Abstract
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) of intact bacteria yields a reproducible spectrum depending upon growth conditions, strain, or species. Using whole viable bacteria we describe here the application of MALDI-TOF-MS to the identification of coagulase-negative staphylococci (CoNS). Our aim was, once a bacterium has been recognized as Micrococcaceae, to identify peaks in the spectrum that can be used to identify the species or subspecies. MALDI-TOF-MS was performed using bacteria obtained from one isolated colony. One reference strain for each of the 23 clinically relevant species or subspecies of Micrococcaceae was selected. For each reference strain, the MALDI-TOF-MS profile of 10 colonies obtained from 10 different passages was analyzed. For each strain, only peaks that were conserved in the spectra of all 10 isolated colonies and with a relative intensity above 0.1 were retained, thus leading to a set of 3 to 14 selected peaks per strain. The MALDI-TOF-MS profile of 196 tested strains was then compared with that of the set of selected peaks of each of the 23 reference strains. In all cases the best hit was with the set of peaks of the reference strain belonging to the same species as that of the tested strain, thus demonstrating that the 23 sets of selected peaks can be used as a database for the rapid species identification of CoNS. Similar results were obtained using four different growth conditions. Extending this strategy to other groups of relevant pathogenic bacteria will allow rapid bacterial identification.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) can examine the profile of proteins detected directly from the intact bacterial cell surface (4, 8, 13, 17). This technique, based on relative molecular masses, is a soft-ionization method, allowing desorption of peptides and proteins from whole different cultured microorganisms (24). Ions are separated and detected according to their molecular masses and charges. Bacteria are identified by their mass/charge ratio (m/z). This approach yields a reproducible spectrum within minutes (4, 13), consisting of a series of peaks from m/z 500 to 11,000. Each peak corresponds to a molecular fragment released from the cell surface during laser desorption.
MALDI-TOF-MS has already been used to characterize bacteria (15, 16, 18, 26, 27). Among the various components identified in a spectra, only a few are specific for a given species; others are either strain specific or vary upon growth conditions (media, incubation time, etc.) and cannot be used to identify a bacterial species. Several reports have addressed this issue of bacterial identification using mass spectrometry. Jarman et al. have developed a statistically based algorithm for bacterial identification with MALDI-TOF-MS of five reference strains cultivated in liquid media, with the aim of differentiating bacteria within a mixture of germs. These authors could obtain the correct identification with a rate of 75% to 95% (15). Keys et al. have developed a technique for rapid characterization of pathogenic bacteria from colonies isolated on solid plates. They tested 293 unknown clinical strains to assess the potential of their database. The percentage of correct identity varied between 33 and 100%, depending upon the number of representative strains per species in the database. These authors pointed out that species- or subspecies-specific markers in the spectra are difficult to identify, as overlapping signal ions increase along with the number of strains registered in the database (16). These drawbacks have hampered the use of MALDI-TOF-MS in clinical microbiology laboratories.
Bacterial identification following isolation is an important step in the management of infectious diseases. For example, identification of coagulase-negative staphylococci (CoNS), frequently isolated in routine clinical microbiology laboratories, is important to establish the role of these bacteria as an infectious agent. Indeed, the repeated isolation of the same microorganism in several samples indicates the clinical significance of CoNS isolates (3). This points out that a rapid identification at the species level of a clinical isolate is therefore required. Routine identification of CoNS appears to be unsatisfactory, unreliable, and irreproducible (3, 12). Commercial identification kits and automated systems are indeed unable to differentiate between the different species of CoNS (2, 9, 23), and molecular methods remain time-consuming and often expensive.
In this work, we engineered a strategy that identifies peaks within the spectrum obtained by MALDI-TOF-MS from intact bacteria that can be used for the identification of bacterial species or subspecies belonging to Micrococcaceae. Extending this strategy to other major groups of pathogenic bacteria will open the path to rapid and inexpensive means of bacterial identification in routine clinical microbiology laboratories.
MATERIALS AND METHODS
Bacterial strains.
Strains used in this study are listed Tables 1 and 2. These strains were obtained from two different origins. Fifty-one characterized strains belonging to various species and subspecies of Micrococcaceae (Tables 1 and 2) were purchased from the collection of the Institut Pasteur (Paris, France). One hundred clinical CoNS isolates were also studied (Table 2): 64 strains isolated from blood cultures, 24 strains isolated from cases of child mediastinitis (skin, mediastinal liquid, electrodes), and 12 strains isolated from bone infections (Hôpital Necker-Enfants Malades, Paris, France; Hôpital Raymond Poincaré, Garches, France). In addition, 68 clinical strains of Staphylococcus aureus isolated from miscellaneous infections were analyzed. The clinical strains of CoNS were differentiated from S. aureus strains by conventional phenotypic tests including the Slidex latex agglutination test (bioMérieux, Marcy l'Etoile, France) and DNase test. Tube coagulase tests were performed in case of discordance between the previous two techniques. The identification of the CoNS at the species level was obtained by sequencing an internal fragment of the sodA gene as previously described (22). Briefly, extraction of genomic DNA from pure cultures of CoNS was performed with a QIAGEN (Courtaboeuf, France) kit. The partial sodA gene was amplified and the PCR product sequenced using an ABI Big Dye Terminator v1.1 cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA). The nucleotide sequences were sent to the GenBank database. This strategy was used to identify the 100 clinical CoNS isolates (Table 2).
TABLE 1.
Selected strains used to establish the MALDI-TOF-MS databases
| Species or subspecies | Strain |
|---|---|
| S. aureus | CIP 7625 |
| Micrococcus luteus | CIP 103664 |
| S. lentus | CIP 103585 |
| S. epidermidis | CIP 103563 |
| S. warneri | CIP 103960 |
| S. xylosus | CIP 8166 |
| S. intermedius | CIP 8177 |
| S. haemolyticus | CIP 81.56 |
| S. saprophyticus subsp. saprophyticus | CIP 104064 |
| S. saprophyticus subsp. bovis | CIP 105260T |
| S. lugdunensis | CIP 103642 |
| S. hominis subsp. hominis | CIP 102642 |
| S. hominis subsp. novobiosepticus | CIP 105721 |
| S. capitis subsp. capitis | CIP 8153T |
| S. capitis subsp. urealyticus | CIP 104191 |
| S. caprae | CIP 104519 |
| S. pasteuri | CIP 103831 |
| S. cohnii subsp. cohnii | CIP 8154T |
| S. cohnii subsp. urealyticus | CIP 104023 |
| S. schleiferi subsp. schleiferi | CIP 103643T |
| S. schleiferi subsp. coagulans | CIP 104370 |
| S. sciuri subsp. sciuri | CIP 103824 |
| S. simulans | CIP 8164T |
TABLE 2.
Strains belonging to the family Micrococcaceae
| Species or subspecies | Strain(s) and/or no. of CIa |
|---|---|
| S. aureus | 68 CI |
| M. luteus | CIP A270 |
| S. epidermidis | 81 CI |
| S. warneri | CIP 106511, CIP 8165, 6 CI |
| S. xylosus | CIP 103720, CIP 104065 |
| S. intermedius | CIP 81.60 |
| S. haemolyticus | CIP 104114, 13 CI |
| S. saprophyticus subsp. saprophyticus | CIP 76.125T, CIP 103545 |
| S. saprophyticus subsp. bovis | CIP 105262, CIP 105261 |
| S. lugdunensis | CIP 103584 |
| S. hominis subsp. hominis | CIP 81.57, CIP 104689 |
| S. hominis subsp. novobiosepticus | CIP 105719T |
| S. capitis subsp. capitis | CIP 103688 |
| S. capitis subsp. urealyticus | CIP 104192T |
| S. caprae | CIP 104000T, CIP 104520 |
| S. pasteuri | CIP 105540T, CIP 103830, CIP 103832 |
| S. cohnii subsp. urealyticus | CIP 104024T, CIP 104025 |
| S. schleiferi subsp. coagulans | CIP 104366 |
| S. sciuri subsp. sciuri | CIP 8162T, CIP 103583, CIP 103825 |
CI, clinical isolates.
MALDI-TOF-MS.
The strains were grown on Mueller-Hinton agar or Columbia agar supplemented with 5% horse blood (bioMérieux) and incubated 24 or 48 h at 37°C. An isolated colony was harvested in 100 μl of sterile water; 1 μl of this mixture was deposited on a target plate (Bruker Daltonics, Bremen, Germany) in three replicates and allowed to dry at room temperature. One microliter of absolute ethanol was then added in each well. After the mixture dried, 1 μl of matrix solution DHB (2,5-dihydroxybenzoic acid, 50 mg/ml, 30% acetonitrile, 0.1% trifluoroacetic acid) was added. Samples were then processed in the MALDI-TOF-MS spectrometer (Autoflex; Bruker Daltonics) with flex control software (Bruker Daltonics). Positive ions were extracted with an accelerating voltage of 20 Hz in linear mode. Each spectrum was the sum of the ions obtained from 200 laser shots performed in five different regions of the same well. The spectra have been analyzed in an m/z range of 1,000 to 11,000. The analysis was performed with the flex analysis software and calibrated with protein calibration standard T (Protein I; Bruker Daltonics). The data obtained with the three replicates were added to minimize random effect. The presence and absence of peaks were considered as fingerprints for a particular isolate. The profiles were analyzed and compared using the software BGP database available on the website http://sourceforge.net/projects/bgp.
RESULTS
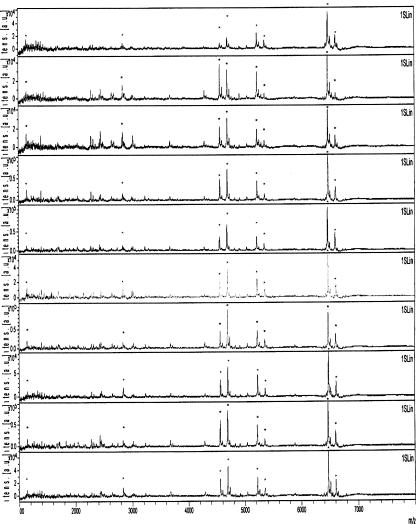
Micrococcaceae are defined as catalase-positive, gram-positive cocci which grow aerobically. Strains listed Table 1 are those selected as being representative of each species or subspecies routinely isolated in clinical microbiology laboratories. Our aim was, once a bacterium has been recognized as being a member of the Micrococcaceae, to identify peaks obtained by MALDI-TOF-MS that are specific for each species or subspecies listed Table 1. Subsequent peaks could then be used for bacterial identification. Ten isolates of each of these selected strains listed Table 1 were analyzed by MALDI-TOF-MS as described in Materials and Methods. For each spectrum, a value corresponding to the intensity was given to each peak. The peak with the highest intensity was arbitrarily set to 1; all the other peaks had a value corresponding to the relative intensity of this highest peak (Fig. 1). It should be pointed out that minor peaks (relative intensity below 0.1) were inconstantly present. We reasoned that peaks that are species specific are likely to correspond to bacterial components produced in high quantity and that such components would therefore generate conserved peaks of high relative intensity. We subsequently concentrated on peaks with a relative intensity above 0.1. As some of these major peaks were missing when various colonies of the same strain were studied (Fig. 1), we retained only those peaks with an intensity above 0.1 that were present in all 10 sets of data obtained for a given strain. In order to determine the impact of growth conditions on bacterial identification, the above-described procedure was performed with all strains listed in Table 1 and grown on Mueller-Hinton agar for 24 or 48 h or Columbia agar supplemented with 5% horse blood for 24 or 48 h. These four growth conditions were the basis for four databases, designated 1 through 4. For a given strain, the standard deviation of the m/z value (normalized data) for each of the conserved peaks was never above 7. For the m/z values of the peaks that had a relative intensity above 0.1 and that were present in all 10 sets of data for each growth condition, see Fig. S1 in the supplementary material. Table 3 shows for each selected strain and each database the number of peaks that have been retained. Some peaks were present in all databases, and others varied with the growth conditions. As shown Fig. 2, for a given database, the set of peaks was specific of each selected strain shown on Table 1.
FIG. 1.
MALDI-TOF-MS profiles of 10 isolates of the same strain of S. hominis subsp. hominis CIP 102642. *, peaks constantly obtained.
TABLE 3.
Number of peaks used to differentiate the Micrococcaceae
| Species or subspecies | No. of peaks for databasea:
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| S. aureus | 11 | 11 (8) | 7 (2) | 8 (4) |
| S. capitis subsp. capitis | 8 | 14 (7) | 4 (4) | 6 (4) |
| S. capitis subsp. urealyticus | 5 | 9 (4) | 6 (3) | 4 (4) |
| S. caprae | 5 | 12 (5) | 6 (2) | 6 (2) |
| S. cohnii subsp. cohnii | 6 | 9 (4) | 10 (2) | 11 (5) |
| S. cohnii subsp. urealyticus | 6 | 12 (6) | 7 (4) | 8 (5) |
| S. epidermidis | 4 | 11 (4) | 4 (2) | 6 (2) |
| S. haemolyticus | 5 | 10 (5) | 4 (4) | 8 (4) |
| S. hominis subsp. hominis | 8 | 9 (4) | 4 (4) | 5 (3) |
| S. hominis subsp. novobiosepticus | 6 | 12 (6) | 8 (2) | 8 (3) |
| S. intermedius | 10 | 12 (9) | 7 (6) | 6 (5) |
| S. lugdunensis | 9 | 12 (5) | 5 (3) | 9 (4) |
| S. pasteuri | 11 | 12 (11) | 10 (10) | 12 (9) |
| S. saprophyticus subsp. bovis | 9 | 12 (8) | 7 (2) | 5 (3) |
| S. saprophyticus subsp. saprophyticus | 8 | 10 (6) | 7 (4) | 11 (4) |
| S. schleiferi subsp. coagulans | 7 | 8 (6) | 4 (4) | 10 (5) |
| S. schleiferi subsp. schleiferi | 9 | 10 (9) | 5 (5) | 9 (5) |
| S. sciuri subsp. sciuri | 8 | 11 (8) | 3 (1) | 4 (3) |
| S. simulans | 8 | 8 (7) | 4 (5) | 10 (7) |
| S. warneri | 9 | 13 (6) | 10 (7) | 14 (6) |
| S. xylosus | 5 | 9 (4) | 8 (4) | 9 (4) |
| M. luteus | 6 | 13 (4) | 10 (4) | 13 (6) |
| S. lentusb | 3 | 7 (3) | ||
The databases are obtained using bacterial cultures grown on Mueller-Hinton agar for 24 h (database 1), Mueller-Hinton agar for 48 h (database 2), Columbia horse blood medium for 24 h (database 3), or Columbia horse blood medium for 48 h (database 4). Numbers in parentheses are the numbers of peaks for genospecies in common with those of database 1.
S. lentus did not grow on horse blood medium.
FIG. 2.
Dendrograms of species of Micrococcaceae. The dendrograms were engineered using databases 1 (spectra obtained from bacteria grown on Mueller-Hinton agar for 24 h) and 2 (spectra obtained from bacteria grown on Mueller-Hinton agar for 48 h). Similar results (not shown) were observed with databases 3 and 4.
We next aimed at determining whether the above databases could be used for bacterial identification, thus demonstrating that the set of peaks of each selected strain is, at least partially, conserved among strains belonging to the same species or subspecies. To address this point, MALDI-TOF-MS was performed using bacteria grown on Mueller-Hinton agar for 24 h at 37°C. The strains used are isolates listed Table 2. For each of these tested strains, the peaks with a value above 0.1 were retained. We next compared the profile obtained for each of these isolates with that of those of database 1. To perform this task, a software (BGP-database, available on http://sourceforge.net/projects/bgp) was developed, allowing the rapid identification of the set of values in the database closest to that for the tested strain. This software chooses the best match between the tested strain and the reference strains of the database, taking into account a possible error of the m/z value. This value was set to 7. All the 196 tested strains listed Table 2 had always the best match with the strain belonging to the same species or subspecies of the database (see Fig. S2 in the supplemental material). Taken together, these data demonstrate that database 1 is suitable for species or subspecies identification of Micrococcaceae grown on Mueller-Hinton agar for 24 h.
We next tested the same set of data, obtained with strains grown on Mueller-Hinton agar for 24 h, using databases 2, 3, and 4, which were obtained using growth conditions different from those used to grow the 196 tested strains. Table 4 shows for each species or subspecies the minimal and maximal numbers of peaks that were conserved between the tested strains and each of the four databases. Even though in average these numbers were lower than those obtained with database 1, which was engineered using the same growth conditions as those used to grow the tested strains, identification at the species level remained possible in all cases. The only difference observed is that identification at the subspecies level was not always possible for S. hominis and S. saprophyticus, unlike results obtained with database 1.
TABLE 4.
Numbers of common peaks (m/z > 0.1) between the tested strains grown on Mueller-Hinton agar for 24 h and the four databases
| Species or subspecies | No. of strains | No. or range of common peaks/total no. of peaks of database:
|
|||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| S. aureus | 68 | 7-11/11 | 7-11/11 | 3-6/7 | 4-7/8 |
| S. capitis subsp. capitis | 1 | 8/8 | 13/14 | 4/4 | 5/6 |
| S. capitis subsp. urealyticus | 1 | 5/5 | 7/9 | 6/6 | 4/4 |
| S. caprae | 2 | 5/5 | 9-11/12 | 6/6 | 4-6/6 |
| S. cohnii subsp. urealyticus | 2 | 6/6 | 9-11/12 | 6-7/7 | 4-8/8 |
| S. epidermidis | 81 | 2-4/4 | 3-8/11 | 2-3/4 | 2-5/6 |
| S. haemolyticus | 14 | 3-5/5 | 4-7/10 | 2-4/4 | 4-7/8 |
| S. hominis subsp. hominis | 2 | 8/8 | 5-8/9 | 4/4 | 4/5 |
| S. hominis subsp. novobiosepticus | 1 | 6/6 | 8/12 | 5/8 | 4/8 |
| S. intermedius | 1 | 10/10 | 11/12 | 7/7 | 6/6 |
| S. lugdunensis | 1 | 7/7 | 9/12 | 5/5 | 7/9 |
| S. saprophyticus subsp. bovis | 2 | 9/9 | 10-11/12 | 4/7 | 4/5 |
| S. saprophyticus subsp. saprophyticus | 2 | 8/8 | 10/10 | 6-7/7 | 8-9/11 |
| S. schleiferi subsp. coagulans | 1 | 7/7 | 7/8 | 4/4 | 8/10 |
| S. pasteuri | 3 | 11/11 | 11-12/12 | 10/10 | 10/12 |
| S. sciuri subsp. sciuri | 3 | 7/7 | 9-11/11 | 2/3 | 3-4/4 |
| S. xylosus | 2 | 5/5 | 9/9 | 5-7/8 | 6-7/9 |
| S. warneri | 8 | 5-9/9 | 6-12/13 | 5-10/10 | 5-10/14 |
| M. luteus | 1 | 7/7 | 9/12 | 5/10 | 11/11 |
Altogether, our data demonstrate that, by selecting an appropriate set of strains and retaining only the conserved peaks with a m/z above 0.1, a database can be engineered and used for species or subspecies identification of Micrococcaceae. Furthermore the specificity of these peaks is such that species identification remained possible even if the strains to be identified were grown using culture conditions different from those used to build the database.
DISCUSSION
Bacterial identification is routinely achieved using phenotypically based techniques. However, those techniques remain time-consuming and sometimes of limited value, as for example for CoNS, where commercial identification kits identified only 37% of 177 CoNS isolates with the API 20 Staph system (3). Ribotyping and PCR amplicon sequencing-based methods for identification of CoNS have been described (1, 3, 5, 6, 10, 14, 20, 22, 25). The methods targeting the sodA or the tuf gene are currently preferred for diagnostic purposes (20, 22, 25). However, they remain time-consuming, expensive, and technically demanding. In addition, differentiation at the subspecies level is not always possible using the sodA sequence.
With MALDI-TOF-MS technique, sample preparation and analysis are simple and can be performed within minutes. No special lysis step is necessary beyond the exposure to the matrix solution, and the instrument does not require a specialist operator. Only a loopful of cells is needed for MALDI-TOF-MS analysis, and the profile is generated with minimal consumables and cost. For one sample, MALDI-TOF-MS analysis is obtained in a few minutes (versus 1 day for API 20 Staph and at least several hours for the molecular biology techniques). Numerous samples can be processed per day, and furthermore the cost of the analysis is inexpensive compared to other techniques (in the range of a few cents).
Surface biomarkers, which are excluded in the description of species, can be used as useful criteria for describing species where there is a paucity of reliable differential characters. The approach establishes a unique system for bacterial identification, as no other phenotypic analysis system currently utilizes surface components for identification. Few studies described this technique in a medical application of species identification. Haag et al. have reported the rapid characterization of pathogenic Haemophilus strains (11). The same authors could determine strain differences from the same species of Haemophilus influenzae in several patients in the same hospital, to establish if their infections were nosocomial. Other authors have detected strain-specific biomarkers based on analysis of six different strains of Helicobacter pylori (21). Lundquist et al. showed that this technique permits the differentiation of the four subspecies of Francisella tularensis, indistinguishable by serological methods (19). In addition, this method may allow the differentiation of methicillin-resistant and -sensitive strains of S. aureus (7).
In this work, we demonstrate that MALDI-TOF-MS is a powerful tool for the identification of clinically relevant species of CoNS. The strategy reported in this paper is currently being extended to other major groups of bacteria isolated in clinical microbiology laboratories, thus allowing for the proposal of a strategy forbacterial identification in clinical laboratories relying on a two-step process. The first one is a rapid classification of the isolated bacteria to be identified based on routine phenotypic analysis such as growth conditions, Gram staining, and morphology, thus allowing classification of the pathogen within a group of bacteria usually isolated in clinical microbiology laboratories, e.g., Streptococacceae, aerobic gram-negative bacteria, aerobic gram-positive bacilli, anaerobic bacteria, etc. The second step relies on a MALDI-TOF-MS analysis, allowing the rapid identification of the species. We believe that such a strategy may allow the replacement in the near future of the traditional methods of identification, which are time-consuming and sometimes not reliable.
Supplementary Material
Acknowledgments
We thank Jean-Louis Gaillard, Claire Poyart, and Valérie Sivadon for providing CoNS strains. We thank Franck Brouillard and Aleksander Edelman from the Proteomic Core Facilities, Faculté de Médecine Paris 5 (Paris, France).
We declare that we have no conflict of interest.
Footnotes
Published ahead of print on 16 May 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Andollina, A., A. De Cesare, G. Bertoni, L. Modeli, and G. Manfreda. 2004. Identification and genetic characterization of orthopaedic Staphylococcus isolates collected in Italy by automated EcoRI ribotyping. FEMS Microbiol. Lett. 234:275-280. [DOI] [PubMed] [Google Scholar]
- 2.Calvo, J., J. L. Hernandez, M. C. Farinas, D. Garcia-Palomo, and J. Aguero. 2000. Osteomyelitis caused by Staphylococcus schleiferi and evidence for misidentification of this Staphylococcus species by an automated identification system. J. Clin. Microbiol. 38:3887-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carretto, E., D. Barbarini, I. Couto, D. De Vitis, P. Marone, J. Verhoef, H. De Lencastre, and S. Brisse. 2005. Identification of coagulase-negative staphylococci other than Staphylococcus epidermidis by automated ribotyping. Clin. Microbiol. Infect. 11:177-184. [DOI] [PubMed] [Google Scholar]
- 4.Claydon, M. A., S. N. Davey, V. Edwards-Jones, and D. B. Gordon. 1996. The rapid identification of intact microorganisms using mass spectrometry. Nat. Biotechnol. 14:1584-1586. [DOI] [PubMed] [Google Scholar]
- 5.Couto, I., S. Pereira, M. Miragaia, I. S. Sanches, and H. de Lencastre. 2001. Identification of clinical staphylococcal isolates from humans by internal transcribed spacer PCR. J. Clin. Microbiol. 39:3099-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Buyser, M. L., A. Morvan, S. Aubert, F. Dilasser, and N. El Sohl. 1992. Evaluation of rRNA gene probe for identification of species and subspecies within the genus Staphylococcus. J. Gen. Microbiol. 138:889-899. [DOI] [PubMed] [Google Scholar]
- 7.Edwards-Jones, V., M. A. Claydon, D. J. Evason, J. Walker, A. J. Fox, and D. B. Gordon. 2000. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 49:295-300. [DOI] [PubMed] [Google Scholar]
- 8.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by maldi mass spectrometry. Mass Spectrom. Rev. 20:157-171. [DOI] [PubMed] [Google Scholar]
- 9.Grant, C. E., D. L. Sewell, M. Pfaller, R. V. Bumgardner, and J. A. Williams. 1994. Evaluation of two commercial systems for identification of coagulase-negative staphylococci to species level. Diagn. Microbiol. Infect. Dis. 18:1-5. [DOI] [PubMed] [Google Scholar]
- 10.Gribaldo, S., B. Cookson, N. Saunders, R. Marples, and J. Stanley. 1997. Rapid identification by specific PCR of coagulase-negative staphylococcal species important in hospital infection. J. Med. Microbiol. 46:45-53. [DOI] [PubMed] [Google Scholar]
- 11.Haag, A., M. S. N. Taylor, K. H. Johnston, and R. B. Cole. 1998. Rapid identification and speciation of Haemophilus bacteria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 33:750-756. [DOI] [PubMed] [Google Scholar]
- 12.Heikens, E., A. Fleer, A. Paauw, A. Florijn, and A. C. Fluit. 2005. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 43:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holland, R. D., J. G. Wilkes, F. Rafii, J. B. Sutherland, C. C. Persons, K. J. Voorhees, and J. O. Lay. 1996. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10:1227-1232. [DOI] [PubMed] [Google Scholar]
- 14.Izard, N. C., G. M. Hächler, and F. H. Kayser. 1992. Ribotyping of coagulase-negative staphylococci with special emphasis on intraspecific typing of Staphylococcus epidermidis. J. Clin. Microbiol. 30:817-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarman, K. H., S. T. Cebula, A. J. Saenz, C. E. Petersen, N. B. Valentine, M. T. Kingsley, and K. L. Wahl. 2000. An algorithm for automated bacterial identification using matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 72:1217-1223. [DOI] [PubMed] [Google Scholar]
- 16.Keys, C. J., D. J. Dare, H. Sutton, G. Wells, M. Lunt, T. McKenna, M. McDowall, and H. N. Shah. 2004. Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterization of bacteria implicated in human infectious diseases. Infect. Genet. Evol. 4:221-242. [DOI] [PubMed] [Google Scholar]
- 17.Krishnamurthy, T., and P. L. Ross. 1996. Rapid identification of bacteria by direct matrix-assisted laser desorption/ionization mass spectrometric analysis of whole cells. Rapid Commun. Mass Spectrom. 10:1992-1996. [DOI] [PubMed] [Google Scholar]
- 18.Lay, J. O. 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172-194. [DOI] [PubMed] [Google Scholar]
- 19.Lundquist, M., M. B. Caspersen, P. Wikström, and M. Forsman. 2005. Discrimination of Francisella tularensis subspecies using surface enhanced laser desorption ionization mass spectrometry and multivariate data analysis. FEMS Microbiol. Lett. 243:303-310. [DOI] [PubMed] [Google Scholar]
- 20.Martineau, F., F. J. Picard, D. Ke, S. Paradis, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2001. Development of a PCR assay for identification of staphylococci at genus and species level. J. Clin. Microbiol. 39:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson, C. L. 1999. Fingerprinting of Helicobacter pylori strains by matrix-assisted laser desorption/ionization mass spectrometric analysis. Rapid Commun. Mass Spectrom. 13:1067-1071. [DOI] [PubMed] [Google Scholar]
- 22.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renneberg, J., J. K. Rieneck, and E. Gutschik. 1995. Evaluation of Staph ID 32 system and Staph-Zym system for identification of coagulase-negative staphylococci. J. Clin. Microbiol. 33:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruelle, V., B. El Moualij, W. Zorzi, P. Ledent, and E. De Pauw. 2004. Rapid identification of environmental bacterial strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 18:2013-2019. [DOI] [PubMed] [Google Scholar]
- 25.Sivadon, V., M. Rottman, S. Chaverot, J.-C. Quincampoix, V. Avettand, P. de Mazancourt, L. Bernard, P. Trieu-Cuot, J.-M. Féron, A. Lortat-Jacob, P. Piriou, T. Judet, and J.-L. Gaillard. 2005. Use of genotypic identification by sodA sequencing in a prospective study to examine the distribution of coagulase-negative staphylococcus species among strains recovered during septic orthopedic surgery and evaluate their significance. J. Clin. Microbiol. 43:2952-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valentine, N., S. Wunschel, D. Wunschel, C. Petersen, and K. Wahl. 2005. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 71:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wahl, K. L., S. C. Wunschel, K. H. Jarman, N. B. Valentine, C. E. Petersen, M. T. Kingsley, K. A. Zartolas, and A. J. Saenz. 2002. Analysis of microbial mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74:6191-6199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.