Abstract
Vibrio cholerae strains from diverse O-antigen groups were evaluated for RTX toxin actin cross-linking activity. This study demonstrates that the actin cross-linking domain sequence is present within rtxA in the majority of clinical and environmental isolates tested, and the RTX toxin produced by these strains catalyzes the covalent cross-linking of cellular actin.
The causative agent of cholera disease is the gram-negative bacterial pathogen Vibrio cholerae. There are more than 200 O-antigen serogroups of V. cholerae, but only the O1 and O139 strains have been linked to pandemic disease (9). However, non-O1, non-O139 isolates have been associated with a cholera-like disease that leads to clinical symptoms indistinguishable from those of patients infected with O1 and O139 strains (1). Current epidemiological data indicate an increase in the number of pathogenic non-O1, non-O139 isolates, and it has been suggested that the sporadic outbreaks of cholera caused by these strains may be due to bacterial virulence factors other than the well-characterized cholera toxin (CT) and toxin-coregulated pilus (TCP) (1, 6, 15).
The V. cholerae RTX (repeats-in-toxin) toxin is encoded by rtxA (12), a gene carried by several non-O1, non-O139 V. cholerae strains linked to cholera disease (3, 5). The activity of the RTX toxin leads to host cell rounding and a novel rearrangement of the actin cytoskeleton—the covalent cross-linking of actin monomers into dimer, trimer, and higher-multimer proteins (8). The region of the toxin responsible for actin cross-linking has been identified as a 412-amino-acid domain present within the 4,545-amino-acid holotoxin (16), and it has been demonstrated that this actin cross-linking domain (ACD) directly catalyzes the cross-linking reaction (4). Actin cross-linking is a fundamental activity of the RTX toxin, and the toxin has been implicated in the pathogenesis of cholera disease (7).
Analysis of the rtxA gene in several V. cholerae isolates revealed that the O1 classical strains harbor a deletion in rtxA that eliminates a large portion of the N terminus and inactivates the toxin (3, 12). A previous study by Chow et al. (3) with DNA primers directed against this region indicates that rtxA is distributed across all of the V. cholerae serogroups other than the O1 classical strains. However, a genome sequence comparison of O1 El Tor strain N16961 and O135 strain RC385 showed extensive variation throughout the RTX toxin structure, and among the many differences, the putative RTX toxin in strain RC385 does not carry the ACD. Interestingly, the ACD sequence has been detected in O1 classical isolate O395, despite the fact that this strain contains the N-terminal deletion in rtxA present in all O1 classical strains (The Institute for Genomic Research [www.tigr.org]). The potential for heterogeneity within the toxin structure suggests that an evaluation for the presence of rtxA should include a more thorough investigation of the rtxA sequence, particularly in the region containing the ACD. In addition, it is important to specifically assess V. cholerae strains for RTX toxin function, as molecular detection of the rtxA gene does not confirm the production of an active toxin protein.
In this study, we analyzed a collection of clinical and environmental non-O1, non-O139 isolates of V. cholerae for the presence of the rtxA gene and the ACD sequence within rtxA, and each strain was monitored for actin cross-linking activity in vivo. We determined that both the ACD and actin cross-linking were detected in a broad range of non-O1, non-O139 serogroups.
The 24 bacterial strains used in this study are listed in Table 1. The V. cholerae non-O1, non-O139 clinical and environmental isolates were collected between 1962 and 1998 from China, India, Iraq, Japan, the Philippines, Thailand, and the United States (10, 11). O1 El Tor strain N16961 was originally isolated in 1975 from a patient with diarrhea in Bangladesh (11), and the presence of the RTX-encoding rtxA gene has previously been established (12). N16961 derivative strain KFV119 was used in place of N16961 in each assay (16). The other O1 El Tor strain used in this study, P27459 (13), has been characterized as rtxA+ by Lin et al. (12), and genome sequence analysis of O139 serogroup isolate MO10 (17) has identified both the rtxA and ACD sequences (The Institute for Genomic Research [www.tigr.org]). Strain MO6-24/O is a Vibrio vulnificus isolate from a patient with septicemia (18). V. vulnificus, a pathogen closely related to V. cholerae, contains an RTX toxin (2), but the protein does not have actin cross-linking activity (K. Sheahan and K. Satchell, unpublished results). The data regarding the presence of tcpA, which encodes TCP, and the CT-encoding ctxAB genes have been previously published (10, 11, 14, 17).
TABLE 1.
Bacterial strains evaluated in this study
| Strain | Serogroup | Country | Yr | Source | Referencea | tcpAb | ctxABb | rtxAc | acdc | Actin cross-linkingc |
|---|---|---|---|---|---|---|---|---|---|---|
| V. cholerae | ||||||||||
| N16961d | O1 El Tor | Bangladesh | 1975 | Diarrhea | 11 | + | + | + | + | + |
| P27459 | O1 El Tor | Bangladesh | 1976 | Diarrhea | 13 | +e | +e | + | + | + |
| MO10 | O139 | India | 1993 | Diarrhea | 17 | +f | +f | + | + | + |
| 153-94 | O8 | Unknown | 1994 | CDC | 10 | + | + | + | + | + |
| 112-68 | O9 | Philippines | 1968 | Diarrhea | 10 | − | − | + | + | + |
| No. 63 | O26 | Japan | 1991 | Diarrhea from travel in Thailand | 10 | + | + | + | + | + |
| 365-96 | O27 | Japan | 1996 | Prawn imported from Thailand | 11 | + | + | + | + | + |
| 5473-62 | O31 | Philippines | 1962 | Diarrhea | 10 | − | − | + | − | − |
| 1311-69 | O35 | India | 1968 | Diarrhea | 10 | − | − | + | − | − |
| 1322-69 | O37 | India | 1969 | Diarrhea | 11 | + | + | + | + | + |
| 506-94 | O44 | Thailand | 1994 | Diarrhea | 10 | + | + | + | + | + |
| AQ1875 | O48 | Japan | 1998 | Tortoise imported from Taiwan | 10 | + | − | + | + | + |
| 507-94 | O49 | Thailand | 1994 | Diarrhea | 10 | + | + | + | + | + |
| 8585 | O53 | Iraq | 1966 | Diarrhea | 11 | + | − | + | + | + |
| 981-75 | O65 | India | 1975 | Diarrhea | 11 | + | − | + | + | + |
| 8-76 | O77 | India | 1976 | Diarrhea | 10 | + | − | + | + | + |
| 1421-77 | O80 | India | 1977 | Diarrhea | 10 | + | − | + | + | + |
| 984-81 | O89 | India | 1981 | Diarrhea | 10 | − | − | + | + | + |
| 571-88 | O105 | China | 1988 | Diarrhea | 10 | + | + | + | + | + |
| 523-80 | O115 | United States | 1980 | Diarrhea | 10 | + | − | − | − | − |
| 203-93 | O141 | India | 1995 | Diarrhea | 10 | + | + | + | + | − |
| 254-93 | O144 | India | 1993 | Diarrhea | 10 | − | − | + | + | + |
| 366-96 | O191 | Japan | 1996 | Prawn imported from Thailand | 10 | + | + | + | + | + |
| V. vulnificus MO6-24/O | NAg | NA | NA | Septicemia | 18 | NA | NA | − | − | − |
Serotype, country, year, and source information was obtained from the referenced publications.
The presence or absence of tcpA and ctxAB was previously described by Li et al. (10, 11), unless otherwise noted.
Data from this study.
Strain KFV119 is a ΔhapA ΔhlyA N16961 derivative that was used in place of N16961 (16).
Data for strain P27549 were previously reported by Nesper et al. (14).
Data for strain MO10 were previously reported by Waldor and Mekalanos (17).
NA, not available.
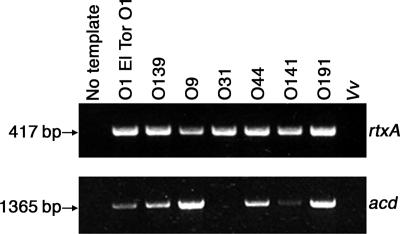
Each strain was assessed for the presence of rtxA by PCR amplification with primers specific for the N-terminal region of rtxA (3). PCR products were analyzed by agarose gel electrophoresis and visualized by staining with ethidium bromide (Fig. 1). The rtxA sequence was identified in both O1 El Tor strains N16961 and P27459 and in the O139 serogroup, which confirms previous results (12). In addition, rtxA was present in all of the non-O1, non-O139 V. cholerae strains tested, with the exception of serogroup O115 (Table 1). These data demonstrate that rtxA is contained within 19 out of the 20 non-O1, non-O139 isolates. To verify whether the negative result for rtxA in the O115 strain was due a deletion similar to that in the O1 classical serogroup, which contains the ACD sequence in a dysfunctional remnant of rtxA, the sequences for both the O1 classical deletion and ACD were examined by PCR with primers specific for the new junction created by the rtx deletion present in O1 classical strain O395 (12) and primers for the ACD region (16). Interestingly, neither the O1 classical deletion nor the ACD was present in the O115 strain, indicating that the O115 serogroup either does not carry the rtxA gene or harbors a deletion distinct from the O1 classical serogroup that eliminates the N-terminal region of rtxA and the ACD (data not shown). The absence of rtxA may also be linked to a larger genome variation, as the adjacent CT-encoding ctxAB genes are absent from this strain as well (Table 1). However, there is not always a correlation between the absence of the ctxAB genes and rtxA since 10 of the 20 non-O1, non-O139 strains are rtxA+ and ctxAB−.
FIG. 1.
The rtxA gene and ACD sequence are present in non-O1, non-O139 serogroups of V. cholerae. The sequences for rtxA and the ACD were amplified by colony PCR for each bacterial strain. The resulting PCR products were separated on a 1% agarose gel, and rtxA and the ACD were detected by ethidium bromide staining. A representative sample of strains is shown, and the complete list of results is displayed in Table 1. Vv, V. vulnificus strain MO6-24/O.
Primers were designed to amplify the ACD sequence within rtxA (16), and the ACD was visualized by agarose gel electrophoresis and ethidium bromide staining (Fig. 1). The ACD was present in the O139 serogroup, which confirms previous genome sequencing results, as well as in O1 El Tor strains N16961 and P27459. As shown in Table 1, the ACD sequence was identified in 17 of the 19 rtxA+ non-O1, non-O139 isolates. The ACD from O37 isolate 1322-69 was selected for comparative sequence analysis and shown to be 100% identical to O1 El Tor reference strain N16961, as well as four other fully sequenced strains representing the O1 classical, O37, O139, and O141 serogroups (data not shown). Therefore, the ACD is highly conserved among these strains and it is possible that the sequence conservation extends to other non-O1, non-O139 serogroups. The strains that were rtxA+ but did not contain the ACD belong to the O31 and O35 serogroups. Further examination of the full-length rtxA sequence may reveal that the ACD region is either absent in the RTX toxins of these strains, similar to that of O135 strain RC385, or contains mutations that inhibit primer binding and detection by PCR. It is interesting that the presence of potential variations within the rtxA sequence may also correlate with the absence of the both the tcpA and ctxAB genes, which could suggest that alternate RTX toxins may be more common in TCP− CT− strains.
However, detection of the ACD region by PCR does not necessarily indicate that the bacteria produce a functional RTX toxin that catalyzes actin cross-linking, because genetic mutations may affect gene expression, toxin processing, or toxin secretion. In addition, as the toxin carries multiple cell-rounding activities (16), RTX toxin actin cross-linking activity must be assessed by the formation of cross-linked actin species, not only via an observation of rounded host cells. Human laryngeal epithelial (HEp-2) cells were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. The cells were incubated for 90 min at a multiplicity of infection of ∼200 with liquid cultures of each strain grown in Luria broth for 18 h at 30°C. Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the presence of cross-linked actin proteins was monitored by immunoblotting with a 1:1,000 dilution of a rabbit polyclonal anti-actin antibody, followed by a 1:5,000 dilution of anti-rabbit immunoglobulin G conjugated to horseradish peroxidase.
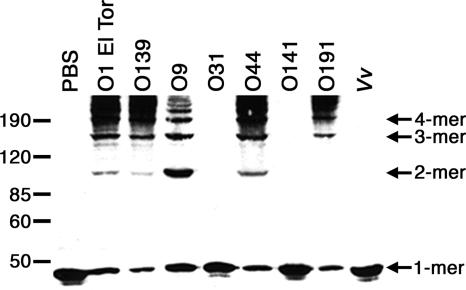
All of the non-O1, non-O139 isolates that contained the sequences for both rtxA and the ACD had actin cross-linking activity, except for those in serogroup O141 (Fig. 2). These data correlate with the PCR results for the ACD (see above) and demonstrate that non-O1, non-O139 isolates that contain the ACD sequence from the RTX toxin also cause the covalent cross-linking of cellular actin. Yet, despite the presence of the ACD within rtxA in serogroup O141, actin cross-linking was not detected. The PCR product from the O141 strain was sequenced to determine whether the defect in actin cross-linking was due to genetic variation in the ACD region. Sequence alignment of the ACD from the O141 and O1 El Tor serogroups revealed several amino acid mutations present in the O141 isolate, including V2066I, P2125S, T2194P, and I2310V (data not shown). These data suggest that the sequence differences in the O141 strain may account for the absence of RTX toxin activity, although it is also possible that mutations in other regions of the rtxA sequence or within the genes responsible for toxin secretion contribute to the lack of actin cross-linking. Ongoing experiments focused on both the crystallization of the ACD and identification of the catalytic residues responsible for actin cross-linking will determine the significance of the O141 mutations in RTX toxin structure and function.
FIG. 2.
V. cholerae non-O1, non-O139 strains cause actin cross-linking in vivo. HEp-2 cells were incubated with phosphate-buffered saline (PBS) or the V. cholerae and V. vulnificus strains listed in Table 1. Cell lysates were separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and actin cross-linking was detected by immunoblotting with an anti-actin antibody. A representative sample of strains is shown, and the complete list of results is displayed in Table 1. Vv, V. vulnificus strain MO6-24/O. The values on the left are molecular sizes in kilodaltons.
Overall, we have shown that the majority of the non-O1, non-O139 strains tested carry the sequences for rtxA and the ACD. In addition, we have demonstrated that the RTX toxins produced by these strains catalyze the formation of cross-linked actin dimers, trimers, and higher multimers. The data presented here have advanced our study of covalent actin cross-linking by the V. cholerae RTX toxin in non-O1, non-O139 strains, and continued investigation will provide insight into the contribution of actin cross-linking activity to the pathogenesis of non-O1, non-O139 serogroups. These data also suggest that a genetic and functional analysis of actin cross-linking activity in clinical and environmental isolates may enhance the epidemiological surveillance of cholera disease, as sequence variation within rtxA can effect both the detection and activity of the RTX toxin.
Acknowledgments
This work was supported by United States Public Health Service grant AI051490 and a Burroughs Wellcome Fund Investigators in Pathogenesis of Infectious Diseases award (to K.J.F.S.). C.L.C. was supported by Ruth L. Kirschstein National Research Service Award fellowship F31-AI52490.
V. vulnificus strain MO6-24/O was acquired from Paul Gulig at the University of Florida.
The views expressed in this article are ours and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, or the U.S. Government.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Bhattacharya, M. K., D. Dutta, S. K. Bhattacharya, A. Deb, A. K. Mukhopadhyay, G. B. Nair, T. Shimada, Y. Takeda, A. Chowdhury, and D. Mahalanabis. 1998. Association of a disease approximating cholera caused by Vibrio cholerae of serogroups other than O1 and O139. Epidemiol. Infect. 120:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, K. H., T. K. Ng, K. Y. Yuen, and W. C. Yam. 2001. Detection of RTX toxin gene in Vibrio cholerae by PCR. J. Clin. Microbiol. 39:2594-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordero, C. L., D. S. Kudryashov, E. Reisler, and K. J. Satchell. 2006. The actin cross-linking domain of the Vibrio cholerae RTX toxin directly catalyzes the covalent cross-linking of actin. J. Biol. Chem. 281:32366-32374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalsgaard, A., O. Serichantalergs, A. Forslund, W. Lin, J. Mekalanos, E. Mintz, T. Shimada, and J. G. Wells. 2001. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J. Clin. Microbiol. 39:4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faruque, S. M., N. Chowdhury, M. Kamruzzaman, M. Dziejman, M. H. Rahman, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2004. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. USA 101:2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fullner, K. J., J. C. Boucher, M. A. Hanes, G. K. Haines III, B. M. Meehan, C. Walchle, P. J. Sansonetti, and J. J. Mekalanos. 2002. The contribution of accessory toxins of Vibrio cholerae O1 El Tor to the proinflammatory response in a murine pulmonary cholera model. J. Exp. Med. 195:1455-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fullner, K. J., and J. J. Mekalanos. 2000. In vivo covalent crosslinking of actin by the RTX toxin of Vibrio cholerae. EMBO J. 19:5315-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaper, J. B., J. G. J. Morris, and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, M., M. Kotetishvili, Y. Chen, and S. Sozhamannan. 2003. Comparative genomic analyses of the vibrio pathogenicity island and cholera toxin prophage regions in nonepidemic serogroup strains of Vibrio cholerae. Appl. Environ. Microbiol. 69:1728-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, M., T. Shimada, J. G. Morris, Jr., A. Sulakvelidze, and S. Sozhamannan. 2002. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect. Immun. 70:2441-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, W., K. J. Fullner, R. Clayton, J. A. Sexton, M. B. Rogers, K. E. Calia, S. B. Calderwood, C. Fraser, and J. J. Mekalanos. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA 96:1071-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mekalanos, J. J. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253-263. [DOI] [PubMed] [Google Scholar]
- 14.Nesper, J., A. Kraiss, S. Schild, J. Blass, K. E. Klose, J. Bockemuhl, and J. Reidl. 2002. Comparative and genetic analyses of the putative Vibrio cholerae lipopolysaccharide core oligosaccharide biosynthesis (wav) gene cluster. Infect. Immun. 70:2419-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma, C., M. Thungapathra, A. Ghosh, A. K. Mukhopadhyay, A. Basu, R. Mitra, I. Basu, S. K. Bhattacharya, T. Shimada, T. Ramamurthy, T. Takeda, S. Yamasaki, Y. Takeda, and G. B. Nair. 1998. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36:756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheahan, K. L., C. L. Cordero, and K. J. Satchell. 2004. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc. Natl. Acad. Sci. USA 101:9798-9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waldor, M. K., and J. J. Mekalanos. 1994. Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype. J. Infect. Dis. 170:278-283. [DOI] [PubMed] [Google Scholar]
- 18.Wright, A. C., L. M. Simpson, J. D. Oliver, and J. G. Morris, Jr. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]