Abstract
Recent studies have suggested that community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections are encroaching upon nosocomial settings. We assessed the performance characteristics of a rule using the antimicrobial phenotype to predict genotype. This rule could be applied for epidemiologic purposes to describe the trend in CA-MRSA infections over time.
Hospitals and nursing homes were once thought to be the exclusive province of methicillin-resistant Staphylococcus aureus (MRSA); however, over the past 9 years, a single strain, USA-300, has emerged as a pathogen in community patients without health care-associated risk factors (4, 5). Community-associated MRSA (CA-MRSA) infections range from minor skin and soft tissue to severe systemic infections, and surveillance for this emerging pathogen has gained importance (3, 10). As the epidemic of CA-MRSA progresses, infections, particularly with the predominant USA-300 clone, may be encroaching upon nosocomial settings (7, 13).
In contrast to its nosocomial counterparts, CA-MRSA is typically susceptible to multiple classes of antibiotics. Several methods could be used to classify MRSA as health care associated or community associated, including (i) genotypic testing, based on the results of pulsed-field gel electrophoresis (PFGE) or other molecular techniques (9), (ii) phenotypic testing, based on antimicrobial susceptibility, and (iii) epidemiologic analysis, based on time from admission to a positive culture (4). While several studies have examined the phenotypic characteristics of CA-MRSA isolates, none to our knowledge have assessed the performance characteristics of a rule using the antimicrobial phenotype to predict genotype.
Our aim was to derive a phenotypic prediction rule for the genotypes of MRSA bloodstream isolates. This rule could be used for epidemiologic purposes to monitor the MRSA isolates over time and to describe the evolution of CA-MRSA, e.g., whether CA-MRSA isolates are becoming prevalent nosocomial strains. To assess the value of the phenotypic susceptibility pattern as a predictor of genotype, we examined the performance characteristics of various combinations of antimicrobial susceptibilities.
PFGE was performed on bloodstream MRSA isolates from 147 patients in the Cook County Bureau of Health Services from 1 January 2004 to 31 July 2006. PFGE, Panton-Valentine leukocidin (PVL) analysis, and staphylococcal cassette chromosome mec (SCCmec) analysis were carried out as described elsewhere (1, 6, 8). The first blood culture from an outpatient clinic visit, emergency visit, or inpatient stay was used; 91% of the patients in this group were inpatients. Genotypes were then used to divide isolates into community genotype (CG) and hospital genotype (HG). USA-300 and USA-400 were classified as CG; USA-100, USA-500, and USA-800 were labeled HG (9). We generated phenotypes based on antibiotic susceptibilities (MicroScan, Dade Behring, West Sacramento, CA) to clindamycin, fluoroquinolones, gentamicin, and tetracycline. Since 95% of isolates in this group were susceptible to trimethoprim-sulfamethoxazole and 91% were resistant to erythromycin, these agents were likely to be nondiscriminatory and were not included in our analysis.
A CG was seen in 86 isolates: 84 USA-300 isolates and 2 USA-400 isolates. Fifty-one isolates had an HG: 30 USA-100 isolates, 12 USA-500 isolates, and 9 USA-800 isolates. Ten isolates had another genotype and were not included in the analysis. PVL and SCCmec analyses were performed on 124 and 122 isolates, respectively. CG strains typically carried PVL (99% of CG strains) and SCCmec type IV (100% of CG strains), while HG strains did not (PVL, 11%, and SCCmec type IV, 35%). We found that susceptibility to clindamycin (sensitivity, 95%; specificity, 80%; likelihood ratio, 4.86; 95% confidence interval [CI], 3.29 to 6.47) or fluoroquinolones (sensitivity, 73%; specificity, 86%; likelihood ratio, 5.34; 95% CI, 2.91 to 10.58) best predicted a CG; the higher likelihood ratio of fluoroquinolones was the result of a higher specificity for CG. The positive predictive values (PPVs) for identifying a CG among all MRSA strains were 62%, 83%, and 94% for clindamycin-susceptible strains and 64%, 84%, and 94% for fluoroquinolone-susceptible strains when the prevalences of such strains were 25%, 50%, and 75%, respectively, i.e., the PPV of each rule for CG strains increased when the prevalence of such strains was higher. For strains that were susceptible to clindamycin or fluoroquinolones, the likelihood ratio of predicting a CG among all MRSA strains was 4.48 (95% CI, 3.15 to 5.54); the PPVs for CG strains were 60%, 82%, and 93% when the prevalences of such strains were 25%, 50%, and 75%, respectively.
In a second analysis, we also assessed the impact of increasing antibiotic resistance among CG strains on the performance of a prediction rule. Since the fluoroquinolone rule predicted CG strains the best, we determined its performance when resistance to ciprofloxacin among CG isolates increased by 10%, 20%, and 30%. With increasing resistance, the likelihood ratio for predicting a CG strain based on ciprofloxacin susceptibility decreased: likelihood ratios were 4.58 (95% CI, 2.43 to 9.27), 3.90 (95% CI, 2.03 to 8.02), and 3.14 (95% CI, 1.60 to 6.56) when ciprofloxacin resistance was increased by 10%, 20%, and 30%, respectively.
In a third analysis, we used the number of antibiotic agents from eight different classes, rifampin, vancomycin, clindamycin, erythromycin, gentamicin, fluoroquinolones, tetracycline, and trimethoprim-sulfamethoxazole, to which isolates expressed antibiotic resistance or the hospital day (0 if outpatient) that culture was obtained to predict genotypes. In logistic regression (SPSS version 10; Chicago, IL), each additional antibiotic class to which MRSA was resistant increased the odds of an HG by 3.91 (P < 0.001; 95% CI, 2.50 to 6.13). Figure 1 shows the receiver operating characteristic (ROC) curve for this model (area under the ROC curve, 0.856; 95% CI, 0.775 to 0.936), which suggests a cutoff point of resistance to ≤2 classes for CG isolates and to ≥3 classes for HG isolates. With this cutoff, the likelihood ratio for an organism with resistance to less than three classes for a CG was 4.63 (95% CI, 3.01 to 7.03).
FIG. 1.
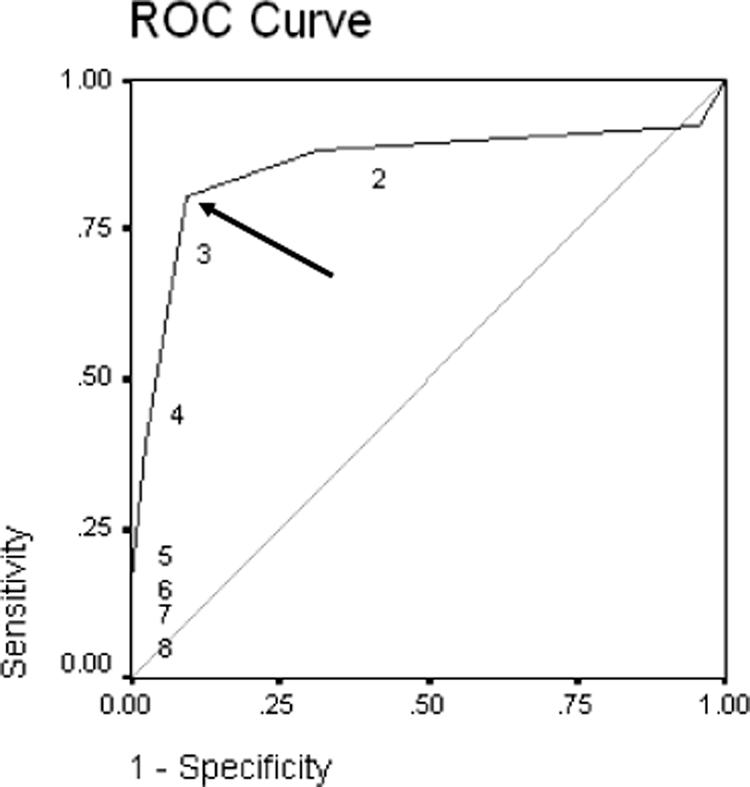
ROC curve for the antibiotic resistance profile of MRSA bloodstream isolates as a predictor of PFGE pattern (area under the ROC curve, 0.856; 95% CI, 0.775 to 0.936). The numbers in the figure refer to the number of antibiotic agents to which isolates expressed antibiotic resistance. A cutoff point of resistance to ≤2 classes for CG isolates and to ≥3 classes for HG isolates is seen; this cutoff point (arrow) maximizes the sensitivity and specificity of genotype prediction by antibiotic susceptibility pattern. With this cutoff, the likelihood ratio for an organism with resistance to less than three classes for a CG was 4.626 (95% CI, 3.007 to 7.025).
Misclassified HG isolates that showed resistance to less than three classes were mostly USA-800 (80%); USA-800 strains were typically PVL negative (75%) but often carried SCCmec type IV (78%). In general, the presence of PVL was highly predictive of CG (likelihood ratio, 9.08; 95% CI, 5.41 to 10.97), while the presence of SCCmec type IV was less so (likelihood ratio, 2.88; 95% CI, 2.33 to 2.88).
In contrast to antibiotic susceptibility, the hospital day that positive culture was obtained was not predictive of the genotype of MRSA isolates. The average numbers of days until positive culture among CG and HG isolates were 4.66 and 5.59, respectively (P = not significant by an independent-sample t test).
In a final analysis, we used a combination of rules to predict genotype. When we combined the cutoff of culture collection <72 h into hospitalization with fluoroquinolone susceptibility, the likelihood ratio for predicting a CG strain was 10.08 (95% CI, 3.79 to 29.74), with a sensitivity of only 59% but a specificity of 94%. The PPVs for a CG strain among all MRSA isolates were 77%, 91%, and 97% when the prevalences of these strains were 25%, 50%, and 75%, respectively. While the likelihood ratio for the day-susceptibility combination is higher than for other rules, the ratio has much wider confidence intervals and the rule has lower sensitivity (i.e., more false-negative predictions).
To summarize, of the various phenotypic rules tested, fluoroquinolone susceptibility was best at predicting genotype, i.e., based on the prevalence of CG isolates among MRSA bloodstream infections in the Cook County Bureau of Health (63%), the positive predictive value of predicting a CG strain by using ciprofloxacin susceptibility is 90%. However, the use of the number of antimicrobial classes to which a strain was susceptible or a combination of fluoroquinolone or clindamycin susceptibility was similar to the use of a single antimicrobial (e.g., susceptibility to fluoroquinolones) to predict CGs.
While a rule combining susceptibility with day of culture did have a high likelihood ratio, this rule had a lower sensitivity and wider confidence intervals, making it less useful for predicting CG strains. An epidemiologic rule, i.e., the time from admission to the collection of a positive culture, provided no help in predicting a CG; the presence of SCCmec type IV was also not as useful as phenotypic rules for predicting CG. While the presence of PVL was predictive of CG, testing requires the use of PCR and is not as generally available as automated susceptibility testing. Therefore, a rule incorporating phenotype (performed as part of routine microbiology practice) would be a more practical approach. However, other virulence factors or genetic markers not yet identified may be helpful in the future, possibly to either determine severity or further describe the epidemiology of CA-MRSA.
A phenotypic rule has specific uses. Foremost, analysis of retrospective data sets for which genotyping is not feasible could be conducted to assess the incursion of CA-MRSA into health care settings (12). The external validity of the rule depends on the local susceptibility of CG to fluoroquinolones. Currently, the USA-300 strains in the Cook County Bureau of Health are mostly fluoroquinolone susceptible (72%), with little change during the study period in the hospital Staphylococcus aureus antibiogram (i.e., fluoroquinolone susceptibility of 76 to 78%); however, other studies have shown that this pattern may be changing (2). With changing trends in antimicrobial resistance, phenotypic rules could be revalidated periodically to assess their effectiveness. A phenotypic rule could also have regional applications to assess the prevalence and describe the epidemiology of CA-MRSA infections.
During the study period, the double-disk diffusion test was not routinely performed by our microbiology laboratory. Common practice during this time period was to report isolates resistant to clindamycin if they were erythromycin resistant and clindamycin resistant. Isolates that were susceptible to both agents were reported as such. Clindamycin susceptibility of discordant isolates was not reported. Despite this limitation, the practice of testing for inducible clindamycin resistance may not be routine at some centers, enhancing the “real-world” effectiveness of the phenotypic rule.
Ongoing surveillance has suggested that CA-MRSA isolates are increasingly responsible for nosocomial infections (13). Our findings support this assertion; the failure of epidemiologic definitions (i.e., the day of positive culture) to predict genotype suggests that nosocomial MRSA infections are increasingly the result of CG strains. A phenotypic prediction rule can be applied for epidemiologic purposes to help describe this trend in CA-MRSA infections. At our facility, as we accumulate more data and as antibiotic susceptibilities change, we will likely need to modify our phenotypic rule. However, to understand better whether these infections result from within-hospital spread of CG strains or from endogenous sources (i.e., importation by colonized patients), admission nasal surveillance cultures, proposed to control nosocomial MRSA (11), could serve an additional role. Understanding the origin of the CA-MRSA strains now appearing in the hospital could improve our knowledge of both the epidemiology of this pathogen and the infection control interventions needed to control it. Furthermore, if these strains are becoming endemic in the hospital, the nomenclature of “community” or “hospital” genotype and onset may need to be revised.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Berglund, C., P. Molling, L. Sjoberg, and B. Soderquist. 2005. Predominance of staphylococcal cassette chromosome mec (SCCmec) type IV among methicillin-resistant Staphylococcus aureus (MRSA) in a Swedish county and presence of unknown SCCmec types with Panton-Valentine leukocidin genes. Clin. Microbiol. Infect. 11:447-456. [DOI] [PubMed] [Google Scholar]
- 2.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 3.Ellis, M. W., D. R. Hospenthal, D. P. Dooley, P. J. Gray, and C. K. Murray. 2004. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin. Infect. Dis. 39:971-979. [DOI] [PubMed] [Google Scholar]
- 4.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 5.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 6.Johnsson, D., P. Molling, K. Stralin, and B. Soderquist. 2004. Detection of Panton-Valentine leukocidin gene in Staphylococcus aureus by LightCycler PCR: clinical and epidemiological aspects. Clin. Microbiol. Infect. 10:884-889. [DOI] [PubMed] [Google Scholar]
- 7.King, M. D., B. J. Humphrey, Y. F. Wang, E. V. Kourbatova, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 144:309-317. [DOI] [PubMed] [Google Scholar]
- 8.Matushek, M. G., M. J. Bonten, and M. K. Hayden. 1996. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J. Clin. Microbiol. 34:2598-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller, L. G., F. Perdreau-Remington, G. Rieg, S. Mehdi, J. Perlroth, A. S. Bayer, A. W. Tang, T. O. Phung, and B. Spellberg. 2005. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N. Engl. J. Med. 352:1445-1453. [DOI] [PubMed] [Google Scholar]
- 11.Muto, C. A., J. A. Jernigan, B. E. Ostrowsky, H. M. Richet, W. R. Jarvis, J. M. Boyce, and B. M. Farr. 2003. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect. Control Hosp. Epidemiol. 24:362-386. [DOI] [PubMed] [Google Scholar]
- 12.Popovich, K. J., R. A. Weinstein, T. Rice, A. Aroutcheva, and B. Hota. 2007. Are community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strains replacing traditional nosocomial MRSA?, p. 107. Abstr. 17th Soc. Healthcare Epidemiol. Am. Meet. Society for Healthcare Epidemiology of America, Baltimore, MD, 14 to 17 April 2007.
- 13.Seybold, U., E. V. Kourbatova, J. G. Johnson, S. J. Halvosa, Y. F. Wang, M. D. King, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin. Infect. Dis. 42:647-656. [DOI] [PubMed] [Google Scholar]


