regulon by deletion of the flagellum master operon flhDC. J. Bacteriol. 184:3214-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 101:7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, P. S., and D. C. Laux. 1995. Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods Enzymol. 253:309-315. [DOI] [PubMed] [Google Scholar]
- 7.Conway, K. A. Krogfelt, and P. S. Cohen. 29 December 2004, posting date. The life of commensal Escherichia coli in the mammalian intestine. In R. Curtiss III, et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egger, L. A., H. Park, and M. Inouye. 1997. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells 2:167-184. [DOI] [PubMed] [Google Scholar]
- 10.Foynes, S., N. Dorrell, S. J. Ward, R. A. Stabler, A. A. McColm, A. N. Rycroft, and B. W. Wren. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freter, R., P. C. O'Brien, and M. S. Macsai. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect. Immun. 34:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girón, J. A. 2005. Role of flagella in mucosal colonization, p. 213-235. In J. P. Nataro, P. S. Cohen, H. L. T. Mobley, and J. N. Weiser (ed.), Colonization of mucosal surfaces. American Society for Microbiology Press, Washington, DC.
- 13.He, G., R. A. Shankar, M. Chzhan, A. Samouilov, P. Kuppusamy, and J. L. Zweier. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. USA 96:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horne, S. M., and B. M. Prüß. 2006. Global gene regulation in Yersinia enterocolitica: effect of FliA on the expression levels of flagellar and plasmid-encoded virulence genes. Arch. Microbiol. 185:115-126. [DOI] [PubMed] [Google Scholar]
- 15.Hoskins, L. 1984. Mucin degradation by enteric bacteria: ecological aspects and implications for bacterial attachment to gut mucosa, p. 51-65. In E. C. Boedecker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 16.Laux, D. C., P. S. Cohen, and T. Conway. 2005. Role of the mucus layer in bacterial colonization of the intestine, p. 199-212. In J. P. Nataro, P. S. Cohen, H. L. T. Mobley, and J. N. Weiser (ed.), Colonization of mucosal surfaces. American Society for Microbiology Press, Washington, DC.
- 17.Leatham, M. P., S. J. Stevenson, E. J. Gauger, K. A. Krogfelt, J. J. Lins, T. L. Haddock, S. M. Autieri, T. Conway, and P. S. Cohen. 2005. The mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licht, T. R., T. Tolker-Nielsen, K. Holmstrøm, K. A. Krogfelt, and S. Molin. 1999. Inhibition of Escherichia coli precursor 16S rRNA processing by mouse intestinal contents. Environ. Microbiol. 1:23-32. [DOI] [PubMed] [Google Scholar]
- 19.Liévin-Le Moal, V., and A. L. Servin. 2006. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin. Microbiol. Rev. 19:315-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology Press, Washington, DC. [Google Scholar]
- 22.Miller, C. P., and M. Bohnhoff. 1963. Changes in the mouse's enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin-treatment. J. Infect. Dis. 113:59-66. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 24.Miranda, R. L., T. Conway, M. P. Leatham, D. E. Chang, W. E. Norris, J. H. Allen, S. J. Stevenson, D. C. Laux, and P. S. Cohen. 2004. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 72:1666-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Møller, A. K., M. P. Leatham, T. Conway, P. J. M. Nuijten, L. A. M. de Haan, K. A. Krogfelt, and P. S. Cohen. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 71:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monday, S. R., S. A. Minnich, and P. C. Feng. 2004. A 12-base-pair deletion in the flagellar master control gene flhC causes nonmotility of the pathogenic German sorbitol-fermenting Escherichia coli O157:H− strains. J. Bacteriol. 186:2319-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neutra, M. R. 1984. The mechanism of intestinal mucous secretion, p. 33-41. In E. C. Boedecker (ed.), Attachment of organisms to the gut mucosa, vol. II. CRC Press, Inc., Boca Raton, FL. [Google Scholar]
- 28.Newman, J. V., R. Kolter, D. C. Laux, and P. S. Cohen. 1994. The role of leuX in Escherichia coli colonization of the streptomycin-treated mouse large intestine. Microb. Pathog. 17:301-311. [DOI] [PubMed] [Google Scholar]
- 29.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine from rRNA in situ hybridization. Infect. Immun. 62:5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prüß, B. M. 2000. FlhD, a transcriptional regulator in bacteria. Recent Res. Dev. Microbiol. 4:31-42. [Google Scholar]
- 31.Prüß, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prüß, B. M., X. Liu, W. Hendrickson, and P. Matsumura. 2001. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol. Lett. 197:91-97. [DOI] [PubMed] [Google Scholar]
- 33.Rang, C. U., T. R. Licht, T. Midtvedt, P. L. Conway, L. Chao, K. A. Krogfelt, P. S. Cohen, and S. Molin. 1999. Estimation of growth rates of Escherichia coli BJ4 in streptomycin-treated and previously germfree mice by in situ rRNA hybridization. Clin. Diagn. Lab. Immunol. 6:434-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Revel, H. R. 1967. Restriction and nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology 31:688-701. [DOI] [PubMed] [Google Scholar]
- 35.Robertson, J. M., G. Grant, E. Allen-Bercoe, M. J. Woodward, A. Pusztai, and H. J. Flint. 2000. Adhesion of Salmonella enterica var. Enteritidis strains lacking fimbriae and flagella to rat ileal explants cultured at the air interface or submerged in tissue culture medium. J. Med. Microbiol. 49:691-696. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt, C. K. J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sweeney, N. J., P. Klemm, B. A. McCormick, E. Moller-Nielsen, M. Utley, M. A. Schembri, D. C. Laux, and P. S. Cohen. 1996. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect. Immun. 64:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweeney, N. J., D. C. Laux, and P. S. Cohen. 1996. Escherichia coli F-18 and K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 64:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takata, T., S. Fujimoto, and K. Amako. 1992. Isolation of nonchemotactic mutants of Campylobacter jejuni and their colonization of the mouse intestinal tract. Infect. Immun. 60:3596-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tominaga, A., R. Lan, and P. R. Reeves. 2005. Evolutionary changes of the flhDC flagellar master operon in Shigella strains. J. Bacteriol. 187:4295-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Twiss, E., A. M. Coros, N. P. Tavakoli, and K. M. Derbyshire. 2005. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 57:1593-1607. [DOI] [PubMed] [Google Scholar]
- 43.Wadolkowski, E. A., D. C. Laux, and P. S. Cohen. 1988. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect. Immun. 56:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, S., R. T. Fleming, E. M. Westbrook, P. Matsumura, and D. B. McKay. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798-808. [DOI] [PubMed] [Google Scholar]
- 45.Worton, K. J., D. C. Candy, T. S. Wallis, G. J. Clarke, M. P. Osborne, S. J. Haddon, and J. Stephen. 1989. Studies on early association of Salmonella typhimurium with intestinal mucosa in vivo and in vitro: relationship to virulence. J. Med. Microbiol. 29:283-294. [DOI] [PubMed] [Google Scholar]