Abstract
Epstein-Barr virus (EBV) infection is associated with a broad spectrum of disease. While quantification of EBV nucleic acid in the peripheral blood has been demonstrated to be useful for diagnosis and patient care, the optimal sample type and reporting format for such testing remain uncertain. Using quantitative real-time PCR (QRT-PCR), we evaluated EBV in whole blood (WB), peripheral blood mononuclear cells (PBMC), and plasma in 249 samples from 122 patients. In WB and PBMC, results were reported both in viral copies/ml and in copies/μg of total DNA. Trendings of quantitative values over time among the different sample types were compared. The sensitivities of QRT-PCR using WB and that using PBMC did not differ significantly (P = 0.33), and both were more sensitive than plasma alone (P < 0.0001). EBV viral load results from WB and PBMC paired sample types also showed a significant correlation (P < 0.05), as did results reported in copies/ml and copies/μg DNA for both WB and PBMC (R2 > 0.93). EBV viral loads detected using WB and PBMC trended very closely for the few patients who had multiple positive samples available for analysis. WB and PBMC show comparable sensitivities and a close quantitative correlation when assayed for EBV by QRT-PCR. The close correlation between copies/ml and copies/μg DNA also suggests that normalization to cell number or genomic DNA in cellular specimens may not be necessary.
Epstein-Barr virus (EBV) is associated with a broad spectrum of epithelial and lymphoproliferative disorders, many associated with altered host immune status. EBV-associated conditions include infectious mononucleosis, Burkitt's lymphoma, Hodgkin's lymphoma, T-cell or NK cell lymphoma, B-cell non-Hodgkin's lymphoma, nasopharyngeal carcinoma, gastric carcinoma, oral hairy leukoplakia, AIDS-related lymphoma, and posttransplantation lymphoproliferative disease (PTLD) (8). Much work has focused on PTLD, with several studies showing that high levels of EBV DNA in peripheral blood from immunocompromised patients predicts the onset of PTLD (1, 2, 10, 11, 16, 19-23), while a decrease in EBV DNA load is associated with response to treatment and regression of PTLD (5, 6, 12, 17, 24).
A variety of molecular diagnostic methods, primarily based on PCR, have been developed to detect and quantify circulating EBV in an effort to predict or detect the onset of EBV-associated disorders and to assess the efficacy of therapeutic intervention (2, 7, 19, 21-23). Over the past few years, such methods have undergone significant improvement (11). In particular, the introduction of real-time amplification and detection methods has reduced the risk of carry-over contamination, shortened the time needed for the postamplification analysis, improved ease of use, and improved quantitative test performance (9, 18, 25). In addition, numerous studies have begun to define the role for such assays in relation to clinical care and predictive value (1, 2, 14, 16, 19, 22, 28). Critical to the accuracy of these methods is the detection of inefficiencies in the specimen preparation or amplification processes. Several authors have addressed this point, using both endogenously and exogenously added internal controls, in an effort to monitor for suboptimal test performance. Some have advocated the normalization of viral load results to a coamplified housekeeping gene as the best means of meeting these goals (9, 25).
Sample type selection often impacts test performance characteristics, including clinical predictive value. Several peripheral blood compartments have been used to measure EBV viral load, including whole blood (WB) (1, 23), serum (3, 4, 13, 14), plasma (15, 18, 32), peripheral blood leukocytes, and mononuclear cells (10, 16, 19, 21, 22). Likewise, reporting units for EBV DNA viral loads have variously included copies/ml, copies/μg DNA, and copies/105 cells. Ongoing uncertainty related to the optimal sample type and reporting format for quantitative EBV detection has contributed to the absence of standardized test guidelines and has complicated the effort to define EBV treatment threshold values. The goal of the present study is to directly compare EBV DNA viral loads for matched specimens from WB, peripheral blood mononuclear cells (PBMC), and plasma obtained from a population of pediatric hematopoietic stem cell transplant recipients tested by quantitative real-time PCR (QRT-PCR).
MATERIALS AND METHODS
Patient samples.
Following Institutional Review Board (IRB) approval, 249 peripheral blood samples from 122 patients treated at St. Jude Children's Research Hospital in Memphis, TN, were included for study. These specimens were received for clinical testing from September 2002 to September 2005.
DNA extracts from 249 paired WB and PBMC samples, 167 paired plasma and WB samples, and 167 paired plasma and PBMC samples were deidentified and tested blindly in duplicate. Either EDTA-treated blood (WB and plasma samples) or sodium citrated blood (PBMC samples) was used for study. All paired samples were collected concurrently (single blood draw), and 200 μl each of WB and plasma samples was used for DNA extraction. PBMC pellets were prepared by centrifuging 4 ml of WB in cell preparation tubes (Becton Dickinson, Franklin Lakes, NJ) for 20 min at 1,800 relative centrifugal force; PBMC pellets were resuspended in 700 μl of phosphate-buffered saline. Two hundred microliters each of the WB, plasma, and PBMC pellet samples was used for DNA extraction using a QIAamp blood mini kit (QIAGEN Inc., Valencia, CA), with final elution in 200 μl 10% AE buffer. Extracts were frozen at −70°C until use. Eight microliters of eluate was used for each PCR, a portion which corresponded to 0.008 ml of the original sample for WB and plasma and 0.032 ml of the original sample for PBMC. Viral control samples were similarly extracted with the QIAamp blood mini kit and serially diluted prior to QRT-PCR.
Determination of EBV DNA load. (i) QRT-PCR.
The PCR standard curve used a 90-bp PCR product from the EBV BALF5 gene, which was cloned into a pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA) and transformed into One Shot Top10 competent Escherichia coli cells. Colonies were screened by sequencing to confirm the correct insert. Plasmid DNA from the confirmed colony was isolated using a QIAprep spin miniprep kit (QIAGEN Inc., Valencia, CA), and the PCR products for the standard curve were quantified spectrophotometrically. Raji (American Type Culture Collection [ATCC] CCL-86) and Namalwa (ATCC CRL-1432) cell lines were used as EBV-positive controls. HL60 cells were used as EBV-negative controls. The acceptable range for the EBV-positive control Namalwa cell line was within a range width of 0.39 viral copies/cell, while that for the Raji cell line was within 2.43 copies/cell. The results from these positive controls fell within these ranges in all the assay runs.
QRT-PCR was performed in a manner similar to that described by Kimura et al. (11). Briefly, a multiplexed PCR targeted a 90-bp region of the BALF5 gene coamplified with the human housekeeping gene RNase P. The assay consisted of a primer set and a dual-labeled (5′ 6-carboxyfluorescein/3′ Black Hole Quencher -1) TaqMan probe specific to EBV. An 8-μl aliquot of patient DNA extract was used in a final reaction volume of 50 μl. RT-PCR consisted of 50 cycles and was performed using an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA). The reaction was run under the following cycling conditions: a temperature of 50°C for 2 min, followed by 95°C for 10 min, 95°C for 15 s, and then 60°C for 1 min. These conditions were repeated for 50 cycles. Results were expressed in copies/ml and copies/μg of total DNA.
(ii) PCR calibration curves and calculation of viral loads.
A regressive standard curve was generated using plasmid DNA, as described above, serially diluted in 10-fold increments from 2 × 106 copies to 2 copies/5 μl. Cycle threshold values from clinical WB, PBMC, and plasma extracts were plotted on this curve to determine copies of EBV genome/reaction (copies/rxn). Copies/ml of sample were then calculated according to the following equations:
 |
and
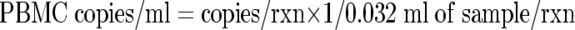 |
Based on dilution factors introduced during DNA extraction and amplification, the lower limit of detection for the assay was 125 viral copies/ml in WB and plasma and 31.25 copies/ml in PBMC. Results of PCR from cellular samples (WB and PBMC) were normalized to the quantity of input genomic DNA. The human RNase P gene (a single-copy housekeeping gene) was coamplified with EBV by use of a TaqMan RNase P control reagent kit (Applied Biosystems, Foster City, CA). Human genomic DNA RNase P (Promega, Madison, WI) was serially diluted from 6 × 105 pg to 6 pg DNA per PCR. Following amplification and generation of a regression curve, RNase P was quantified in a manner similar to that described above for the primary amplification target. Quantification of the RNase P gene was then used to calculate EBV copies per microgram of input DNA as follows:
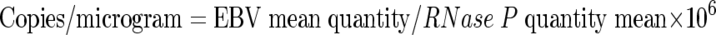 |
Statistical analysis.
Only one sample per patient was included in the comparative analysis in order to minimize potential bias. In those patients with multiple positive samples, a single sample was randomly chosen for the analysis using a computer program by which each sample has the same chance to be chosen. This resulted in 122 sample pairs for WB and PBMC, 79 pairs for WB and plasma, and 79 pairs for PBMC and plasma samples. McNemar's test was used to examine the sensitivity and concordance of EBV viral load levels among any two of the three blood sources, i.e., WB, PBMC, and plasma. A threshold of “zero” was chosen for positive virus detection for the qualitative comparison. In an effort to better understand the relationship between the levels of virus detected among the two sample types that were positive for EBV, regression analysis was performed after transforming to log10 (load plus 1) using ordinary least squares. Only results of ≥125 copies/ml in WB and plasma samples and 31.25 copies/ml in PBMC were included for analysis. For patients with three or more positive samples, trendings of quantitative values over time were also compared among the different sample types.
RESULTS
Qualitative sensitivities of different sample types for EBV detection.
A total of 122 paired samples of WB and PBMC were available for analysis. In 39 of 122 (32%) pairs, EBV genome was detected in both sample types. EBV was detected in 1 of the 2 samples (either in WB or PBMC) in 17 out of 122 (14%) samples. Of these 17 samples, EBV was detected in only 6 WB samples whose matching PBMC was negative, while it was picked up in 11 PBMC samples whose matching WB was negative (Table 1). Among the discordant sample pairs, EBV loads in the 6 positive WB samples ranged from ≤125 to 237.5 copies/ml, while EBV loads in the 11 positive PBMC samples were all ≤31.25 copies/ml (lower limit of detection). McNemar's test showed no significant difference in test sensitivities between WB and PBMC samples (P = 0.33).
TABLE 1.
Comparison of EBV detection using PCR in clinical samples
| Parameter | No. tested or no. indicated results for:
|
||
|---|---|---|---|
| WB and PBMC | WB and plasma | PBMC and plasma | |
| Total no. of sample pairs tested | 122 | 79 | 79 |
| No. of concordant paired results | 105 | 57 | 54 |
| Positive results from discordant pairs | |||
| WB | 6 | 21 | |
| PBMC | 11 | 25 | |
| Plasma | 1 | 0 | |
Test results from 79 samples of plasma were compared to results for matched WB. EBV was detected in both WB and plasma samples in 9 out of 79 (11.4%) sample pairs, while it was detected only in WB in 21 of 79 (26.6%) and only in plasma in 1 of 79 (1.3%) samples (Table 1). EBV loads in the 21 positives detected using only WB samples ranged from ≤125 to 2,280 copies/ml. The one viral load that was positive in plasma but negative in WB samples showed <125 copies/ml. The use of plasma resulted in significantly reduced sensitivity compared to WB in detecting EBV viral load (P < 0.0001).
Analysis of the 79 available pairs of PBMC and plasma showed concordant EBV-positive results in 54 out of 79 (12.7%) pairs. Twenty-five of 79 (31.6%) PBMC samples were positive for EBV viral copies, while their matched plasma samples were negative. None of the negative PBMC samples had EBV viral copies detected in matched plasma samples (Table 1). The 25 positive PBMC samples with paired negative plasma samples had viral loads ranging from ≤31.25 to 3,228.13 copies/ml that were missed in plasma. Using plasma for EBV detection resulted in reduced sensitivity compared to that obtained with PBMC (P < 0.0001).
Correlation of viral loads in different sample types.
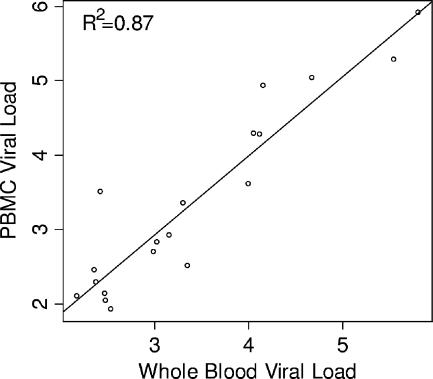
In the quantitative analysis, only four plasma samples had EBV loads greater than the threshold of 125 copies/ml. Therefore, regression analysis was applied only to viral load results of paired WB and PBMC samples which were above the threshold. The correlation was significant (P < 0.05) with an R2 of 0.87, a y intercept approximating zero (−0.27; 95% confidence interval [95% CI], −0.98, 0.44), and slope close to 1 (1.06; 95% CI, 0.87, 1.26) (Fig. 1).
FIG. 1.
Comparison between EBV viral loads in WB and PBMC expressed in log10.
The EBV loads quantified for the four positive plasma samples are shown in Table 2 with their matching WB and PBMC samples.
TABLE 2.
EBV loads in the four positive plasma samples with corresponding WB and PBMC results
| Sample no. | EBV load (copies/ml) in:
|
||
|---|---|---|---|
| Plasma | WB | PBMC | |
| 1 | 9,640 | 13,128 | 19,013 |
| 2 | 130 | 1,453 | 845 |
| 3 | 9,151 | 640,223 | 827,483 |
| 4 | 1,901 | 47,501 | 110,885 |
Correlation between reporting units.
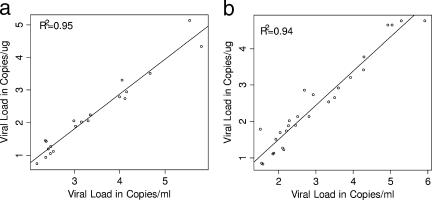
Regression analysis showed strong correlations between results reported as copies/ml and copies/μg DNA for both WB (R2 = 0.95) and PBMC (R2 = 0.94). Linear regression using positive WB samples showed a y intercept of −1.39 (95% CI, −1.77, −1.01) and a slope of 1.07 (95% CI, 0.96, 1.17). Mean EBV viral concentrations reported in copies/μg DNA were 1.39 log10 units lower than those reported in copies/ml (Fig. 2a). Similar results were obtained from the analysis of positive PBMC samples. Mean EBV viral loads reported in copies/μg DNA from PBMC were 0.38 log10 units lower than viral loads reported in copies/ml (intercept, −0.38 [95% CI, −0.70, −0.07]; slope of 0.94 [95% CI, 0.85, 1.04]) (Fig. 2b).
FIG. 2.
Comparison of the two reporting formats in WB and PBMC, with all viral loads expressed in log10. (a) Comparison between EBV viral loads of copies/μg and copies/ml for WB. (b) Comparison between EBV viral loads of copies/μg and copies/ml for PBMC.
Relative trending within patients.
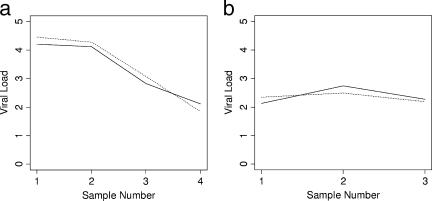
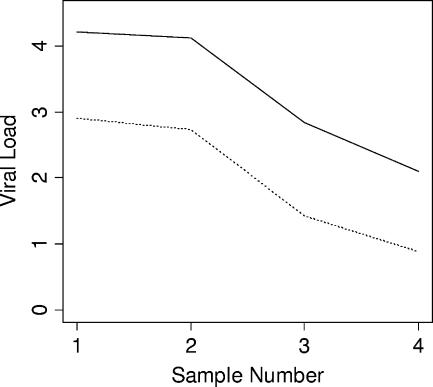
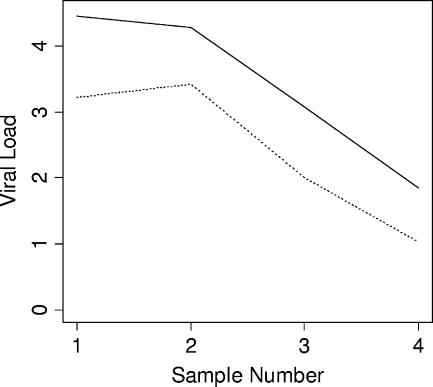
Comparative dynamic trending of EBV viral loads using WB and PBMC sample types was examined for four patients, each of whom had at least three consecutive samples with all results within the linear dynamic range of the assay (Fig. 3). Viral loads tracked closely among both sample types. Similar findings were obtained when comparing within-patient trends using different reporting units (copies/ml and copies/μg DNA) (Fig. 4 and 5). Results from 13 patients were available for the latter analysis. Among all 17 cases in which comparisons were made, no differences were seen in dynamic viral load trends.
FIG. 3.
Comparative trends of EBV viral loads in different sample types from two individual patients (a and b). Solid line, WB; dotted line, PBMC.
FIG. 4.
Comparative trends of EBV viral loads in WB from a patient by use of two different reporting units. Solid line, copies/ml; dotted line, copies/μg DNA.
FIG. 5.
Comparative trends of EBV viral loads in PBMC from a patient by use of two different reporting units. Solid line, copies/ml; dotted line, copies/μg DNA.
DISCUSSION
The findings in this study support the value of WB for quantitative detection of EBV by RT-PCR relative to that for paired PBMC and plasma specimens from pediatric hematopoietic stem cell transplant recipients. The use of WB maintained test sensitivity and close correlation of viral load results comparable to those for PBMC, while the use of plasma resulted in a significant loss of test sensitivity compared with results from the other (cellular) compartments. Dynamic quantitative trending for WB was similar to that for PBMC, although this analysis was limited by the small number of patients with multiple positive samples in the linear detection range of the assay.
Previous studies have demonstrated increased sensitivity for EBV detection with the use of cellular compartments compared to serum or plasma. PBMC has been shown to have advantages in this respect, and many investigators have used circulating lymphocytes as the specimen of choice (9, 10, 11, 19, 20, 22). Fewer investigators have examined the use of WB. Among those studies, results appear largely consistent with the data presented here (24, 25, 27). Stevens et al. (24, 25) demonstrated improved sensitivity of WB compared to serum and plasma using quantitative competitive EBV PCR, with acellular samples yielding negative results despite high viral loads in corresponding WB samples (25). Similarly, in a semiquantitative comparative study, Wadowsky and colleagues (27) showed a strong correlation of EBV DNA load values in WB (TaqMan PCR) and peripheral blood lymphocytes (competitive PCR), while correlation was poor between plasma and peripheral blood lymphocyte viral loads. In contrast, Wagner et al. (29) showed a DNA amplification efficiency and sensitivity lower for WB than for PBMC or B-cell samples. Although results were different from those described above, the latter study included blood from only a limited number of healthy subjects (11 subjects). The relative disadvantage exhibited using WB in that study was felt to be due largely to the presence of inhibitors, an issue not encountered in our series, possibly as a result of different specimen preparation methodologies. None of the above-described series compared WB, PBMC, and plasma using a single real-time quantitative method, and most included very limited numbers of patients and did not address additional issues related to sample reporting format. Some of the previous studies (23-25, 30, 31) included both pediatric and adult age groups, while the present study was limited to samples from pediatric patients.
The relative benefits of various reporting formats for EBV viral load assays are not clear from the literature. Some investigators have used copies per unit volume (25, 27, 31), and others (9, 11, 29-31) have reported in copies/μg of genomic DNA (when testing cellular compartments, such as WB or PBMC). The present study showed a close correlation between results reported as copies/μg DNA and copies/ml, similar to the findings of Wadowsky et al. (27) in a semiquantitative comparison. Normalizing viral load results to micrograms of input DNA (9) or to the number of cells present requires additional processing steps, increased expense, and increased volume of blood. The findings here suggest that little value is gained in this process. Dynamic trends of viral load within individual patients tracked very closely irrespective of the reporting units used. This study was limited in scope and design to a comparison of analytical findings using various specimen types and reporting formats. Further work will be needed to correlate these findings with clinical patient status and the presence or progression of lymphoproliferative diseases. Additional studies should also address the application of findings to the adult patient population.
The use of WB appears to offer potential advantages as the specimen of choice for QRT-PCR detection of EBV. Compared with PBMC, WB requires less blood volume and fewer processing steps. In addition, WB shows a significantly improved sensitivity and linear dynamic range compared to plasma. The findings in this study may help address the lack of standardization commonly encountered among quantitative molecular diagnostic assays. Further work will be needed to confirm these findings for other patient populations and using other methods. Other studies will also be needed to clarify the implications of these results for the prediction of clinical disease and therapeutic response in both the transplant and nontransplant populations in the context of their respective EBV-related disorders.
Acknowledgments
This work was supported in part by the American Lebanese Syrian Associated Charities (ALSAC).
The generous support of this work by Phill and Liz Gross is also deeply appreciated. Statistical support and advice provided by S. Pounds is greatly appreciated.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Bai, X., et al. 1997. Quantitative polymerase chain reaction for human herpesvirus diagnosis and measurement of Epstein-Barr virus burden in posttransplant lymphoproliferative disorder. Clin. Chem. 43:1843-1849. [PubMed] [Google Scholar]
- 2.Baldanti, F., et al. 2000. High levels of Epstein-Barr virus DNA in blood of solid-organ transplant recipients and their value in predicting posttransplant lymphoproliferative disorders. J. Clin. Microbiol. 38:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallagher, A., et al. 1999. Detection of Epstein-Barr virus (EBV) genomes in the serum of patients with EBV-associated Hodgkin's disease. Int. J. Cancer 84:442-448. [DOI] [PubMed] [Google Scholar]
- 4.Gan, Y. J., J. L. Sullivan, and J. W. Sixbey. 1994. Detection of cell-free Epstein-Barr virus DNA in serum during acute infectious mononucleosis. J. Infect. Dis. 170:436-439. [DOI] [PubMed] [Google Scholar]
- 5.Green, M., et al. 2000. Predictive negative value of persistent low Epstein-Barr virus viral load after intestinal transplantation in children. Transplantation 70:593-596. [DOI] [PubMed] [Google Scholar]
- 6.Green, M., et al. 1998. Serial measurement of Epstein-Barr viral load in peripheral blood in pediatric liver transplant recipients during treatment for posttransplant lymphoproliferative disease. Transplantation 66:1641-1644. [DOI] [PubMed] [Google Scholar]
- 7.Gustafsson, A., et al. 2000. Epstein-Barr virus (EBV) load in bone marrow transplant recipients at risk to develop posttransplant lymphoproliferative disease: prophylactic infusion of EBV-specific cytotoxic T cells. Blood 95:807-814. [PubMed] [Google Scholar]
- 8.Hanto, D. W., et al. 1982. Epstein-Barr virus-induced B-cell lymphoma after renal transplantation: acyclovir therapy and transition from polyclonal to monoclonal B-cell proliferation. N. Engl. J. Med. 306:913-918. [DOI] [PubMed] [Google Scholar]
- 9.Jabs, W. J., et al. 2001. Normalized quantification by real-time PCR of Epstein-Barr virus load in patients at risk for posttransplant lymphoproliferative disorders. J. Clin. Microbiol. 39:564-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenagy, D. N., et al. 1995. Epstein-Barr virus DNA in peripheral blood leukocytes of patients with posttransplant lymphoproliferative disease. Transplantation 60:547-554. [DOI] [PubMed] [Google Scholar]
- 11.Kimura, H., et al. 1999. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 37:132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krieger, N. R., et al. 2000. Significance of detecting Epstein-Barr-specific sequences in the peripheral blood of asymptomatic pediatric liver transplant recipients. Liver Transplant. 6:62-66. [DOI] [PubMed] [Google Scholar]
- 13.Limaye, A. P., et al. 1999. Detection of Epstein-Barr virus DNA in sera from transplant recipients with lymphoproliferative disorders. J. Clin. Microbiol. 37:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo, Y. M., et al. 1999. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 59:1188-1191. [PubMed] [Google Scholar]
- 15.Lo, Y. M., et al. 1999. Quantitative and temporal correlation between circulating cell-free Epstein-Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res. 59:5452-5455. [PubMed] [Google Scholar]
- 16.Lucas, K. G., et al. 1998. Semiquantitative Epstein-Barr virus polymerase chain reaction analysis of peripheral blood from organ transplant patients and risk for the development of lymphoproliferative disease. Blood 92:3977-3978. [PubMed] [Google Scholar]
- 17.McDiarmid, S. V., et al. 1998. Prevention and preemptive therapy of postransplant lymphoproliferative disease in pediatric liver recipients. Transplantation 66:1604-1611. [DOI] [PubMed] [Google Scholar]
- 18.Niesters, H. G., et al. 2000. Development of a real-time quantitative assay for detection of Epstein-Barr virus. J. Clin. Microbiol. 38:712-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riddler, S. A., M. C. Breinig, and J. L. McKnight. 1994. Increased levels of circulating Epstein-Barr virus (EBV)-infected lymphocytes and decreased EBV nuclear antigen antibody responses are associated with the development of posttransplant lymphoproliferative disease in solid-organ transplant recipients. Blood 84:972-984. [PubMed] [Google Scholar]
- 20.Rooney, C. M., et al. 1995. Early identification of Epstein-Barr virus-associated post-transplantation lymphoproliferative disease. Br. J. Haematol. 89:98-103. [DOI] [PubMed] [Google Scholar]
- 21.Rowe, D. T., et al. 1997. Use of quantitative competitive PCR to measure Epstein-Barr virus genome load in the peripheral blood of pediatric transplant patients with lymphoproliferative disorders. J. Clin. Microbiol. 35:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savoie, A., et al. 1994. Direct correlation between the load of Epstein-Barr virus-infected lymphocytes in the peripheral blood of pediatric transplant patients and risk of lymphoproliferative disease. Blood 83:2715-2722. [PubMed] [Google Scholar]
- 23.Stevens, S. J., et al. 1999. Monitoring of Epstein-Barr virus DNA load in peripheral blood by quantitative competitive PCR. J. Clin. Microbiol. 37:2852-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens, S. J., et al. 2002. Comparison of quantitative competitive PCR with LightCycler-based PCR for measuring Epstein-Barr virus DNA load in clinical specimens. J. Clin. Microbiol. 40:3986-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens, S. J., I. Pronk, and J. M. Middeldorp. 2001. Toward standardization of Epstein-Barr virus DNA load monitoring: unfractionated whole blood as preferred clinical specimen. J. Clin. Microbiol. 39:1211-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Wadowsky, R. M., et al. 2003. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J. Clin. Microbiol. 41:5245-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner, H. J., et al. 1992. Detection and quantification of latently infected B lymphocytes in Epstein-Barr virus-seropositive, healthy individuals by polymerase chain reaction. J. Clin. Microbiol. 30:2826-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner, H. J., et al. 2000. Real-time polymerase chain reaction (RQ-PCR) for the monitoring of Epstein-Barr virus (EBV) load in peripheral blood mononuclear cells. Klin. Padiatr. 212:206-210. [DOI] [PubMed] [Google Scholar]
- 30.Wagner, H. J., et al. 2002. Longitudinal analysis of Epstein-Barr viral load in plasma and peripheral blood mononuclear cells of transplanted patients by real-time polymerase chain reaction. Transplantation 74:656-664. [DOI] [PubMed] [Google Scholar]
- 31.Wagner, H. J., et al. 2001. Patients at risk for development of posttransplant lymphoproliferative disorder: plasma versus peripheral blood mononuclear cells as material for quantification of Epstein-Barr viral load by using real-time quantitative polymerase chain reaction. Transplantation 72:1012-1019. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto, M., et al. 1995. Detection and quantification of virus DNA in plasma of patients with Epstein-Barr virus-associated diseases. J. Clin. Microbiol. 33:1765-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]