Abstract
Regulation of bacterial gene expression by small RNA (sRNA) molecules is an increasingly recognized phenomenon but one that is not yet fully understood. We show that the sRNA RyhB suppresses several virulence-associated phenotypes of Shigella dysenteriae, a causative agent of bacillary dysentery in humans. The virulence genes repressed by S. dysenteriae RyhB include those encoding the type III secretion apparatus, its secreted effectors, and specific chaperones. Suppression of Shigella virulence occurs via RyhB-dependent repression of the transcriptional activator VirB, leading to reduced expression of genes within the VirB regulon. Efficient repression of virB is mediated by a single-stranded region of RyhB that is distinct from the region required for repression of Shigella sodB. Regulation of virB by RyhB implicates iron as an environmental factor contributing to the complex regulation of Shigella virulence determinants.
Shigella species, which are closely related to Escherichia coli, cause bacillary dysentery, a disease associated with invasion of the colonic epithelium and provocation of an intense inflammatory response (12). Using cultured epithelial cells to measure bacterial invasion (invasion assay) (8) and intercellular spread (plaque assay) (22), investigators have identified a number of the genes required for Shigella pathogenesis. Many Shigella virulence-associated genes, including those encoding the type III secretion apparatus (mxi and spa genes), its secreted effectors (ipaA-D, ipgD, icsB, and virA), and specific chaperones (ipgC, ipgA, ipgE, and spa15), map to a 220-kbp virulence plasmid. Expression of these Shigella virulence determinants is highly regulated in response to environmental signals, such as temperature, osmolarity, and pH (12). This complex regulation is accomplished primarily by the transcriptional activators VirF and VirB (1). Transcription of many Shigella virulence-associated genes, including those encoding the type III secretion system (TTSS), is positively regulated by direct binding of VirB to the regulated promoters. Transcription of virB, in turn, is controlled primarily by the opposing activities of VirF, a transcriptional activator, and HNS, a transcriptional repressor. (For a review of the regulation of Shigella virulence gene expression, see reference 6.) VirF and VirB are both required to induce expression of virulence determinants that allow efficient invasion of epithelial cells by Shigella, an essential step in disease initiation. While the regulation of Shigella virulence has been the focus of intense investigation, the identification and characterization of all contributing environmental factors and regulatory elements have not yet been achieved.
Noncoding RNA molecules (ncRNAs) control diverse cellular functions in organisms ranging from bacteria to humans. Although it is known that ncRNAs employ a variety of mechanisms, including methylation of rRNA, inhibition of translation or transcription, and sequestration of regulatory proteins (11, 13, 21), the full spectrum of ncRNAs, their mechanism of action, and their impact on cellular activities remain to be determined.
One type of ncRNA is the regulatory small RNA (sRNA). sRNA molecules play an important role in the regulation of bacterial gene expression (33). One well-characterized sRNA is RyhB, which was first identified in E. coli and is involved in the iron-responsive regulation of genes required for metabolism and iron storage in this organism (17). Expression of E. coli ryhB is repressed by Fur, a global iron-responsive transcriptional repressor (17). Thus, ryhB is repressed in high-iron conditions and is derepressed in low-iron conditions. RyhB has been shown to decrease the stability of specific transcripts when, together with Hfq, it binds to a complementary nucleic acid sequence within the target mRNA molecule (17, 32). Subsequent degradation of both the target mRNA and RyhB is mediated by RNase E and RNase III (2, 16). In this report, we describe the RyhB-dependent regulation of Shigella virulence mediated by repression of virB, a gene encoding a virulence-associated transcriptional activator.
MATERIALS AND METHODS
Growth conditions.
Bacterial strains and plasmids used in this study are shown in Table 1. E. coli was cultured in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl) or on LB agar plates at 37°C. Shigella dysenteriae was cultured in LB broth, in modified M9 medium (26), or on tryptic soy broth (Becton, Dickinson and Company, Sparks, MD) agar plates containing 0.01% (wt/vol) Congo red at 37°C. Unless otherwise noted, antibiotics were used at the following final concentrations: carbenicillin, 250 μg/ml; chloramphenicol, 30 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | Life Technologies | |
| DH5α(λpir) | J. Kaper | |
| Shigella dysenteriae strains | ||
| O-4576S1 | 20 | |
| ND100 | Spontaneous Strr mutant of O-4576S1 | N. Davies |
| ND100ryhB | ryhB deletion | This study |
| ND100fur | fur deletion | This study |
| ND100fur,ryhB | fur and ryhB deletions | This study |
| Plasmids | ||
| pCVD442N2 | Suicide vector | E. Wyckoff |
| pQE-2 | Expression vector | QIAGEN |
| pryhB | ryhB in pQE-2 | This study |
| pryhBΔ | ryhBΔ in pQE-2 | This study |
| pAlt-leftryhB | Alt-leftryhB in pQE-2 | This study |
| pAlt-rightryhB | Alt-rightryhB in pQE-2 | This study |
Paraquat sensitivity assay.
A single colony was used to inoculate 3 ml of LB broth containing antibiotics, and the culture was grown to the stationary phase at 30°C. Ten microliters of the stationary-phase culture was used to inoculate 1 ml of modified M9 medium supplemented with FeSO4 (40 μM), isopropyl-β-d-thiogalactoside (IPTG) (200 μM), antibiotics, and paraquat (1 μM) where indicated. The optical density at 650 nm of each culture was measured following growth for 24 h at 37°C.
Microarray analysis.
Synthetic oligonucleotides specific for each gene in the E. coli K-12 and enterohemorrhagic E. coli genomes were purchased from QIAGEN (Valencia, CA). Additionally, oligonucleotides specific for genes located on the Shigella flexneri virulence plasmid, several S. dysenteriae-specific genes, and putative sRNA sequences (10) were designed and synthesized by QIAGEN. Glass microscope slides were coated with polylysine (www.microarray.org) prior to spotting of each oligonucleotide using a robotic arrayer and Array Maker 2.4 (MGuide at smgm.stanford.edu/pbrown/mguide). Hydration and postprocessing were performed as described by MGuide. RNA was isolated from S. dysenteriae containing the IPTG-inducible ryhB gene grown to the mid-logarithmic phase in the presence or absence of IPTG using an RNeasy midi kit (QIAGEN) prior to cDNA generation and labeling as follows. Fifteen micrograms of RNA was combined with 5 μg of oligonucleotide PdN6 and incubated for 10 min at 65°C, followed by incubation on ice for 10 min. The RNA was then incubated for 1 h at 42°C in the presence of 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dGTP, 0.2 mM dTTP, 0.3 mM amino allyl-modified dUTP (Sigma-Aldrich, St. Louis, MO), 0.01 M dithiothreitol, 120 U RNasin (Promega, Madison, WI), 800 U SuperScript II reverse transcriptase, and 1X SuperScriptII RT buffer (Invitrogen, Carlsbad, CA). Following this incubation, 400 U of SuperScriptII reverse transcriptase was added, and the reaction mixture was incubated at 42°C for an additional hour. Next, the cDNA solution was incubated for 5 min at 65°C, snap cooled on ice, and hydrolyzed by addition of 50 mM NaOH. Following incubation at 65°C for 15 min, the reaction mixture was neutralized by addition of 0.6 M HEPES (pH 7.5), bringing the final volume to 85 μl. The modified cDNA was washed and concentrated to 9 μl using a MinElute PCR purification kit (QIAGEN) and was coupled to Cy3 or Cy5 dye (Amersham Biosciences, Little Chalfont, United Kingdom) as follows. Concentrated cDNA samples were combined with desiccated dye following addition of 1 μl of 1 M sodium bicarbonate to a dye pellet (pH 10.0) and incubated in the dark at room temperature for 60 min. The Cy3- and Cy5-labeled cDNAs were washed and concentrated to 32 μl each using the MinElute PCR purification kit (QIAGEN). The labeled cDNA samples were combined with 3× SSC and 0.25% sodium dodecyl sulfate (SDS) prior to hybridization on a microarray slide for 6 h at 65°C (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Following hybridization, each slide was washed once in 0.6× SSC with 0.03% SDS and twice in 0.06× SSC. Microarrays were dried by centrifugation and were scanned using a Genepix Array Scanner 4000A (Axon Instruments, Union City, CA). Data were analyzed using Genepix 5.0 and were normalized using the Longhorn Array Database (powered by the Stanford Microarray Database).
Secreted protein analysis.
Bacterial proteins secreted into the culture supernatant were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (26) following trichloroacetic acid precipitation as follows. Each strain was grown at 37°C to an optical density at 650 nm of 0.9 in LB broth supplemented with 250 μg/ml carbenicillin, 200 μM IPTG, and 0.1% (wt/vol) deoxycholate. Cultures were centrifuged (2 min at 16,000 × g), and 900 μl of each supernatant was added to 100 μl of 100% trichloroacetic acid and incubated overnight at −80°C. Precipitated proteins were harvested by centrifugation (15 min at 16,000 × g at 4°C) and washed in 300 μl cold acetone. The samples were centrifuged again as described above, each supernatant was removed, and the pellets were dried. The proteins were solubilized in 200 μl protein solubilization buffer (26) and boiled for 10 min. The proteins present in 10 μl of each sample were separated on a 7.5% SDS-polyacrylamide gel and stained with Coomassie blue (26).
Tissue culture.
Henle cell monolayers were cultured in six-well polystyrene tissue culture plates (Corning Inc. Costar, Corning, NY) in Gibco minimum essential medium (MEM) (Invitrogen Corp.) supplemented with 10% fetal bovine serum and 2 mM glutamine. The plates were incubated at 37°C in an atmosphere containing 5% CO2.
The plaque assay procedure was a modification of the procedure of Oaks et al. (22). Briefly, a single Congo red-positive colony was used to inoculate a 3-ml LB broth culture containing antibiotics and grown overnight at 30°C. The culture was diluted 1:100 into LB broth containing antibiotics, 0.1% deoxycholate, and IPTG (200 μM or the indicated concentration) and grown to the mid-logarithmic phase at 37°C. The bacteria were diluted in phosphate-buffered saline (PBS), and 104 bacteria were added to Henle cell monolayers in six-well tissue culture plates containing 2 ml MEM supplemented with 250 μg/ml carbenicillin and 200 μM IPTG per well. The plates were centrifuged for 10 min at 669 × g, incubated at 37°C for 1.5 h, washed with 2 ml PBS, and then overlaid with 2 ml MEM supplemented with 0.3% glucose, 250 μg/ml carbenicillin, 200 μM IPTG, and 20 μg/ml gentamicin. Following incubation for 72 h at 37°C, the plates were washed with PBS and stained with Wright-Giemsa stain (Camco, Ft. Lauderdale, FL).
Invasion assays were performed like the plaque assay, with the following modifications. The monolayers were infected with 2 × 108 bacteria per well and incubated for 30 min prior to washing and addition of gentamicin. The monolayers were stained with Wright-Giemsa stain (Camco) after 2 h (total time) of incubation, and cells were scored positive for invasion if they contained three or more bacteria, as determined by microscopy.
Cloning of wild-type ryhB and the altered ryhB molecules.
Wild-type ryhB was amplified by PCR using oligonucleotides ryhB-14EcoRI and ryhB-5HindIII. The PCR conditions were denaturation for 30 s at 95°C, annealing for 45 s at 50°C, and extension for 30 s at 72°C for 30 cycles in a Peltier thermal cycler (MJ Research, Watertown, MA). The resulting 183-bp product was purified using the MinElute PCR purification kit (QIAGEN), digested with the EcoRI and HindIII restriction endonucleases, and ligated into the MfeI and HindIII restriction endonuclease recognition sites of pQE-2 (QIAGEN), which carries lacI. In the resulting plasmid, pryhB, expression of ryhB is under the control of the IPTG-inducible T5 promoter.
Oligonucleotides encoding the entire functional molecule were used to clone ryhBΔ (ryhBsodB1 and ryhBsodB2), Alt-rightryhB (altrightryhB1 and altrightryhB2), and Alt-leftryhB (tfelryhB1 and tfelryhB2) behind the inducible T5 promoter of pQE-2. Each oligonucleotide was diluted to obtain a concentration of 100 mM in STE (10 mM Tris-Cl [pH 8.0], 0.1 M NaCl, 1 mM EDTA; pH 8.0). Five microliters of each complementary oligonucleotide solution was combined and boiled for 1 min. The solution was slowly cooled to room temperature prior to 1:10 dilution in STE. The annealed primer products were cloned directly into pQE-2 digested with both the MfeI and HindIII restriction endonucleases, thereby placing the altered ryhB gene under the control of the IPTG-inducible T5 promoter. The resulting plasmids were designated pryhBΔ, pAlt-rightryhB, and pAlt-leftryhB, respectively. The nucleic acid sequences of all oligonucleotides used for cloning and real-time PCR, described below, are available upon request.
Real-time PCR.
RNA was isolated using an RNeasy midi kit (QIAGEN) according to the product directions from bacteria cultured for approximately 20 h at 37°C on tryptic soy broth agar plates supplemented with 0.01% Congo red, 200 μg/ml carbenicillin, and IPTG (200 μM or the indicated concentration). Each RNA sample was then treated with 16 U of amplification grade DNase I (Invitrogen), ethanol precipitated, and dried. The RNA pellet was resuspended in diethyl pyrocarbonate-treated water, and the nucleic acid was quantitated using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). No more than 10 μg of total RNA was used to generate cDNA with a High Capacity cDNA archival kit (Applied Biosystems, Foster City, CA) according to the product directions. Each cDNA sample was diluted 1:10 in water, and 2.5 μl was used as the template for each 25-μl reaction mixture. All probes were minor groove binding, 6-carboxyfluorescein-labeled probes. Probes and oligonucleotides were designed using Primer Express (Applied Biosystems, Foster City, CA) and were synthesized by Applied Biosystems. TaqMan universal master mixture (Applied Biosystems) was used for all reactions. rrsA was used as the normalizer for each sample. Reactions were performed in a 7300 real-time PCR system (Applied Biosystems) under standard reaction conditions.
Construction of ryhB, fur, and fur ryhB deletion strains.
S. dysenteriae ryhB was replaced with a chloramphenicol resistance cassette as described previously for S. flexneri (23). S. dysenteriae fur was replaced with a kanamycin resistance cassette by splice overlap (26), using oligonucleotides Sdfur-1, Sdfur-2, Sdfur-3, and Sdfur-4.
RESULTS
RyhB represses the expression of several genes within the S. dysenteriae VirB regulon.
To identify RyhB-regulated genes in S. dysenteriae, the expression profile of wild-type S. dysenteriae was compared to that of S. dysenteriae expressing ryhB from an IPTG-inducible plasmid promoter (pryhB) by microarray analysis. Based on the known mechanism of RyhB regulation, increasing the level of RyhB was predicted to result in decreased levels of target mRNA molecules. Although Shigella ryhB expression is regulated by Fur and iron levels (23), these factors also influence the expression of many other genes. Thus, introducing a plasmid encoding ryhB under the control of an inducible promoter (pryhB) into wild-type S. dysenteriae provided consistent, reproducible levels of RyhB.
Induction of ryhB resulted in reduced mRNA levels for several genes, including the known E. coli RyhB target gene sodB, the expression of which was reduced 7.7-fold in the presence of increase levels of RyhB (Table 2). To confirm that the reduced expression of sodB resulted in phenotypic differences, paraquat sensitivity was measured. Repression of E. coli sodB has been shown to result in increased sensitivity to paraquat (4). Therefore, if, as predicted by microarray analysis, RyhB represses the expression of S. dysenteriae sodB, an increase in sensitivity to paraquat would be expected upon induced expression of ryhB. Induced expression of ryhB resulted in significant growth inhibition of wild-type S. dysenteriae in the presence of 1 μM paraquat compared to the growth of the strain carrying the vector control or to the growth of either strain grown in the absence of paraquat (Table 3). These data confirm the RyhB-dependent repression of sodB measured in the microarray analysis.
TABLE 2.
Effect of RyhB on the expression of virulence-associated genes in S. dysenteriae
| Gene | Function | Fold repression upon ryhB expressiona |
|---|---|---|
| sodB | Superoxide dismutase | 7.7 |
| ipaC | Secretion apparatus | 6.6 |
| ipgC | Chaperone | 5.8 |
| ipgB | Secreted effector | 4.6 |
| ipaA | Secreted effector | 4.2 |
| icsB | Secreted effector | 4.2 |
| ipgA | Chaperone | 4.1 |
| mxiM | Secretion apparatus | 3.6 |
| mxiD | Secretion apparatus | 3.2 |
| mxiL | Secretion apparatus | 3.1 |
| mxiE | Secretion apparatus | 3.1 |
| ipgE | Chaperone | 3.0 |
| spa15 | Chaperone | 2.8 |
| ipaD | Secreted effector | 2.8 |
| mxiA | Secretion apparatus | 2.7 |
| mxiC | Secretion apparatus | 2.4 |
| mxiJ | Secretion apparatus | 2.1 |
Representative data from a microarray analysis comparing the mRNA produced by wild-type S. dysenteriae carrying pryhB grown in the presence of IPTG to that produced when the organism was grown in the absence of IPTG.
TABLE 3.
Effect of ryhB expression on paraquat sensitivity of S. dysenteriae
| Strain | Plasmid | Growth (optical density at 650 nm)a
|
|
|---|---|---|---|
| Without paraquat | With paraquat | ||
| ND100 (wild type) | Vector | 1.16 ± 0.03 | 0.88 ± 0.07 |
| ND100 (wild type) | pryhB | 0.87 ± 0.03 | 0.24 ± 0.03b |
| ND100fur | Vector | 0.62 ± 0.03 | 0.37 ± 0.02b |
| ND100fur/ryhB | Vector | 1.18 ± 0.02 | 1.02 ± 0.07 |
| ND100ryhB | Vector | 1.12 ± 0.03 | 1.06 ± 0.07 |
An endpoint growth analysis of S. dysenteriae wild-type strain ND100, S. dysenteriae fur mutant ND100fur, S. dysenteriae fur ryhB double mutant ND100fur/ryhB, and S. dysenteriae ryhB mutant ND100ryhB containing the vector control or pryhB in the presence or in the absence of 1 μM paraquat was performed. The data are the averages of three independent experiments. All strains were cultured in the presence of 200 μM IPTG to induce expression of ryhB from pryhB.
Significantly different than the growth of the wild-type strain in the presence of paraquat (P ≤ 0.004).
In addition to sodB, induction of ryhB expression resulted in reduced mRNA levels of several genes within the VirB regulon as measured by microarray analysis. Specifically, the levels of mRNA corresponding to ipaACD, ipgABCE, mxiACDEJL, and virA were reduced between 2.1- and 6.6-fold in the presence of increased levels of RyhB (Table 2). The microarray results indicating reduced expression of genes within the VirB regulon were validated by real-time PCR analysis of one of these virulence genes, mxiE, measured in the presence and in the absence of induced ryhB expression (Fig. 1A). To confirm that the reduced expression of genes within the VirB regulon resulted in phenotypic changes, the amount of secreted Ipa was determined in supernatants of wild-type S. dysenteriae cultured in the presence of deoxycholate, with and without induced expression of ryhB. When Shigella is grown in the presence of deoxycholate, the majority of the proteins present in the culture supernatant are encoded by genes within the VirB regulon (24). These proteins are secreted by the TTSS, the components of which are also encoded by genes within the VirB regulon. Therefore, if, as predicted by microarray analysis, RyhB represses the expression of genes within the VirB regulon, a marked reduction in the prevalence of secreted proteins in the S. dysenteriae supernatant would be expected upon induced expression of ryhB. This prediction was confirmed by SDS-PAGE analysis of the proteins secreted by wild-type S. dysenteriae in the presence and in the absence of induced ryhB expression (Fig. 1B). Western blot analysis with a monoclonal antibody against IpaC confirmed that this VirB-regulated effector protein was among the proteins whose level in the culture supernatant was reduced upon induced expression of ryhB (data not shown). These data are consistent with the RyhB-dependent repression of genes within the VirB regulon as measured by microarray analysis.
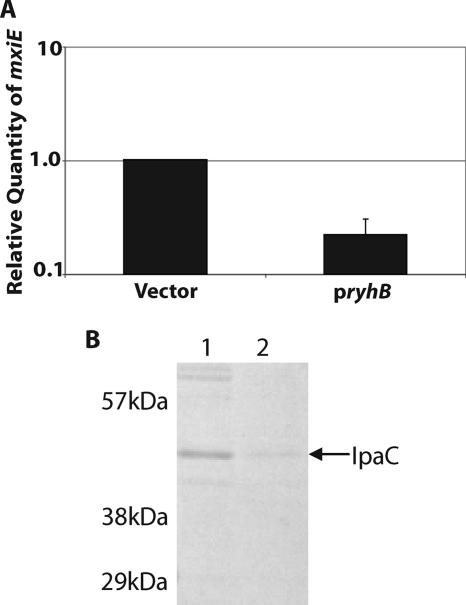
FIG. 1.
Induced expression of S. dysenteriae ryhB results in increased sensitivity to paraquat and decreased secretion of Ipa protein. (A) Real-time PCR analysis of mxiE mRNA levels in S. dysenteriae wild-type strain ND100 carrying the vector control and pryhB. All values were normalized to the level of rrsA mRNA in each sample, and the results are expressed relative to the value obtained for the strain carrying the vector control. The data are the averages of three independent experiments, and the error bars represent one standard deviation. (B) SDS-PAGE analysis of proteins secreted by an equivalent number of S. dysenteriae wild-type strain ND100 cells containing the vector control (lane 1) or pryhB (lane 2). The arrow indicates the location of IpaC as determined by Western blotting. All assays were carried out following growth of each strain in the presence of 200 μM IPTG to induce expression of ryhB from pryhB.
RyhB suppresses plaque formation by inhibiting S. dysenteriae invasion of eukaryotic cells.
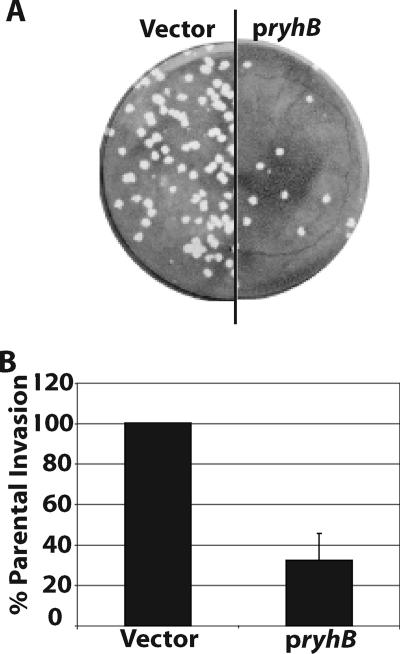
Since expression of genes within the VirB regulon is associated with Shigella virulence (12), analysis of the invasion of eukaryotic cell monolayers and plaque formation by wild-type S. dysenteriae was performed as a means of assessing the effect of RyhB on these virulence-associated phenotypes (8, 22). Induced expression of ryhB from pryhB in wild-type S. dysenteriae severely inhibited plaque formation in epithelial cell monolayers compared to the plaque formation by the strain containing the vector control (Fig. 2A) and compared to the plaque formation by both strains cultured under noninducing conditions (data not shown). When cultured under inducing conditions, the wild-type strain carrying the vector control produced an average of 150 ± 26 plaques, while the wild-type strain carrying pryhB produced an average of 30 ± 12 plaques.
FIG. 2.
RyhB suppresses virulence-associated phenotypes of S. dysenteriae. (A) Plaques formed in a Henle cell monolayer infected with an equal number of S. dysenteriae wild-type strain ND100 cells carrying either the vector control or pryhB. (B) Invasion efficiencies of S. dysenteriae wild-type strain ND100 carrying pryhB and the strain carrying the vector control. The invasion efficiency is expressed relative to that of the strain carrying the vector control, which was defined as 100%. The data are the averages of five independent experiments, and the error bars represent one standard deviation. All assays were carried out following growth of each strain in the presence of 200 μM IPTG to induce expression of ryhB from pryhB.
A reduced ability to form plaques can result from any one of multiple defects in the Shigella life cycle, including an inability to invade the eukaryotic cell, a lack of bacterial growth within the eukaryotic cell, or a defect in cell-to-cell spread. Invasion assays were used to determine the percentages of infected Henle cells following infection with wild-type S. dysenteriae in the presence and in the absence of induced ryhB expression. Expression of ryhB from pryhB significantly reduced the invasion efficiency of wild-type S. dysenteriae compared to that of the strain containing the vector control (Fig. 2B) or compared to that of either strain cultured under noninducing conditions (data not shown). Taken together, these data are consistent with the microarray analysis and support the model that RyhB represses the expression of one or more virulence-associated genes in S. dysenteriae.
Inactivation of fur results in increased expression of ryhB and inhibition of plaque formation by S. dysenteriae.
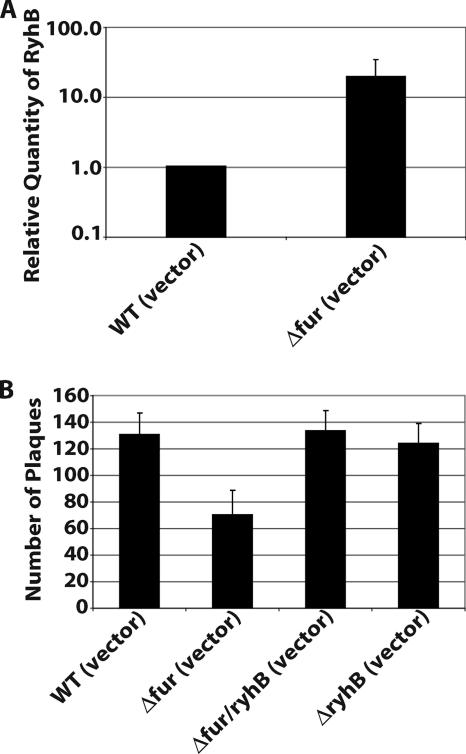
Expression of ryhB is regulated by Fur in response to environmental iron levels such that ryhB is expressed when iron levels are low and Fur is inactive (18, 23). To confirm that the observed regulation of virulence-associated genes by RyhB occurs under conditions normally encountered when Fur repression is relieved, a fur mutant of S. dysenteriae was constructed (ND100fur) and characterized with respect to both ryhB expression and virulence-associated phenotypes. Quantification of RyhB in the S. dysenteriae fur mutant strain by real-time PCR confirmed that loss of Fur resulted in increased expression (19-fold) of ryhB compared to the expression in the wild-type strain (Fig. 3A). Characterization of the S. dysenteriae fur mutant allowed analysis of the effects of ryhB expression from the chromosome at levels that would be expected to occur under conditions where Fur is not active. Since Fur regulates a large number of genes in Shigella (23), a ryhB mutation was introduced into the S. dysenteriae fur mutant (ND100fur/ryhB) and used to confirm that phenotypes observed in the fur mutant are a result of ryhB overexpression and are not the result of aberrant expression of other Fur-regulated genes.
FIG. 3.
Inactivation of S. dysenteriae fur results in increased expression of ryhB and inhibition of plaque formation. (A) Real-time PCR analysis of RyhB levels in S. dysenteriae wild-type strain ND100 (WT) and S. dysenteriae fur mutant strain ND100fur, each carrying the vector control. All values were normalized to the level of rrsA mRNA in each sample, and the results are expressed relative to the value obtained for the wild-type strain. The data are the averages of four independent experiments, and the error bars represent one standard deviation. (B) Numbers of plaques formed in Henle cell monolayers infected with equal numbers of cells of S. dysenteriae wild-type strain ND100, S. dysenteriae fur mutant ND100fur, S. dysenteriae fur ryhB double mutant ND100fur/ryhB, and S. dysenteriae ryhB mutant strain ND100ryhB, each carrying the vector control. The data are the averages of four independent experiments, and the error bars represent one standard deviation.
Paraquat sensitivity of the fur mutant was measured and compared with the results obtained following induced expression of ryhB in wild-type S. dysenteriae. Although to a lesser extent, the increased sensitivity to paraquat of the fur mutant compared to that of the wild-type strain (Table 3) is similar to the sensitivity observed upon expression of ryhB from an inducible plasmid promoter (Table 3). Inactivation of ryhB in the fur mutant restored paraquat resistance to wild-type levels, while inactivation of ryhB alone had no significant effect (Table 3), indicating that the increased paraquat sensitivity of the fur mutant results from increased expression of ryhB.
Plaque formation by S. dysenteriae was decreased significantly in the fur mutant compared to plaque formation by the wild-type strain (Fig. 3B), although to a lesser extent than was observed upon expression of ryhB from the inducible promoter of pryhB (Fig. 2A). Inactivation of ryhB in the S. dysenteriae fur mutant restored plaque formation to wild-type levels (Fig. 3B), while inactivation of ryhB alone had no significant effect (Fig. 3B). These data are consistent with the conclusion that RyhB inhibits the expression of one or more genes required for plaque formation by S. dysenteriae and that this regulation occurs under conditions normally encountered when Fur repression is relieved.
Inhibition of plaque formation is directly related to the level of ryhB expression.
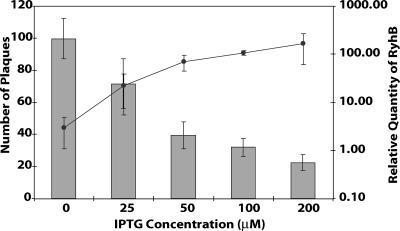
The amount of RyhB produced in the S. dysenteriae ryhB mutant strain carrying pryhB can be controlled by altering the amount of inducer added, allowing analysis of the biological effects of ryhB expression at physiological levels without the pleiotropic effects of fur inactivation. The amount of RyhB produced by the ryhB mutant strain carrying pryhB was proportional to the concentration of inducer added, as determined by real-time PCR (Fig. 4). The level of RyhB synthesized in response to IPTG also correlated with the degree of inhibition of plaque formation (Fig. 4). Addition of 25 μM IPTG induced levels of RyhB very similar to the RyhB levels produced upon fur inactivation (compare Fig. 3A and 4) and inhibited plaque formation to the same extent (compare Fig. 3B and 4). High concentrations of inducer had no intrinsic detrimental effects on plaque formation, as demonstrated by the wild-type level of plaque formation seen in the S. dysenteriae ryhB mutant strain carrying the vector control (data not shown). These data further confirm that inhibition of plaque formation by S. dysenteriae is a direct result of the activity of RyhB and that the phenotypes of the S. dysenteriae fur mutant can be reproduced by expression of ryhB from the inducible plasmid promoter of pryhB.
FIG. 4.
Increased expression of ryhB results in increased inhibition of plaque formation by S. dysenteriae. Real-time PCR analysis of RyhB levels in S. dysenteriae ryhB mutant strain ND100ryhB carrying pryhB cultured in the presence of different concentrations of IPTG was performed (line). All values were normalized to the level of rrsA mRNA in each sample, and the results are expressed relative to the value obtained for wild-type S. dysenteriae carrying the vector control. The numbers of plaques formed in Henle cell monolayers infected with equal numbers of cells of S. dysenteriae ryhB mutant ND100ryhB carrying pryhB were also determined (bars). Plaque assays were carried out in the presence of, and following the growth of, cultures with the indicated concentrations of IPTG. All data are the averages of three independent experiments, and the error bars represent one standard deviation.
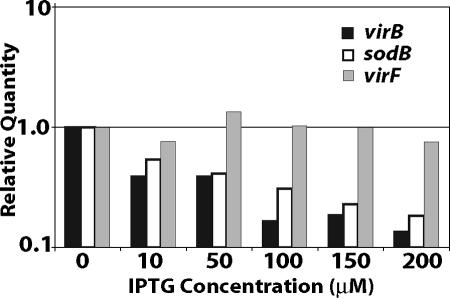
RyhB inhibits the expression of virB.
While virF has not been annotated in the S. dysenteriae strains sequenced to date (34, 35), the presence of virF in the clinical isolate used in this study was confirmed by PCR and DNA sequencing analysis (data not shown). Therefore, reduced Ipa secretion and decreased plaque formation and invasion efficiency upon increased ryhB expression are consistent with RyhB repression of either virF or virB. Increasing levels of ryhB expression from an inducible promoter resulted in a corresponding reduction in the level of virB mRNA, as determined by real-time PCR (Fig. 5). A similar reduction in the level of sodB mRNA was also observed, but virF expression was not affected (Fig. 5). These data indicate that RyhB represses the expression of virB either directly or via a pathway independent of VirF, the primary transcriptional activator of virB.
FIG. 5.
RyhB represses the expression of S. dysenteriae virB. Real-time PCR analysis of virB mRNA, virF mRNA, and sodB mRNA levels in S. dysenteriae ryhB mutant strain ND100ryhB carrying pryhB was performed. ND100ryhB (pryhB) was cultured in the presence of the indicated concentrations of IPTG. All values were normalized to the level of rrsA mRNA in each sample, and the results are expressed relative to the value obtained following growth in the absence of IPTG.
RyhB-dependent suppression of S. dysenteriae virulence-associated phenotypes is mediated by a specific nucleic acid sequence within RyhB.
RyhB-dependent repression of E. coli sodB is mediated by a sequence-specific interaction between the central loop of RyhB and sodB mRNA (17, 32). E. coli sodB is the only target to date for which the specific RyhB sequence required to mediate repression has been experimentally determined (32). Deletion of the central loop of RyhB (RyhBΔ) abolishes the ability of RyhB to repress the expression of sodB (32). To investigate and compare the ability of RyhBΔ to repress S. dysenteriae sodB and Shigella virulence-associated phenotypes, paraquat sensitivity and plaque assays were carried out following expression of ryhBΔ from an inducible plasmid promoter (pryhBΔ) (Fig. 6A). To avoid interference from wild-type RyhB, expression of ryhBΔ and other ryhB genes with an altered nucleic acid sequence was carried out in a ryhB deletion strain of S. dysenteriae. Consistent with the previously reported inability of RyhBΔ to repress sodB in E. coli (32), expression of S. dysenteriae ryhBΔ did not result in increased sensitivity to paraquat (Fig. 6B). In contrast, overexpression of ryhBΔ inhibited plaque formation by S. dysenteriae (Fig. 6C), indicating that the altered RyhB molecule retains the ability to suppress this virulence-associated phenotype. These data indicate that suppression of Shigella plaque formation is mediated by a region of RyhB that is distinct from the region required for repression of sodB.
FIG. 6.
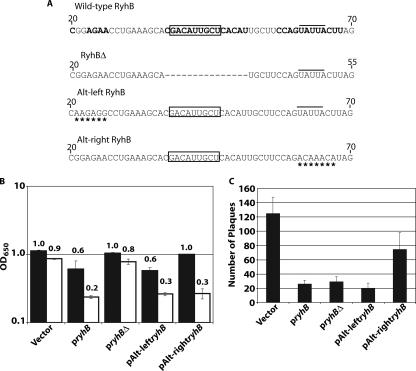
Repression of virB is mediated by a portion of RyhB distinct from the portion required for repression of sodB. (A) Partial sequences of wild-type, RyhBΔ, and altered RyhB molecules. Regions of wild-type RyhB predicted both experimentally (7) and by computer analysis (17) to be single stranded are in bold type. The RyhB sequence known to mediate repression of sodB is boxed (17, 32). The dashed line indicates the nucleic acid sequence deleted from wild-type ryhB to form ryhBΔ, while a line above the sequence indicates the Hfq binding site (7). Asterisks indicate nucleic acids that are altered in Alt-leftRyhB and Alt-rightRyhB. (B) Endpoint growth analysis of S. dysenteriae ryhB mutant strain ND100ryhB carrying the vector control, pryhB, pryhBΔ, pAlt-leftryhB, or pAlt-rightryhB in the absence (solid bars) or in the presence (open bars) of paraquat. The data are the averages of three independent experiments, and each the bars represent one standard deviation. OD650, optical density at 650 nm. (C) Numbers of plaques formed in Henle cell monolayers infected with an equal number of cells of S. dysenteriae ryhB mutant strain ND100ryhB carrying the vector control, pryhB, pryhBΔ, pAlt-leftryhB, or pAlt-rightryhB. The data are the averages of four independent experiments, and the error bars represent one standard deviation. All assays were carried out following growth of each strain in the presence of 200 μM IPTG to induce expression of the indicated ryhB gene.
To determine the sequence within RyhB responsible for efficient repression of Shigella virulence-associated phenotypes, regions of RyhB predicted both experimentally (7) and by computer analysis (17) to be single stranded or partially single stranded were systematically altered (Fig. 6A). Each altered ryhB was cloned under the control of an inducible promoter, and the ability of the altered RyhB molecule to affect sodB function and virulence properties of Shigella was investigated using paraquat sensitivity and plaque assays, respectively.
An altered RyhB molecule containing changes in the nucleic acid sequence in the predicted partially single-stranded region between bases 21 and 26 was constructed. The altered RyhB molecule (Alt-leftRyhB) retained the ability to suppress paraquat resistance (Fig. 6B) and to decrease plaque formation (Fig. 6C) compared to the properties of the strain carrying the vector control. A second altered RyhB containing changes in the nucleic acid sequence between bases 61 and 67 (Alt-rightRyhB) also retained the ability to suppress paraquat resistance (Fig. 6b) but lost the ability to inhibit plaque formation (Fig. 6C). These data indicate that the predicted single-stranded region between bases 61 and 67 of RyhB mediates the suppression of plaque formation by Shigella but is not required for the repression of sodB. While the nucleic acid changes between bases 61 and 67 are within the Hfq binding site of RyhB (7), the changes retain the predicted structure and the AU-rich nature of the region. The increase in paraquat sensitivity, indicating repression of sodB by Alt-rightRyhB (Fig. 6B), suggests that the interaction with Hfq essential for RyhB-dependent repression of sodB (7, 17, 32) is not disrupted. These data demonstrate that the specific nucleic acid sequence between bases 61 and 67 of RyhB, but not the nucleic acid sequence between bases 21 and 26, is critical for repression of plaque formation by S. dysenteriae.
DISCUSSION
Although RyhB has been shown to regulate important cellular functions, such as metabolism and iron storage in E. coli (17) and biofilm formation in Vibrio cholerae (19), this small regulatory RNA had not been implicated in the direct regulation of bacterial virulence determinants previously. In this report, we demonstrate that several virulence-associated phenotypes of S. dysenteriae, including effector protein secretion, plaque formation, and invasion of eukaryotic epithelial cells, are suppressed by RyhB and that this suppression is due to RyhB-dependent repression of virB.
The molecular mechanism of RyhB-dependent repression has been detailed only for E. coli sodB and has been shown to require complementarity between the sodB mRNA and RyhB (16, 17, 32). While no significant complementarity between RyhB and the virB mRNA was found, it is possible that limited complementarity, not identified in our screen, between these RNA molecules is sufficient to mediate repression or that the RyhB-dependent repression of virB is indirect via a pathway excluding VirF.
As shown for E. coli sodB (7, 17, 32), RyhB-mediated repression of S. dysenteriae sodB requires the RNA binding protein Hfq (data not shown); however, the role of Hfq in the RyhB-mediated repression of virB remains unknown. Inactivation of S. dysenteriae hfq (data not shown) and S. flexneri hfq (29) resulted in decreased expression of genes within the VirB regulon independent of RyhB. Thus, it was not possible to determine the effect of Hfq on RyhB-dependent regulation of virulence gene expression.
Expression of Shigella virulence determinants is regulated in response to environmental conditions such as temperature, osmolarity, and pH, primarily by the activity of the transcriptional activators VirB and VirF and the transcriptional repressor HNS (1, 31). Expression of several Shigella virulence-associated genes is activated following binding of VirB to the regulated promoters. Expression of virB in turn is controlled primarily by the opposing activities of the transcriptional activator VirF and the transcriptional repressor HNS (for a review, see reference 6). Since ryhB is regulated by iron and Fur (17, 23), iron has now been identified as an environmental factor contributing to the regulation of Shigella virulence determinants. Thus, the role of RyhB in this complex regulatory pathway may be to coordinate the expression of Shigella virulence factors with the local concentration of environmental iron. In the relatively iron-rich environment of the gut, S. dysenteriae Fur would repress the expression of ryhB, ensuring maximal expression of virB and, consequently, genes within the VirB regulon. It is during this initial stage of infection that proteins encoded by genes within the VirB regulon are required for invasion of the eukaryotic cell. Once the bacterium is intracellular, repression of the S. dysenteriae VirB regulon would be advantageous to the bacterium since aberrant expression of the systems could result in premature lysis of the eukaryotic cell. Analysis of proteins produced by Shigella growing in the intracellular environment has shown that there is a decrease in the amount of IpaB and IpaC during the initial phase of intracellular growth (9). Additionally, Lucchini et al. (15) showed that there was significant down-regulation of the ipa, spa, and mxi genes during intracellular replication of Shigella. It is during this stage of infection that expression of ryhB is expected to increase due to the lack of Fur repression in the less iron-replete intracellular environment. While the intracellular iron concentration is not low enough to result in derepression of all Fur-regulated genes, the iron limitation is sufficient to induce expression of a subset of Fur-responsive genes in Shigella (25). Thus, RyhB may provide a mechanism to quickly repress the expression of virB and allow progression of the disease. As the bacteria multiply within the epithelial cell and begin the process of intracellular spread, environmental signals other than iron may diminish or relieve RyhB repression and allow the transient expression of TTSS genes needed for transit to the adjacent eukaryotic cells (28). The RyhB mutant, in which iron-mediated repression of virulence-associated genes should not occur, did not have a phenotype in the invasion and plaque assays. This suggests that the role of RyhB in virulence involves fine-tuning the expression of virulence genes. Because RyhB is one of a number of factors that regulate expression of the virulence genes, there may be sufficient redundancy of control to prevent significant overexpression of virulence genes during intracellular replication.
Regulation of virB by RyhB is reminiscent of other, horizontally acquired virulence factors whose regulation has been incorporated into existing regulatory networks (5). S. dysenteriae and E. coli Shiga toxin (slt) (3, 30), Corynebacterium diphtheriae diphtheria toxin (tox) (27), and Salmonella enterica serovar Typhimurium SPI1 genes (14) are regulated by Fur, DtxR, and PhoPQ, respectively. Fur, DtxR, and PhoPQ are global regulators that control the bacterial cell response to environmental signals such as iron or magnesium levels. Integrating the regulation of virulence genes into networks that respond to specific environmental signals, such as the local iron concentration, is an elegant mechanism to coordinate the expression of specific bacterial virulence factors during the course of infection.
Acknowledgments
We thank members of the Payne lab for helpful discussions and critical reading of the manuscript. In particular, we thank Megan Boulette and Ashima Sharma for sharing unpublished data.
This work was funded in part by Ruth L. Kirschstein National Research Service Award NIH 8-FGM069312A, by NIH grant AI16935, and by a grant from The Foundation for Research.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Adler, B., C. Sasakawa, T. Tobe, S. Makino, K. Komatsu, and M. Yoshikawa. 1989. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627-635. [DOI] [PubMed] [Google Scholar]
- 2.Afonyushkin, T., B. Vecerek, I. Moll, U. Blasi, and V. R. Kaberdin. 2005. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 33:1678-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 6.Dorman, C. J., and M. E. Porter. 1998. The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol. Microbiol. 29:677-684. [DOI] [PubMed] [Google Scholar]
- 7.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23:396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale, T. L., and S. B. Formal. 1981. Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect. Immun. 32:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Headley, V., M. Hong, M. Galko, and S. M. Payne. 1997. Expression of aerobactin genes by Shigella flexneri during extracellular and intracellular growth. Infect. Immun. 65:818-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hershberg, R., S. Altuvia, and H. Margalit. 2003. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 31:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huttenhofer, A., P. Schattner, and N. Polacek. 2005. Non-coding RNAs: hope or hype? Trends Genet. 21:289-297. [DOI] [PubMed] [Google Scholar]
- 12.Jennison, A. V., and N. K. Verma. 2004. Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol. Rev. 28:43-58. [DOI] [PubMed] [Google Scholar]
- 13.Johansson, J., and P. Cossart. 2003. RNA-mediated control of virulence gene expression in bacterial pathogens. Trends Microbiol. 11:280-285. [DOI] [PubMed] [Google Scholar]
- 14.Jones, B. D. 2005. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 43(Spec. No.):110-117. [PubMed] [Google Scholar]
- 15.Lucchini, S., H. Liu, Q. Jin, J. C. Hinton, and J. Yu. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 73:88-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masse, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masse, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mey, A. R., S. A. Craig, and S. M. Payne. 2005. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73:5706-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mills, M., and S. M. Payne. 1997. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect. Immun. 65:5358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson, P., M. Kiriakidou, A. Sharma, E. Maniataki, and Z. Mourelatos. 2003. The microRNA world: small is mighty. Trends Biochem. Sci. 28:534-540. [DOI] [PubMed] [Google Scholar]
- 22.Oaks, E. V., M. E. Wingfield, and S. B. Formal. 1985. Plaque formation by virulent Shigella flexneri. Infect. Immun. 48:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oglesby, A. G., E. R. Murphy, V. R. Iyer, and S. M. Payne. 2005. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol. Microbiol. 58:1354-1367. [DOI] [PubMed] [Google Scholar]
- 24.Pope, L. M., K. E. Reed, and S. M. Payne. 1995. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 63:3642-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Runyen-Janecky, L. J., and S. M. Payne. 2002. Identification of chromosomal Shigella flexneri genes induced by the eukaryotic intracellular environment. Infect. Immun. 70:4379-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schmitt, M. P., and R. K. Holmes. 1991. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59:1899-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuch, R., R. C. Sandlin, and A. T. Maurelli. 1999. A system for identifying post-invasion functions of invasion genes: requirements for the Mxi-Spa type III secretion pathway of Shigella flexneri in intercellular dissemination. Mol. Microbiol. 34:675-689. [DOI] [PubMed] [Google Scholar]
- 29.Sharma, A. K., and S. M. Payne. 2006. Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol. Microbiol. 62:1498. [DOI] [PubMed] [Google Scholar]
- 30.Svinarich, D., and S. Palchaudhuri. 1992. Regulation of the SLT-1A toxin operon by a ferric uptake regulatory protein in toxinogenic strains of Shigella dysenteriae type 1. J. Diarrhoeal Dis. Res. 10:139-145. [PubMed] [Google Scholar]
- 31.Tobe, T., M. Yoshikawa, T. Mizuno, and C. Sasakawa. 1993. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J. Bacteriol. 175:6142-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vecerek, B., I. Moll, T. Afonyushkin, V. Kaberdin, and U. Blasi. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 50:897-909. [DOI] [PubMed] [Google Scholar]
- 33.Wassarman, K. M. 2002. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell 109:141-144. [DOI] [PubMed] [Google Scholar]
- 34.Yang, F., J. Yang, X. Zhang, L. Chen, Y. Jiang, Y. Yan, X. Tang, J. Wang, Z. Xiong, J. Dong, Y. Xue, Y. Zhu, X. Xu, L. Sun, S. Chen, H. Nie, J. Peng, J. Xu, Y. Wang, Z. Yuan, Y. Wen, Z. Yao, Y. Shen, B. Qiang, Y. Hou, J. Yu, and Q. Jin. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33:6445-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, J., L. Chen, J. Yu, L. Sun, and Q. Jin. 2006. ShiBASE: an integrated database for comparative genomics of Shigella. Nucleic Acids Res. 34:D398-D401. [DOI] [PMC free article] [PubMed] [Google Scholar]