Abstract
A member of the Trypanosoma cruzi gp82 family, expressed on metacyclic trypomastigote surface and identified by monoclonal antibody (MAb) 3F6, plays a key role in host cell invasion. Apart from the gp82 defined by MAb 3F6, no information is available on members of this protein family. From cDNA clones encoding gp82 proteins sharing 59.1% sequence identity, we produced the recombinant proteins J18 and C03, the former containing and the latter lacking the epitope for MAb 3F6. Polyclonal antibodies to J18 and C03 proteins were generated and used, along with MAb 3F6, to analyze the expression and cellular localization of gp82 family members in metacyclic forms of CL and G strains, which belong to highly divergent genetic groups. By two-dimensional gel electrophoresis and immunoblotting, molecules of 82 to 86 kDa, focusing at pH 4.6 to 5.4, and molecules of 72 to 88 kDa, focusing at pH 4.9 to 5.7, were visualized in CL and G strains, respectively. Flow cytometry and microscopic analysis revealed in both strains similar expression of MAb 3F6-reactive gp82 in live and permeabilized parasites, indicating its surface localization. The reaction of live parasites of both strains with anti-J18 antibodies was weaker than with MAb 3F6 and was increased by permeabilization. Anti-C03 antibodies bound predominantly to flagellar components in permeabilized G strain parasites, but in the CL strain the flagellum was not the preferential target for these antibodies. Host cell invasion of metacyclic forms was inhibited by J18 protein, as well as by MAb 3F6 and anti-J18 antibodies, but not by C03 protein or anti-C03 antibodies.
Metacyclic trypomastigotes of different Trypanosoma cruzi strains may engage different surface molecules to invade host cells (32). In the highly infective CL strain, the metacyclic stage-specific surface glycoprotein gp82 identified by monoclonal antibody (MAb) 3F6 promotes target cell invasion by triggering bidirectional signaling cascades leading to Ca2+ mobilization in both parasite and target cells (17, 22, 34), which is an event essential for parasite internalization (12, 26). Binding of gp82 to target cells induces a Ca2+-dependent disruption of actin microfilaments (10), a process reported to facilitate parasite entry (21). gp82 also has the ability to bind to gastric mucin (18), and this is crucial for the establishment of T. cruzi infection by the oral route since the binding to mucin represents the first step toward the invasion of gastric mucosal epithelium (8, 9). The poorly invasive G strain metacyclic forms express MAb 3F6-reactive gp82 molecules, but they preferentially use the mucin-like surface glycoproteins gp35/50 to enter host cells (22, 24, 33).
The MAb 3F6-reactive gp82 molecule is a member of a multigene family, which is part of the large trans-sialidade/gp85 gene family (1). According to T. cruzi proteome analysis, 30 of the 50 top-scoring proteins detected exclusively in the infective trypomastigote forms are trans-sialidase (TS) family members (2). The repertoire of metacyclic trypomastigote gp82 molecules, the degree to which they are heterogeneous, the expression of members lacking the MAb 3F6 epitope, and their presence in locations other than on the parasite surface are matters that remain to be determined. The gp82 molecules characterized so far are highly conserved in T. cruzi strains CL and G, which belong to highly divergent genetic groups (5), and show 97.9% peptide sequence identity overall and 100% identity with regard to the cell binding site and the epitope for MAb 3F6 (32).
In this study we isolated and characterized a new member of the gp82 family and performed a global analysis on the expression as well as the cellular localization of gp82 proteins in metacyclic forms of the CL and G strains. The strategy consisted of the following steps: (i) isolation of a cDNA clone encoding a member of the gp82 family lacking the epitope for MAb 3F6, (ii) production of recombinant proteins with and without the MAb 3F6 epitope, (iii) generation of antibodies against the referred recombinant proteins, and (iv) two-dimensional (2D) gel electrophoresis of metacyclic trypomastigote extracts and immunoblotting in parallel with analysis by flow cytometry and fluorescent microscope visualization of live as well as permeabilized parasites, using MAb 3F6 and anti-gp82 polyclonal antibodies.
MATERIALS AND METHODS
Parasites.
The following T. cruzi strains were used: CL, isolated from the insect Triatoma infestans in the state of Rio Grande do Sul (4), and G, isolated from an opossum in the Amazon (31). Parasites were maintained cyclically in mice and in liver infusion tryptose. Before purification, in some cases the parasites were grown in Grace's medium. Metacyclic forms from cultures in liver infusion tryptose or Grace's medium at the stationary growth phase were purified by passage through a DEAE-cellulose column, as described previously (27).
Purification of RNA, RT-PCR, and cloning in pGEM-T.
Purified CL strain metacyclic trypomastigotes (1 × 108) were lysed with 1 ml of Trizol reagent (Invitrogen). Following complete dissolution and the addition of 0.2 ml of chloroform, the parasite preparation was centrifuged at 14,000 × g for 15 min at 4°C. The aqueous phase was collected, and an equal volume of isopropyl alcohol was added to precipitate the total RNA. After washing with 75% ethanol, the precipitate was resuspended in RNase-free water. Reverse transcription-PCR (RT-PCR) was performed using the Access Quick RT-PCR system (Promega). The reaction mixture contained the following: total T. cruzi RNA; deoxynucleoside triphosphate mix; avian myeloblastosis virus reverse transcriptase; Taq polymerase; the forward primer SL (5′-GATACAGTTTCTGTACTATATTGAG-3′), which is specific for the spliced leader sequence (GenBank accession no. M30787); and the reverse primer 1775R (5′-GTTCCATTCGAAAGCATCCAGTT-3′), which is specific for a sequence of the 3′ region of the gp82 gene (GenBank accession no. L14824). Production of cDNA was carried out at 48°C for 45 min. Amplification was performed by running 45 cycles of denaturing, annealing, and elongation at 94°C for 15 s, 44°C for 30 s, and 72°C for 1 min, respectively. After purification with a GenClean kit (Bio 101), the PCR product was cloned in the plasmid vector pGEM using a pGEM-T Easy Vector kit (Promega). Following ligation to the vector, the product was transformed in Escherichia coli strain DH5α, and the colonies were grown in LB broth. Inserts released from cDNA clones were screened with a 32P-labeled probe containing the full-length T. cruzi gp82 gene. The selected clones were sequenced using a Big Dye terminator cycle sequencing ready reaction kit (Perkin-Elmer).
Production and purification of recombinant proteins J18 and C03.
The recombinant protein J18, containing the full-length T. cruzi gp82 in frame with glutathione S-transferase, was produced in E. coli DH5α by transforming the bacteria with a pGEX-3 construct comprising the gp82 gene (23). All steps for induction of the recombinant protein J18 and its purification are detailed elsewhere (10). The recombinant protein C03, containing the full-length gp82 in fusion with six histidine residues, was produced in E. coli BL21(DE3) by transforming the bacteria with the construct pHIS-C03. This construct was generated by PCR using the following primers: one containing the third ATG initiation codon plus an artificial BamHI site (5′-ATTGGATCCGATGTGCTGCGCCACC-3′) and the other containing the stop codon plus an artificial HindIII site (5′-GGAAGCTTTCTCAGTAAAGGGCCGC-3′). As a template, we used the plasmid pGEM-T-C03. Following cloning in the vector pET-22(b+) (pHIS; Novagen, Madison, WI), the clones were digested with BamHI/HindIII in order to release the insert. Upon confirmation of the sequence, the clone pHIS-C03 was selected for further characterization. The transformed bacteria were grown in LB medium and induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C, treated with 15 mg of lysozyme in phosphate-buffered saline (PBS) for 30 min at room temperature, and then sonicated for 20 min and centrifuged at 12,000 × g for 30 min a 4°C. After three washings with 10 ml of CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) buffer at 1%, the precipitate was collected, resuspended in binding buffer (20 mM Na2HPO4, 0.5 M NaCl, 20 mM imidazole, 8 mM urea, pH 7.4), and incubated at room temperature under agitation for 2 h. Thereafter, the sample in binding buffer was centrifuged at 2,600 × g for 10 min, and the supernatant was added to a 5-ml Ni3+ column (Ni Sepharose 6 Fast Flow; Amersham Biosciences). Following incubation for 30 min under agitation at room temperature, the bound protein containing the histidine tail was eluted with the elution buffer (20 mM Na2HPO4, 0.5 M NaCl, 250 mM imidazole, 8 M urea, pH 7.4) and dialyzed against double-distilled water for 48 h at 4°C. The amount of purified protein was quantified by reaction with Coomassie Plus (Pierce) in 96-well plates, and readings were taken at 620 nm. To certify that the desired protein was obtained, the purified samples were analyzed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels stained with Coomassie blue and by immunoblotting using anti-His antibodies (Amersham Biosciences).
Generation of antibodies to recombinant proteins J18 and C03.
BALB/c mice were injected by intraperitoneal route with recombinant protein (5 μg/mouse) in the presence of aluminum hydroxide (0.5 mg/mouse) as adjuvant. Fourteen days later, all animals received the same amount of antigen plus adjuvant. Thereafter, at 1-week intervals mice were given two more doses. Ten days after the last immunizing dose, the mice were bled, and sera were collected and stored at −20°C until used.
Isoelectric focusing, SDS-PAGE, and immunoblotting.
The standard Western blot analysis was performed, as previously described (33), by applying NP-40-solubilized parasite extracts corresponding to 3 × 107 cells into each well of 10% SDS-PAGE gels. For 2D electrophoresis, samples containing 5 × 108 metacyclic trypomastigotes were washed three times in 25 mM HEPES, pH 7.4, containing 0.9% NaCl; samples were boiled for 5 min in 0.2% SDS lysis buffer and maintained on ice before incubation with 2D buffer (7 M urea, 2 M thiourea, 1% dithiothreitol [DTT], 2% Triton X-100, 0.5% immobilized pH gradient buffer, pH 4 to 7) containing protease inhibitors (100 μM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 100 μM leupeptin, and 5 mM EDTA) for 30 min at room temperature. After centrifugation at 23,100 × g for 10 min, the supernatant was applied to immobilized pH gradient gel strips with 0.5% (pH 4.0 to 7.0) ampholites (Amersham Biosciences). Isoelectric focusing was performed in an Ettan IPGphor system (Amersham Biosciences) by applying 500 V, 1,000 V, 8,000 V, and 7,400 V, sequentially at 1-h intervals. Thereafter, the gel strips were incubated for 15 min in a solution containing 6 M urea, 50 mM Tris, pH 8.8, 30% glycerol, 2% SDS, and 25 mM DTT. Following another 15-min incubation in this solution without DTT but containing 125 mM iodoacetamide plus 0.02% bromophenol blue, the samples were subjected to electrophoresis in 10% SDS-PAGE gels. The proteins were then transferred to a nitrocellulose membrane, which was processed for reaction with anti-gp82 antibodies.
Flow cytometry.
Live metacyclic trypomastigotes (3 × 107) were incubated for 1 h on ice with anti-gp82 antibodies. After washings in PBS and fixation with 2% paraformaldehyde for 30 min, the parasites were incubated with anti-mouse immunoglobulin G (IgG) conjugated to fluorescein at room temperature for 1 h. Following two more washes, the number of fluorescent parasites was estimated with a Becton Dickinson FACscan cytometer. Assays with fixed and permeabilized parasites were carried out as follows: fixation with 2% paraformaldehyde for 30 min, washings in PBS, treatment with 0.1% saponin in PBS at room temperature for 30 min, washings in PBS, and incubation with antibodies as described above.
Microscopic visualization of fluorescent parasites.
Live metacyclic forms were incubated for 1 h on ice with anti-gp82 antibodies, washed, fixed with 3.5% formaldehyde in PBS, and placed onto glass slides and dried. Afterwards, the parasites were incubated sequentially with fluorescein-conjugated anti-mouse IgG diluted 1:40 in PGN (0.15% gelatin in PBS containing 0.1% sodium azide) for 1 h, and 10 μM DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes) for visualization of kinetoplast and nucleus. Images were acquired on a Nikon E600 fluorescence microscope coupled to a Nikon DXM 1200F digital camera using ACT-1 software. In parallel, the parasites were first fixed with 3.5% formaldehyde, washed, and then processed as above, except that a 1-h incubation with anti-gp82 antibodies was carried out in the presence of 0.1% saponin in PGN for parasite permeabilization.
Host cell invasion assay.
HeLa cells, the human carcinoma-derived epithelial cells, were grown at 37°C in Dulbecco's minimum essential medium, supplemented with 10% fetal calf serum, streptomycin (100 μg/ml), and penicillin (100 U/ml) in a humidified 5% CO2 atmosphere. Cell invasion assays were carried out as detailed elsewhere (33) by seeding the parasites onto each well of 24-well plates containing 13-mm-diameter round glass coverslips coated with 1.5 × 105 HeLa cells. After a 1-h incubation with parasites at a parasite:cell ratio of 10:1, the coverslips were washed in PBS and stained with Giemsa, and the numbers of intracellular parasites were counted.
Nucleotide sequence accession number.
The sequence of the recombinant protein C03 has been deposited in the GenBank database under accession number EF445668.
RESULTS
Isolation of a new member of the gp82 family.
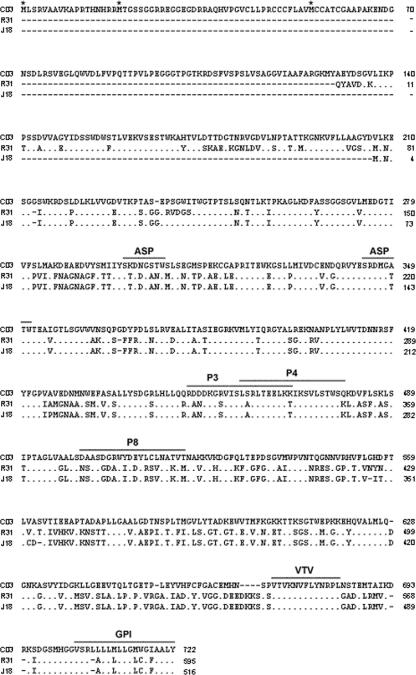
We cloned a full-length gp82 cDNA (C03) by RT-PCR, using a set of primers based on the sequence of clone J18, which codes for a gp82 protein containing the epitope for MAb 3F6 (1) and the miniexon sequence (29) present in all trypanosomatid mRNAs. Translation of C03 cDNA in each of the six possible reading frames indicated only one large open reading frame with three in-frame ATG initiator codons. Within this open reading frame the first potential start codon is separated from the spliced leader sequence by 89 bp. The second and third start codons are in the same reading frame as the first ATG and they are 51 and 159 bp downstream from it, respectively. Analysis of the 5′ sequences flanking the three potential start codons indicates that the third ATG best fits the Kozak eukaryotic consensus sequences (15). This argues for the use of the third ATG as the initiating methionine. However, we cannot exclude the possibility of translation from the first or the second in-frame methionine, provided that the amino acid sequence of the amino-terminal end of the native gp82 has not yet been determined. Assuming the third codon as the initiating methionine, C03 cDNA encodes a polypeptide of 668 amino acids with a predicted molecular mass of 72.54 kDa and pI 5.03 (Fig. 1). It has two highly conserved copies of the sialidase motif SXDXGXTW (Asp box) and a complete copy of the subterminal motif VTVKNVFLYNR characteristic of all members of the TS superfamily (13). Although C03 does not display an N-terminal sequence predicted to be a signal peptide by the SignalP 3.0 program (3), it has a typical glycosylphosphatidylinositol anchor signal sequence at the carboxy terminus (Fig. 1).
FIG. 1.
Sequences of proteins of T. cruzi metacyclic trypomastigote gp82 family members. Shown are the proteins C03 (CL strain), R31 (CL strain), and J18 (G strain). In proteins R31 and J18 are the sequences (indicated by lines above the sequences) identified as the epitope for MAb 3F6 (P3) and as the conformational cell binding site (P4 and P8). The alternative initiator methionines in the C03 are indicated by asterisks. The conserved motif VTV, characteristic of all members of the gp85/TS family, and the potential glycosylphosphatidylinositol (GPI) anchor consensus sequence are indicated (lines above sequences). Points represent residues that are conserved in the three proteins, nonconserved amino acids are indicated, and dashes represent residues that are lacking in the indicated sequence. Note the high degree of identity between protein R31 and J18, as opposed to the much lower sequence identity of these proteins in relation to protein C03.
The amino acid sequence deduced from cDNA clone C03 displayed considerable differences from the sequences deduced from clones J18 and R31 (GenBank accession no. AF128843) isolated in previous studies from cDNA libraries from G and CL strain metacyclic trypomastigotes, respectively. The overall amino acid sequence identity was 59.1% between C03 and J18 proteins and 49% between C03 and R31 proteins.
Comparative analysis of recombinant proteins C03 and J18.
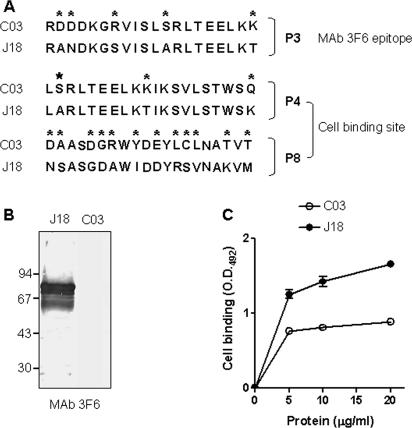
Using synthetic peptides based on the J18 protein sequence, the epitope for MAb 3F6 has been mapped, and it corresponds to the sequence represented by peptide P3 (16). As shown in Fig. 2A, compared to the sequence of J18 corresponding to P3, the most conspicuous difference in the C03 protein was the substitution of three uncharged residues by charged ones (Fig. 2A). Reflecting this difference, the C03 protein failed to be recognized by MAb 3F6 (Fig. 2B). Considerable differences were also seen in the sequence corresponding to the cell binding site, which in the J18 protein is formed by juxtaposition of two sequences, P4 and P8, containing several charged residues separated by a more hydrophobic stretch (16); comparison of the sequences of the two proteins revealed substitutions of charged residues for uncharged amino acids and vice versa (Fig. 2A). Accordingly, the binding capacity of the C03 protein to HeLa cells was lower than that of the J18 protein (Fig. 2C).
FIG. 2.
Properties of recombinant proteins J18 and C03. (A) The sequences identified as the epitope for MAb 3F6 (P3) and as the conformational cell binding site (P4 and P8) are aligned to show the differences between J18 and C03 proteins, with asterisks indicating the changed residues. (B) Immunoblot of purified J18 and C03 proteins using MAb 3F6. Note the lack of reactivity of C03 protein. (C) Binding of proteins J18 and C03 to HeLa cells, assayed as described in Materials and Methods. The reaction was revealed by sequential incubation with polyclonal antiserum directed to J18 or to C03 and anti-mouse IgG conjugated to peroxidase. OD492, optical density at 492 nm.
Expression and cellular localization of molecules of the gp82 family in G and CL strain metacyclic forms.
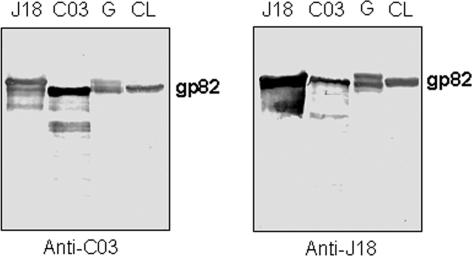
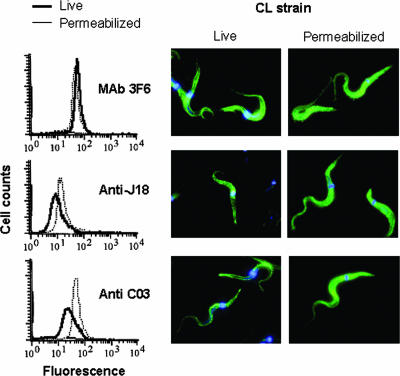
We generated antibodies against C03 and J18 recombinant proteins by immunizing groups of mice with either of these proteins. The anti-C03 antibody also reacted with J18 protein, and anti-J18 antibodies recognized C03 protein as well, and in both cases the reaction was stronger with the protein that served as immunogen (Fig. 3). Both antibodies reacted similarly with gp82 molecules from metacyclic trypomastigotes of G and CL strains (Fig. 3). Immunoblots of CL strain parasite extracts subjected to 2D gel electrophoresis showed the MAb 3F6-reactive gp82 molecules focusing at a pH range of 4.6 to 5.1 (data not shown). We confirmed the presence of these gp82 molecules on the parasite surface by flow cytometry analysis and observation with fluorescence microscopy, which revealed no difference in the reactivity of live and permeabilized parasites with MAb 3F6 (Fig. 4). As regards the gp82 molecules recognized by anti-J18 or anti-C03 antibodies, with isoelectric points ranging from pH 4.6 to 5.4 (data not shown), their location was at least in part intracellular, as deduced from the high reactivity of permeabilized parasites compared to their live counterparts, and this was clearly visible by observations with fluorescence microscopy (Fig. 4). We also noted that in live parasites the distribution of molecules recognized by anti-C03 antibodies was not homogeneous, particularly in the posterior end, where they were more concentrated at some spots (Fig. 4).
FIG. 3.
Cross-reactivity between anti-J18 and anti-C03 antibodies. Purified recombinant proteins J18 and C03, as well as the metacyclic trypomastigote extract of T. cruzi strains G and CL, were analyzed by immunoblotting using polyclonal antibodies to J18 or C03 protein.
FIG. 4.
Cellular localization of proteins of the gp82 family in metacyclic forms of the T. cruzi CL strain. Live or permeabilized parasites were reacted with MAb 3F6 or polyclonal antibodies to J18 or C03 and processed for analysis by flow cytometry or for visualization by fluorescence microscopy. DAPI staining of parasite kinetoplast and nucleus appears in blue. Note the similar expression patterns of MAb 3F6-reactive molecules in live and permeabilized parasites, whereas the molecules recognized by anti-J18 and anti-C03 antibodies were in part localized intracellularly, as indicated by the high fluorescence intensity of permeabilized parasites compared to their live counterparts.
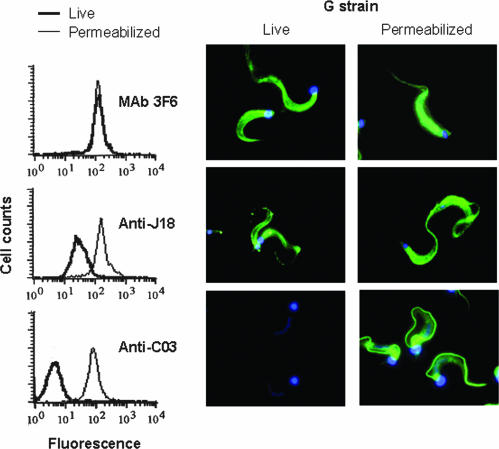
In G strain metacyclic forms, 2D gel electrophoresis and immunoblotting revealed a similar profile of gp82 bands focusing at pH 4.9 to 5.7 when MAb 3F6 or anti-J18 antibodies were used (data not shown). As in the CL strain, the MAb 3F6-reactive gp82 molecules were on the surface, and no difference in reactivity was observed between live and permeabilized parasites (Fig. 5). On the other hand, a portion of gp82 molecules reacting with anti-J18 antibodies was intracellularly located, as indicated by the considerably increased reactivity when the parasites were permeabilized (Fig. 5). The most striking observation in the G strain was the reaction of anti-C03 antibodies with permeabilized metacyclic forms. These antibodies, which reacted with gp82 bands focusing at pH 4.9 to 5.5 (data not shown), barely recognized live parasites but reacted intensely with flagellar components of permeabilized cells (Fig. 5), which means that these are intraflagellar molecules. In order to determine whether the flagellar location of proteins recognized by anti-C03 antibodies was a characteristic of strains of group 1 T. cruzi, to which the G strain belongs (5), two other strains of this group were examined. The same profile of the G strain, with the predominant reaction with flagella, was revealed in permeabilized metacyclic forms of these strains (data not shown).
FIG. 5.
Cellular localization of proteins of the gp82 family in metacyclic forms of the T. cruzi G strain. Live or permeabilized parasites were reacted with MAb 3F6 or polyclonal antibodies to J18 or C03 and processed for analysis by flow cytometry or for visualization by fluorescence microscopy. Note the similar expression patterns of MAb 3F6-reactive molecules in live and permeabilized parasites, whereas the molecules recognized by anti-J18 antibodies were in part located intracellularly, and those reacting with anti-C03 antibodies were associated with the flagellum and did not react with live parasites.
Effect of different proteins and antibodies on host cell invasion by metacyclic trypomastigotes.
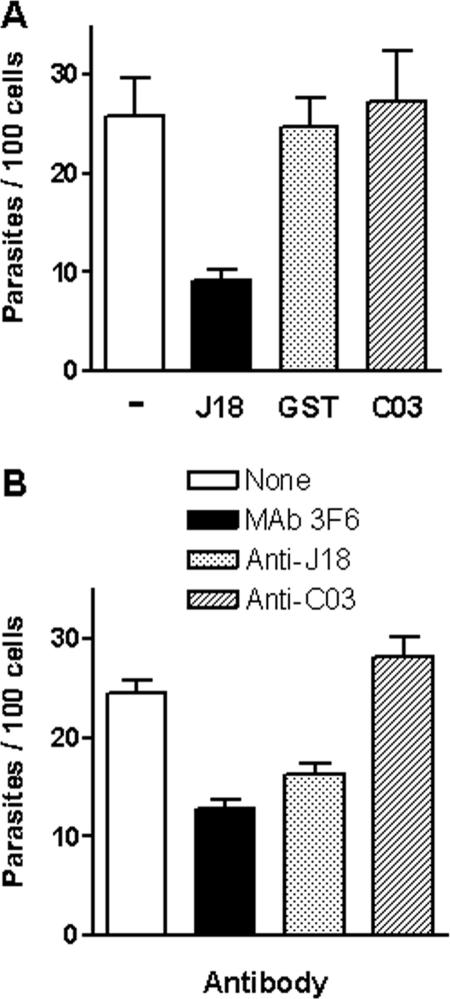
To test the effect of proteins and antibodies generated in this study, we used CL strain metacyclic forms and epithelial HeLa cells. Parasites were incubated with HeLa cells in the absence or presence of purified recombinant protein, J18, C03, or glutathione S-transferase, at 40 μg/ml for 1 h. In contrast to the J18 protein, which inhibited parasite internalization by ∼65%, C03 protein had no inhibitory effect (Fig. 6A). Why the C03 protein does not inhibit T. cruzi invasion even though it binds to host cells, albeit to a lesser extent than J18 (Fig. 2C), is not known. One possibility is that the C03 protein binds to a target receptor distinct from the J18 receptor, which recognizes a sequence different from that corresponding to P4/P8. We also examined the effect of anti-C03 and anti-J18 antibodies on HeLa cell invasion. Parasites were incubated for 30 min with anti-J18 or anti-C03 antiserum diluted 1:10 or with ascitic fluid containing MAb 3F6 diluted 1:10. After the antibody was washed out, the parasites were seeded onto HeLa cells. As shown in Fig. 6B, parasite entry was inhibited ∼50% by MAb 3F6 and ∼35% by anti-J18 antibodies, whereas anti-C03 antibodies showed no effect.
FIG. 6.
Effect of different proteins and antibodies on host cell invasion by T. cruzi metacyclic trypomastigotes (CL strain). (A) HeLa cells were incubated with parasites in the absence or presence of the indicated protein at 40 μg/ml. (B) Parasites were incubated with the indicated antibodies for 30 min. After antibodies were washed out, the parasites were incubated with HeLa cells. In both panels, the incubation time was 1 h, the cells were stained with Giemsa, and the numbers of intracellular parasites were counted in at least 500 cells. Values in panel A are the means ± standard deviation of three independent assays performed in duplicate. The difference between the control and the cells incubated with J18 protein was significant (P < 0.0005). In panel B the values are the means ± standard deviation of triplicates of one representative experiment. A significant difference between the control and the parasites incubated with MAb 3F6 (P < 0.0005) or with anti-J18 antibodies (P < 0.005) was found.
DISCUSSION
Our study provides novel information on the expression and cellular localization of gp82 family members in T. cruzi metacyclic trypomastigotes. This was achieved through the isolation of a cDNA clone encoding a member of the gp82 family distinct from the previously characterized cDNA clone selected for its reactivity with MAb 3F6 (1). From these cDNA clones encoding proteins with 59.1% identity, we generated recombinant proteins C03 and J18 as well as the corresponding antibodies (Fig. 1 and 3), which served to identify and localize the gp82 molecules in the parasites.
As a common characteristic, the members of the gp82 family recognized by MAb 3F6, anti-J18, or anti-C03 antibodies had isoelectric points at the acidic pH range, with the CL and G strain molecules focusing at pH 4.6 to 5.4 and at 4.9 to 5.7, respectively. Otherwise, the gp82 molecules reacting with any of these antibodies showed considerable differences, particularly in their cellular localization. We confirmed that the MAb 3F6-reactive molecules are present predominantly on the surface of metacyclic trypomastigotes of both the CL and G strains (Fig. 4 and 5). In T. cruzi strains that enter host cells in a gp82-mediated manner, the MAb 3F6-reactive molecules contribute to the invasion process (20), primarily by binding to the host cell surface molecules and triggering the activation of signal transduction pathways in both the parasite and target cells (32). On the other hand, the gp82 molecules recognized by anti-J18 antibodies appear to be expressed at low levels on the parasite surface and therefore may have a more marginal role, if any, in parasite internalization. In CL strain metacyclic trypomastigotes, which engage surface gp82 molecules to invade target cells (20, 22), the intensity of the reaction of live parasites with anti-J18 antibodies was much reduced compared to that with MAb 3F6 (Fig. 4). If MAb 3F6-reactive components are included in the gp82 repertoire identified by anti-J18 antibodies, then the gp82 population present on the surface that is specifically recognized by these polyclonal antibodies may be even smaller. Although to a lesser degree, the intensity of reaction of live metacyclic forms of G strain with anti-J18 antibodies was also found to be weaker than with MAb 3F6, but in permeabilized parasites the intensity of reaction was comparable to that with the MAb (Fig. 5). Taking these results together, we provide experimental evidence for the existence of several subpopulations among the gp82 metacyclic trypomastigote proteins, including those that are not targeted to the cell membrane.
What was particularly striking and unpredicted was the finding that members of the gp82 family may be localized to the flagellum in T. cruzi in a strain-dependent manner. That the profile of reactivity of metacyclic trypomastigotes with anti-C03 antibodies could be quite different from that with anti-J18 antibodies was expected, as they were elicited by proteins with considerable sequence diversity (Fig. 1). However, that gp82 family members could have such different cellular localizations and that this distribution varied depending on the T. cruzi strain (Fig. 4 and 5) came as a surprise. Anti-C03 antibodies did not react with live G strain metacyclic forms but reacted strongly with components associated with the flagellum in permeabilized parasites (Fig. 5), indicating that they recognize intraflagellar molecules. In the CL strain, the profile of the reaction of live as well as permeabilized parasites with anti-C03 antibodies was similar to that with anti-J18 antibodies, with no preferential association with flagellum being observed (Fig. 4).
Several T. cruzi molecules associated to flagellum have been previously described, among them a group of surface proteins of the 160-kDa family, belonging to the TS/gp85 superfamily and sharing 48% identity with gp85 proteins (25, 30). These proteins were detected on the cell membrane in the flagellar pocket of bloodstream trypomastigotes (28, 30). Comparison of C03 and Fl-160 amino acid sequences shows 29% identity, suggesting that they could share epitopes. Other proteins associated with T. cruzi flagellum are the flagellar calcium-binding protein (6), the glycoprotein gp72 (19), and a high-molecular-mass (300 kDa) protein built up mostly by nearly identical repeats of 68 amino acids arranged in tandem (11). The flagellar calcium-binding protein, which is a flagellum-specific calcium sensor, is associated with the flagellar membrane via its N-terminal myristate and palmitate moieties in a calcium-modulated, conformation-dependent manner (6). In epimastigotes, gp72 is predominantly membrane associated and located on the cell surface, being evenly distributed over the cell body and somewhat concentrated in the proximal region of the flagellum (14). The morphology of gp72-null mutants was found to be dramatically different from wild-type parasites, and the normal attachment of the flagellum to the cell membrane was lost (7). What role is played by intraflagellar gp82 molecules recognized by anti-C03 antibodies remains to be determined.
We also confirmed here the involvement of MAb 3F6-reactive gp82 molecules in target cell invasion. Purified J18 protein, as well as MAb 3F6 and anti-J18 antibodies, inhibited metacyclic trypomastigote entry into host cells, whereas C03 protein and anti-C03 antibodies showed no inhibitory effect, as expected (Fig. 6). This and the new data provided in this study contribute to our knowledge of T. cruzi gp82 family members, which may have functions other than interacting with target cells. Additionally, the differential cellular localization of gp82 molecules recognized by anti-C03 antibodies in G and CL strains (Fig. 4 and 5) further reminds us of the remarkable differences between these two strains that have been described in previous studies.
Acknowledgments
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo and Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Araya, J. E., M. I. Cano, N. Yoshida, N., and J. Franco da Silveira. 1994. Cloning and characterization of a gene for the stage-specific 82-kDa surface antigen of metacyclic trypomastigotes of Trypanosoma cruzi. Mol. Biochem. Parasitol. 65:161-169. [DOI] [PubMed] [Google Scholar]
- 2.Atwood, J. A., D. B. Weatherly, T. A Minning, B. Bundy, C. Cavola, F. R. Operdoes, R. Orlando, and R. L. Tarleton. 2005. The Trypanosoma cruzi proteome. Science 309:473-476. [DOI] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Brener, Z., and E. Chiari. 1963. Variações morfológicas observadas em diferentes amostras de Trypanosoma cruzi. Rev. Inst. Méd. Trop. São Paulo 5:220-224. [PubMed] [Google Scholar]
- 5.Briones, M. R. S., R. P. Souto, B. S. Stolf, and B. Zingales. 1999. The evolution of two Trypanosoma cruzi subgroups inferred from rRNA genes can be correlated with the interchange of American mammalian faunas in the Cenozoic and has implications to pathogenicity and host specificity. Mol. Biochem. Parasitol. 104:219-232. [DOI] [PubMed] [Google Scholar]
- 6.Buchanan, K. T., J. B. Ames, S. H. Asfaw, J. N. Wingard, C. L. Olson, P. T. Campana, A. P. Araújo, and D. M. Engman. 2005. A flagellum-specific calcium sensor. J. Biol. Chem. 280:40104-40111. [DOI] [PubMed] [Google Scholar]
- 7.Cooper R., A. R. de Jesus, and G. A. Cross. 1993. Deletion of an immunodominant Trypanosoma cruzi surface glycoprotein disrupts flagellum-cell adhesion. J. Cell Biol. 122:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortez, M., I. Neira, D. Ferreira, A. O. Luquetti, A. Rassi, and N. Yoshida. 2003. Infection by Trypanosoma cruzi metacyclic forms deficient in gp82 but expressing a related surface molecule, gp30. Infect. Immun. 71:6184-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortez, M., M. R. Silva, I. Neira, D. Ferreira, G. R. S. Sasso, A. O. Luquetti, A. Rassi, and N. Yoshida. 2006. Trypanosoma cruzi surface molecule gp90 downregulates invasion of gastric mucosal epithelium in orally infected mice. Microbes Infect. 8:36-44. [DOI] [PubMed] [Google Scholar]
- 10.Cortez, M., V. Atayde, and N. Yoshida. 2006. Host cell invasion mediated by Trypanosoma cruzi surface molecule gp82 is associated with F-actin disassembly and is inhibited by enteroinvasive Escherichia coli. Microbes Infect. 8:1502-1512. [DOI] [PubMed] [Google Scholar]
- 11.Cotrim, P. C., G. Paranhos-Baccala, M. R. Santos, C. Mertensen, M. I. Cano, M. Jolivet, M. E. Camargo, R. A. Mortara, and J. F. Silveira. 1995. Organization and expression of the gene encoding an immunodominant repetitive antigen associated to the cytoskeleton of Trypanosoma cruzi. Mol. Biochem. Parasitol. 71:89-98. [DOI] [PubMed] [Google Scholar]
- 12.Docampo, R., and S. J. Moreno. 1996. The role of Ca2+ in the process of cell invasion by intracellular parasites. Parasitol. Today 12:61-65. [DOI] [PubMed] [Google Scholar]
- 13.Frasch, A. A. C. 2000. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol. Today 16:282-286. [DOI] [PubMed] [Google Scholar]
- 14.Haynes, P. A., D. G. Russell, and G. A. Cross. 1996. Subcellular localization of Trypanosoma cruzi glycoprotein gp72. J. Cell Sci. 109:2979-2988. [DOI] [PubMed] [Google Scholar]
- 15.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manque, P. M., D. Eichinger, M. A. Juliano, L. Juliano, J. Araya, and N. Yoshida. 2000. Characterization of the cell adhesion site of Trypanosoma cruzi metacyclic stage surface glycoprotein gp82. Infect. Immun. 68:478-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neira, I., A. T. Ferreira, and N. Yoshida. 2002. Activation of distinct signal transduction pathways in Trypanosoma cruzi isolates with differential capacity to invade host cells. Int. J. Parasitol. 32:405-414. [DOI] [PubMed] [Google Scholar]
- 18.Neira, I., F. A. Silva, M. Cortez, and N. Yoshida. 2003. Involvement of Trypanosoma cruzi metacyclic trypomastigote surface molecule gp82 in adhesion to gastric mucin and invasion of epithelial cells. Infect. Immun. 71:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nozaki, T., and G. A. Cross. 1994. Functional complementation of glycoprotein 72 in a Trypanosoma cruzi glycoprotein 72 null mutant. Mol. Biochem. Parasitol. 67:91-102. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez, M. I., R. C. Ruiz, J. E. Araya, J. Franco da Silveira, and N. Yoshida. 1993. Involvement of the stage-specific 82-kilodalton adhesion molecule of Trypanosoma cruzi metacyclic trypomastigotes in host cell invasion. Infect. Immun. 61:3636-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez, A., M. G. Rioult, A. Ora, and N. N. Andrews. 1995. A trypanosome-soluble factor induces IP3 formation, intracellular Ca2+ mobilization and microfilament rearrangement in host cells. J. Cell Biol. 129:1263-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz, R. C., S. Favoreto, M. L. Dorta, M. E. M. Oshiro, A. T. Ferreira, P. M. Manque, and N. Yoshida. 1998. Infectivity of Trypanosoma cruzi strains is associated with differential expression of surface glycoproteins with differential Ca2+ signaling activity. Biochem. J. 330:505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santori, F. R., M. L. Dorta, L. Juliano, M. A. Juliano, J. Franco da Silveira, R. C. Ruiz, and N. Yoshida. 1996. Identification of a domain of Trypanosoma cruzi metacyclic trypomastigote surface molecule gp82 required for attachment and invasion of mammalian cells. Mol. Biochem. Parasitol. 78:209-216. [DOI] [PubMed] [Google Scholar]
- 24.Schenkman, S., M. A. J. Ferguson, N. Heise, M. L. Cardoso de Almeida, R. A. Mortara, and N. Yoshida. 1993. Mucin-like glycoproteins linked to the membrane by glycosylphosphatidylinositol anchor are the major acceptors of sialic acid in a reaction catalyzed by trans-sialidase in metacyclic forms of Trypanosoma cruzi. Mol. Biochem. Parasitol. 59:293-304. [DOI] [PubMed] [Google Scholar]
- 25.Takle, G. B., J. O'Connor, A. J. Young, and G. A. Cross. 1992. Sequence homology and absence of mRNA defines a possible pseudogene member of the gp85/sialidase multigene family. Mol. Biochem. Parasitol. 56:117-128. [DOI] [PubMed] [Google Scholar]
- 26.Tardieux, I., M. H. Nathanson, and N. W. Andrews. 1994. Role in host cell invasion of Trypanosoma cruzi-induced cytosolic free Ca2+ transients. J. Exp. Med. 179:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teixeira, M. M. G., and N. Yoshida. 1986. Stage-specific surface antigens of metacyclic trypomastigotes of Trypanosoma cruzi identified by monoclonal antibodies. Mol. Biochem. Parasitol. 18:271-282. [DOI] [PubMed] [Google Scholar]
- 28.Van Voorhis, W. C., and H. Eisen. 1989. Fl-160. A surface antigen of Trypanosoma cruzi that mimics mammalian nervous tissue. J. Exp. Med. 169:641-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walder, J. A., P. S. Eder, D. M. Engman, S. T. Brentano, R. Y. Walder, D. S. Knutzon, D. M. Dorfman, and J. E. Donelson. 1986. The 35-nucleotide spliced leader sequence is common to all trypanosome messenger RNAs. Science 233:569-571. [DOI] [PubMed] [Google Scholar]
- 30.W. C. Van Voorhis, L. Barrett, R. Koelling, and A. G. Farr. 1993. FL-160 proteins of Trypanosoma cruzi are expressed from a multigene family and contain two distinct epitopes that mimic nervous tissues. J. Exp. Med. 178:681-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, N. 1983. Surface antigens of metacyclic trypomastigotes of Trypanosoma cruzi. Infect. Immun. 40:836-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida, N. 2006. Molecular basis of mammalian cell invasion of Trypanosoma cruzi. An. Acad. Bras. Ciênc. 78:87-111. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida, N., R. A. Mortara, M. F. Araguth, J. C. Gonzalez, and M. Russo. 1989. Metacyclic neutralizing effect of monoclonal antibody 10D8 directed to the 35- and 50-kilodalton surface glycoconjugates of Trypanosoma cruzi. Infect. Immun. 57:1663-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida, N., S. Favoreto, Jr., A. T. Ferreira, and P. M. Manque. 2000. Signal transduction induced in Trypanosoma cruzi metacyclic trypomastigotes during the invasion of mammalian cells. Braz. J. Med. Biol. Res. 33:269-278. [DOI] [PubMed] [Google Scholar]