Abstract
The human bocavirus (hBoV) was first described in 2005 in respiratory tract samples. The clinical relevance of hBoV is still unclear. The aim of our study was to establish a real-time PCR assay for the detection and quantification of hBoV DNA, to apply the real-time assay for the analysis of stool and serum samples for the presence of hBoV DNA, and to perform a phylogenetic analysis of the hBoV positive samples. A total of 834 nasopharyngeal aspirates (NPA), 10 serum samples, and 31 stool samples of children with acute respiratory diseases were retrospectively tested. For phylogenetic analysis, 968 bp of the VP2 gene were sequenced from 69 hBoV-positive NPA samples. The qualitative results of the real-time hBoV PCR were in good agreement with a conventional hBoV PCR. We found that 12% of the NPA were positive for hBoV DNA. The median viral load in the NPA was 4.9 × 103 copies/ml (range, 2.7 × 10° to 1.5 × 1011 copies/ml). There was no difference of the hBoV load in NPA between children with or without known coinfection, but the load was significantly higher in children with bronchitis than in children with the diagnosis of febrile seizures. hBoV DNA was found in 1 of 10 serum samples and in 14 of 31 stool samples. hBoV sequence identity was >99% in the VP2 region. In conclusion, hBoV DNA can be found in NPA samples at very high titers. In addition to being found in the respiratory tract, hBoV was found in stool samples. The clinical relevance of these findings remains to be determined.
Respiratory tract infections are caused by a broad spectrum of microbial agents, mostly viruses. The classical spectrum of respiratory viruses includes influenza virus A and B, parainfluenza viruses, adenoviruses, respiratory syncytial virus (RSV), rhinoviruses, and coronaviruses. In recent years, the use of molecular biology methods led to the discovery of the human metapneumovirus, several coronaviruses (SARS, NL63, and HKU1), and the human bocavirus (hBoV) in respiratory tract specimens (1, 7, 25). hBoV is the first known human pathogen in the genus Bocavirus within Parvoviridae (21).
Since its first description in 3.1% of 540 Swedish children with lower respiratory tract disease (1), hBoV has been reported worldwide with frequencies ranging from 1.5 to 18.3% in respiratory samples, mostly from children (2, 3, 5, 9, 10, 14, 18, 24). hBoV has been found in patients with a broad variety of both upper and lower respiratory tract diseases and, in several studies, a large number of coinfections with other respiratory pathogens have been reported (1, 5, 6, 9, 15, 18, 24). Thus, at present it is still unclear whether hBoV is a relevant respiratory pathogen or whether it is only an innocent bystander.
The data available thus far on hBoV phylogeny indicate that there is only limited genetic variation between different hBoV strains. Sequences of PCR products located in the NP-1 region have generally shown nucleotide identity of >98% (1-3, 6, 9, 11, 14, 15, 18). However, sequence heterogeneity in the VP1/VP2 region appears to be greater than in the NP-1 region (1, 11). Therefore, a detailed analysis of hBoV phylogeny in the VP1/VP2 region should provide further knowledge on hBoV DNA variation.
The initial studies on hBoV infections have used conventional single-round PCR for hBoV detection, frequently according to the method originally described by Allander et al. (1). In order to facilitate hBoV DNA analysis and to quantify hBoV DNA in clinical samples, we designed a real-time PCR assay. In the present study, we describe the evaluation of the hBoV real-time PCR and its application for the detection of hBoV DNA in nasopharyngeal aspirates (NPA). In addition, we studied stool and serum samples of hBoV-infected children with the real-time assay in order to investigate which body compartments are affected by hBoV infection in addition to the respiratory tract. Furthermore, we sequenced the VP2 gene of hBoV-positive samples with the aim of analyzing the molecular phylogeny of hBoV infections.
MATERIALS AND METHODS
Samples.
The samples tested for hBoV infection consisted of unselected stored NPA that were originally received between April 1998 and September 2005 from the Children's Hospital of the University of Würzburg for screening of respiratory viruses. All samples had been tested for the presence of antigens of adenoviruses, influenza viruses A and B, parainfluenza viruses 1 to 3, and RSV by immunofluorescence assay and for the presence of hBoV DNA by a qualitative hBoV PCR as described previously (24). In addition, archived stool and serum samples of children with positive hBoV DNA in NPA samples were also tested if they were obtained no longer than 16 days before or after the NPA. All samples had been stored at −20°C or below until hBoV testing. The study was carried out in compliance with the Helsinki declaration and was approved by the ethics committee of the medical faculty at the University of Würzburg.
Qualitative hBoV PCR.
DNA was extracted from 200 μl of the NPA and serum samples using the HighPure viral nucleic acid kit (Roche, Mannheim, Germany) and from 140 μl of stool suspensions using the QIAamp viral RNA minikit (QIAGEN, Hilden, Germany). As shown previously, the viral RNA minikit can be used for the simultaneous extraction of RNA and DNA (16, 23). In exceptional cases when the volume of the archived samples was otherwise insufficient, less than the stated starting material was used for the extraction procedures. Both kits were used according to the instructions of the manufacturers. The elution volume of the extractions was 50 μl in case of the NPA and serum samples and 60 μl for the stool samples.
Amplification of hBoV DNA by conventional qualitative PCR was performed with the NP-1 primers BoV188F and BoV542R (Table 1) (1). Reaction conditions and cycling parameters have been described previously (24). Briefly, the PCR was carried out using HotStarTaq DNA polymerase (QIAGEN), and the cycling conditions were 50 cycles (94°C for 30 s, 53°C for 40 s, and 1 min for 72°C) after a preheating step of 10 min at 95°C. Positive PCRs by agarose gel electrophoresis were sequenced for the confirmation of sequence identity. The lower limit of detection with 95% probability was determined to be 11.7 copies/reaction (95% confidence interval, 8.5 to 21.7 copies/reaction) by probit analysis.
TABLE 1.
Primer and probes used for the amplification of hBoV
| Primer or probea | Sequence (5′-3′) | Gene | Positionb | Polarity | Purpose |
|---|---|---|---|---|---|
| BoV188Fa | GAGCTCTGTAAGTACTATTAC | NP-1 | 2351-2371 | + | Qualitative PCR |
| BoV542Ra | CTCTGTGTTGACTGAATACAG | NP-1 | 2704-2684 | − | Qualitative PCR |
| BoV2391s | GCACAGCCACGTGACGAA | NP-1 | 2391-2408 | + | Real-time PCR |
| BoV2411s-TM | 6FAM-TGAGCTCAGGGAATATGAAAGACAAGCATCG-TMRc | NP-1 | 2411-2441 | + | Real-time PCR |
| BoV2466a | TGGACTCCCTTTTCTTTTGTAGGA | NP-1 | 2466-2443 | − | Real-time PCR |
| BoV3885s | ACAATGACCTCACAGCTGGCGT | VP-2 | 3885-3906 | + | Phylogenetic analysis |
| BoV4287s | CAGCCAGCACAGGCAGAATT | VP-2 | 4287-4306 | − | Phylogenetic analysis |
| BoV4456a | TCCAAATCCTGCAGCACCTGTG | VP-2 | 4456-4435 | + | Phylogenetic analysis |
| BoV4939a | TGCAGTATGTCTTCTTTCTGGACG | VP-2 | 4939-4916 | − | Phylogenetic analysis |
Real-time hBoV PCR.
Primers and probe for the real-time PCR were selected by Primer Express 2.0 software (Applied Biosystems, Darmstadt, Germany) from the region of the NP-1 gene that was used for the qualitative PCR. Sequencing of 83 hBoV-positive samples as well as data from the literature (1) had revealed high sequence conservation in this region. The sequences of the primers (BoV2391s and BoV2466a) and of the dual-labeled probe (BoV2411s-TM) used for the real-time PCR are presented in Table 1. Blasting of primers and probe against GenBank to exclude unspecific binding did not reveal any significant homologies with other organisms.
The real-time PCR was carried out in a final volume of 20 μl consisting of 5 μl of extracted DNA, primers, and probe at a final concentration of 200 nM each, and 1× Quantitect probe master mix (QIAGEN). Amplification was performed on an ABI7500 real-time PCR system (Applied Biosystems, Darmstadt, Germany). The cycling conditions were 50 cycles with 30 s at 95°C and 60 s at 60°C after a preheating step of 15 min at 95°C.
The plasmid pBoV1 containing the PCR product of the qualitative PCR in the vector pCR2.1-TOPO (Invitrogen, Karlsruhe, Germany) was used as a positive control and for the standard curve. Viral loads were calculated from the threshold cycle (CT) values of the individual samples with respect to the standard curve. To minimize background fluorescence, the baseline was set two cycles below the first logarithmic amplification cycle.
General laboratory procedures to prevent PCR contamination were strictly adhered to. One negative control was extracted and amplified for every five NPA samples. All negative controls were found to be negative for hBoV DNA.
Phylogenetic analysis of the VP2 gene.
Positive hBoV samples were amplified with the two overlapping primer pairs BoV3885s-BoV4456a and BoV4287s-BoV4939a (Table 1). The PCR was carried out using HotStarTaq DNA polymerase (QIAGEN) with the following cycling conditions: 95°C for 12 min, followed by 52 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s, with a final elongation step at 72°C for 7 min. Positive PCRs were sequenced in both directions using BigDye terminator 3.1 chemistry and the ABI Prism 3100 (Applied Biosystems). The sequences were aligned by CLUSTAL W, and a topology tree was constructed with MEGA version 3.1 (12) by using the Kimura two-parameter estimation to generate the nucleotide distance matrix. To evaluate the phylogenetic relevance, bootstrapping analysis with 1,000 replicates was performed by the neighbor-joining method and the p-distance parameter.
Statistical analysis.
Statistical analysis was carried out by using GraphPad Prism version 3.0c for Mac (GraphPad Software, San Diego, CA) and SPSS version 13.01 for windows (SPSS, Chicago, IL).
RESULTS
Validation of the quantitative real-time PCR.
In order to validate the quantitative real-time PCR, the linearity of the assay, the lower limit of detection, intra-assay and interassay variation, and the specificity of the assay were all investigated. The curve was linear over the range from 3.8 × 10° to at least 3.8 × 108 copies/reaction. For standard curves of 17 independent runs, the mean value of the slope was −3.56 with a standard deviation of 0.1. The mean of the coefficient of correlation was 0.993, with a standard deviation of 0.006.
The lower limit of detection was determined by using probit analysis. It was based on three independent runs with a total of 24 replicates at the dilutions 37.5, 18.8,15.0, 9.4, 7.5, 3.8, 3.0, 1.5, 0.8, and 0.4 copies/reaction. A concentration of 14 copies/reaction (95% confidence interval, 10 to 26 copies/reaction) was detectable with 95% probability (data not shown). This corresponds to a lower limit of detection of 700 copies per ml of starting material (serum or NPA).
The reliability of the real-time PCR was assessed by analyzing the intra-assay and interassay variation (Table 2). The intra-assay coefficient of variation of the CT values for three replicates was 2.6% or lower for standard concentrations of 3.8 × 108 to 3.8 × 103. The interassay coefficient of variation of a run control (nominal concentration, 3.8 × 105) that was regularly included in each run was 15.21% (data not shown).
TABLE 2.
Intra-assay variation of the hBoV real-time PCR assay for different concentrations of the plasmid pBoV1
| No. of copies/ reaction |
CT
|
|||||
|---|---|---|---|---|---|---|
| Value for replicate:
|
Mean | SD | CV (%)a | |||
| 1 | 2 | 3 | ||||
| 3.8 E + 08 | 12.82 | 12.37 | 12.19 | 12.46 | 0.32 | 2.60 |
| 3.8 E + 07 | 15.94 | 15.90 | 15.93 | 15.92 | 0.02 | 0.13 |
| 3.8 E + 06 | 19.19 | 19.17 | 19.08 | 19.15 | 0.06 | 0.31 |
| 3.8 E + 05 | 22.60 | 22.62 | 22.62 | 22.61 | 0.01 | 0.05 |
| 3.8 E + 04 | 26.02 | 25.97 | 26.03 | 26.01 | 0.03 | 0.12 |
| 3.8 E + 03 | 29.41 | 29.47 | 29.20 | 29.36 | 0.14 | 0.48 |
CV, coefficient of variation.
To test the primers and probe for unspecific binding, samples known to be positive for parvovirus B19 and adenovirus DNA were tested in the hBoV real-time PCR assay. All samples were negative in the hBoV assay.
Quantitative real-time PCR of NPA.
For clinical evaluation of the hBoV real-time PCR, 11 samples from the period of 1998 to 2001 and 823 samples from 2002 to September 2005 NPA that had previously been tested by the conventional qualitative hBoV DNA (24) were retested with the real-time assay. A total of 84 samples were positive, and 724 samples were negative with both assays (Table 3). Discordant results in initial testing were obtained for 26 samples. All discordant samples were retested twice with both assays. An initially discordant sample was regarded as positive for the qualitative or real-time PCR assay, if at least two of the three results obtained with the assay were positive. After retesting, 16 samples remained with discordant results. In order to resolve the discrepancies, the discordant samples were further tested using the two primer pairs designed for the phylogenetic analysis VP2. In all samples positive only by real-time PCR, hBoV DNA was amplified by at least one additional primer pair, thus confirming the positive real-time PCR result. One of the three samples positive only by conventional PCR was confirmed by the additional primer pairs. Overall, the frequency of hBoV detection in our patient population increased from 10.8 to 12.0% with the real-time PCR assay. However, the proportions of positive results obtained with the two methods were not significantly different (P = 0.44 [chi-square test]).
TABLE 3.
Comparison of qualitative and real-time PCR for hBoV DNA
| Real-time PCR result | Qualitative PCR resulta
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 87 (84) | 13 (20) | 100 (104) |
| Negative | 3 (6) | 731 (724) | 734 (730) |
| Total | 90 (90) | 744 (744) | 834 (834) |
Values indicate the number of final results obtained after retesting discordant samples. For details, see the text. The numbers in parentheses indicate the results from the initial testing.
The median value of the viral load of all hBoV-positive NPA samples in the real-time PCR assay was 4.9 × 103 copies/ml (range, 2.7 × 10° to 1.5 × 1011 copies/ml). The median viral load of the samples that were positive only in the real-time PCR was 1.4 × 102 copies/ml, which was significantly lower than the median viral load of the samples that were positive in both assays (P < 0.0001 [Mann-Whitney test]).
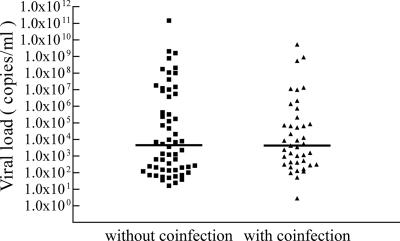
Because it was found previously that a large number of hBoV DNA-positive children were coinfected with other respiratory pathogens [RSV, adenovirus, influenza virus A/B, parainfluenza virus 1/2/3(24)], viral loads in NPA samples of children with coinfections were compared to viral loads of children without detectable coinfection. Of the children positive for hBoV DNA with the real-time PCR assay, 41% (n = 41) were found to be coinfected with RSV (n = 16), adenoviruses (n = 13), influenza A virus (n = 9), parainfluenza viruses (n = 4), or influenza B virus (n = 1), including two triple infections of RSV, parainfluenza virus 3, and hBoV. The hBoV loads in the NPA samples of these children and in the NPA samples of children without coinfection are presented in Fig. 1. Median values between both groups were not significantly different (median with coinfection = 4.3 × 103 copies/ml; median without coinfection = 4.5 × 103 copies/ml; P = 0.759 [Mann-Whitney test]).
FIG. 1.
Viral load in NPA of children with or without respiratory coinfections. In addition to hBoV DNA, all samples were tested for antigens of RSV, influenza virus A/B, adenoviruses, and parainfluenza viruses 1 to 3 by immunofluorescence assay. Median values are indicated by horizontal bars.
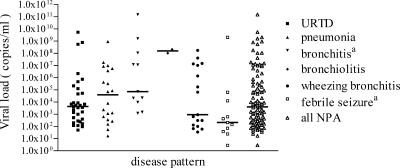
In order to examine possible indications for an association of hBoV with clinical disease, the viral loads in NPA samples were compared with respect to the final clinical diagnosis (Fig. 2). A broad range of viral loads was observed in most of the disease groups. The median values of the hBoV loads were significantly different between the NPA of children with bronchitis and febrile seizures (P = 0.0195 by Kruskal-Wallis analysis with Dunn's post test). The lowest median of viral loads was found in children with febrile seizures, whereas the highest median was observed in children with bronchiolitis. However, the number of hBoV DNA-positive children with bronchiolitis was small (n = 2). Coinfections were not identified in these two children.
FIG. 2.
Bocavirus load in NPA according to the clinical diagnosis. The median values (horizontal bars) were 4.3 × 103 copies/ml for upper respiratory tract disease (URTD), 4.0 × 104 copies/ml for pneumonia, 7.1 × 104 copies/ml for bronchitis, 1.6 × 108 copies/ml for bronchiolitis, 2.1 × 102 copies/ml for febrile seizures, 9.3 × 102 copies/ml for wheezing bronchitis, and 4 × 103 copies/ml for all NPA. The bocavirus load difference between the NPA of children with bronchitis and febrile seizures was statistically significant as determined by Kruskal-Wallis analysis and Dunn's post test (indicated by superscript a; P = 0.0195).
hBoV PCR of serum and stool samples.
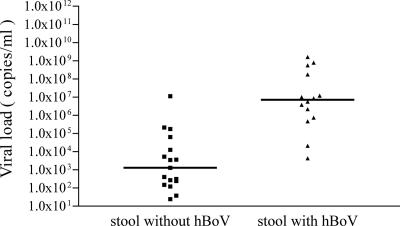
For some of the patients with hBoV-positive NPA samples, archived stool and serum samples from the same hospitalization episode were available. These samples were also tested by hBoV real-time PCR. One of ten serum samples was weakly positive for hBoV DNA (<700 copies/ml). The serum sample was received on the same day as the hBoV-positive NPA sample. Of 31 stool samples tested, 14 (45.2%) were found to be positive for hBoV DNA. Inhibition of the hBoV-negative stool samples was excluded by spiking experiments with hBoV plasmid DNA (data not shown). In order to determine factors associated with the presence of hBoV DNA in stool samples, the hBoV load in NPA was analyzed according to the hBoV status of the stool samples (Fig. 3). The median viral load of hBoV in NPA was significantly higher in children with hBoV-positive stool samples than in children with negative hBoV DNA stool samples (7.2 × 106copies/ml versus 1.3 × 103 copies/ml; P < 0.001 [Mann-Whitney test]). An association of the hBoV DNA detection in stool samples and the time span between the acquisition of the NPA and stool samples was not apparent (data not shown).
FIG. 3.
Comparison of hBoV loads in NPA according to the hBoV status of stool samples from the same patients. Median values as indicated by horizontal bars were significantly different between the two groups (P < 0.001 [Mann Whitney test]).
Phylogenetic analysis of hBoV-positive samples.
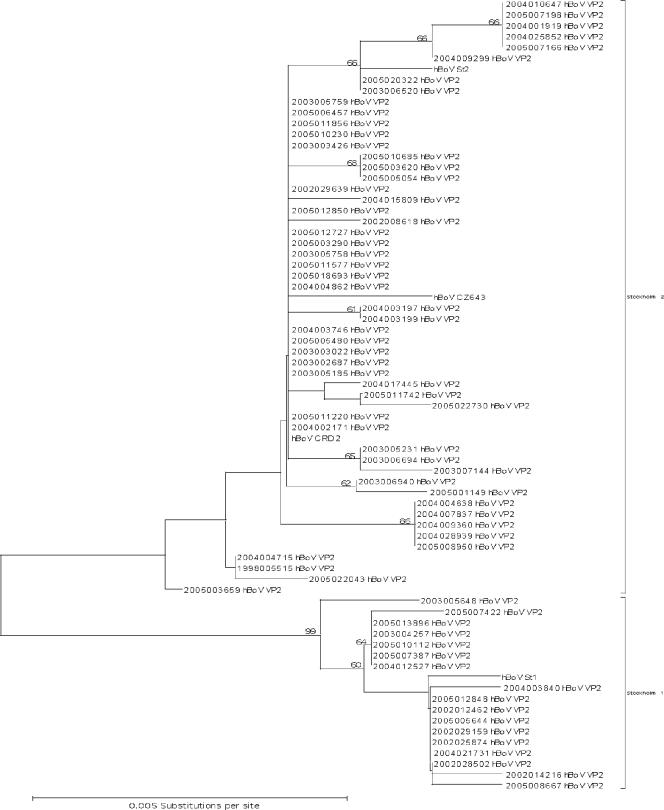
In order to investigate the divergence of hBoV genomes, we sequenced 968 bp of the VP2 gene (nucleotides 465 to 1432) from 69 hBoV-positive NPA samples. Overall, there was high nucleotide identity of >99% between all hBoV sequences. Nevertheless, the phylogenetic analysis resulted in two clusters of sequences with a bootstrap value of 99% (Fig. 4). Overall, 34 of the 968 nucleotide positions were variable, with 93% transitions and 7% transversions. Most of the nucleotide variations were conservative. Thus, at the amino acid level, there was 99.7% sequence identity. No apparent association was observed between phylogenetic clustering and seasonal distribution or disease manifestation (data not shown).
FIG. 4.
Phylogenetic analysis of the VP2 gene. Phylogenetic analysis of hBoV VP2 gene sequences of Germany, Sweden (st1 DQ000495; st2 DQ000496), China (CZ643 DQ457413), and the United States (CRD2 DQ340570). The topology tree was constructed in MEGA3.1. Kimura two-parameter estimation was used to generate the nucleotide distance matrix. The phylogenetic relevance was analyzed by bootstrapping (n = 1,000) and is indicated in nodal confidence values. Only values above 60 are displayed. The scale gives the percent nucleotide substitution with respect to branch length.
DISCUSSION
Following the first description of the hBoV by Allander et al. (1), several reports have confirmed the presence of hBoV infections in patient populations from different continents, suggesting that hBoV is a virus of worldwide distribution (3, 6, 8, 10, 18, 19, 24). The initial studies on hBoV infections have used conventional PCR methods with agarose gel electrophoresis. In order to improve the diagnostic tools for hBoV detection, we have established a real-time PCR method. In our validation experiments, this assay proved to be sensitive, specific, and reliable for hBoV DNA amplification, and quantification was possible over a broad linear range. In comparison to the conventional PCR method used previously (24), the sensitivities determined by probit analysis of both assays were very similar. When NPA samples of children with respiratory tract diseases were retested with the real-time assay, slightly more samples than with the conventional PCR method were positive, and the frequency of hBoV detection in our patient population increased from 10.8 to 12.0%. However, this difference was not significant. Because the amplified PCR fragment in the real-time PCR is shorter than in the conventional PCR (76 bp versus 354 bp), the real-time PCR may appear more sensitive when extracted DNA from the patient samples is fragmented.
One other study using real-time PCR and probe detection for hBoV amplification has been published thus far (13). Lu et al. established two real-time PCR assays with primers and probes located in the NS1 and in the NP-1 region. Sensitivity, amplification efficiency, and reproducibility were similar to the data obtained with our assay. A total of 1,178 throat swab samples from Thai patients with pneumonia were analyzed with their assay. Most of the samples contained only low copy numbers of hBoV DNA as judged by the CT values. However, a thorough quantitative analysis and a comparison of different disease groups was not performed by Lu et al. (13).
The frequency of hBoV infections in our population remains one of the highest reported thus far and is similar to the 11.3% positive samples in a study from Korea (5). Several other studies have described frequencies ranging from 1.5 to 18.3% (2, 3, 5, 9, 10, 14, 18, 24). Differences in the positivity rate for hBoV DNA may be accounted for by different patient characteristics and seasonal and geographical variation. In addition, the results may be influenced by the sensitivity of the hBoV PCR assays used. Because information about this aspect has been provided only in two studies (13, 15), it is not possible at present to estimate how much the reported detection frequencies for hBoV DNA may be affected by assay sensitivity.
Real-time PCR provides the possibility of quantification of DNA copies per reaction with reference to a standard curve. In contrast to serum or plasma, calculation of a viral load in the original patient sample is only of limited value for NPA because of technical variations in the sample acquisition procedure. For example, the volume of the NPA on arrival in the lab ranged approximately from 0.5 and 10 ml. Although this should be borne in mind, the observed viral loads in patient samples differed by a factor of 108, a range much larger than could be accounted for by variation in NPA volumes obtained.
Surprisingly, the median values of the bocavirus loads in NPA of children with or without known coinfection with other viral pathogens were almost identical. This is in contrast to the situation with coronavirus NL63. In children who were only infected with NL63, the viral load for NL63 was significantly higher than in children who were coinfected with NL63 and other viruses (22). With the limitation that additional coinfections remained undetected in our study because antigen-based methods were used for screening of respiratory viruses other than hBoV, and because several respiratory pathogens such as coronaviruses, rhinoviruses, enteroviruses, and the human metapneumovirus were not tested for, our data do not indicate that hBoV replication is influenced by the presence of other respiratory viruses. Also, the finding of comparatively low viral loads in NPA of more than half of the children without coinfection does not support the assumption that hBoV causes respiratory disease in these children.
Thus, whether hBoV is associated with clinical manifestations or is merely an innocent bystander is still an unresolved question. Based on the hypothesis that high viral loads may be an indication of pathogenic relevance, we compared the hBoV load in NPA according to the final diagnosis. A broad range of viral loads, including values of 108 copies/ml and higher, was observed for children with upper respiratory tract disease, pneumonia, bronchitis, and wheezing bronchitis. In contrast, in children with febrile seizures and without prominent respiratory findings the highest viral load was only about 105 copies/ml, and the median of 2.1 × 103 copies/ml was the lowest of all groups. High viral loads were observed in the two children with bronchiolitis. However, because of the small number these results should be interpreted with much caution until more data from children with bronchiolitis are available. Nevertheless, an association between hBoV and bronchiolitis based on qualitative PCR findings has been indicated in other studies (2, 5, 9, 17).
Because hBoV is distantly related to parvovirus B19, a virus highly viremic for several weeks after primary infection, we analyzed serum samples of hBoV-infected children for the presence of hBoV DNA. Only one of ten samples was weakly positive. However, detection of hBoV in NPA samples is not proof of a primary infection, and therefore, we may have missed a viremic phase with the serum samples examined in the present study. Serological studies to detect hBoV-specific immunoglobulin M and immunoglobulin G seroconversion will be necessary to determine the time point of primary hBoV infection. Once this information is available, elucidation of the question of whether hBoV is generally viremic or not will become possible.
hBoV is most closely related to the other two viruses in the genus Bocavirus, the minute virus of canines (MVC) and the bovine parvovirus (BPV). MVC and BPV are known to cause gastrointestinal infections in dogs and calves, respectively (4, 20). Therefore, we also examined available stool samples of children with hBoV-positive NPA. In contrast to the serum results, the stool samples were frequently positive for hBoV DNA (14 of 31 samples). This finding may be explained by replication of hBoV in gastrointestinal epithelium. The observation that the hBoV load in the NPA samples of the children with hBoV-positive stool samples was significantly higher than the hBoV load in the children with hBoV-negative stool samples, leads to an alternative explanation for the presence of hBoV DNA in stool samples. hBoV may be swallowed during respiratory tract infection and subsequently excreted in the feces without further replication steps in the gastrointestinal tract. Whether the hBoV DNA detected in stool samples is derived from infectious or from degraded virus particles and if hBoV may be a cause of diarrhea, remains to be determined once cell culture and animal model systems for hBoV become available.
Phylogenetic analysis of the hBoV-positive NPA samples in our study population confirmed previous findings that hBoV is highly conserved. Although this finding may be biased by the fact that most studies published thus far have used PCR screening methods based on the NP-1 sequence, which is highly conserved, there is currently no evidence that more than one hBoV genotype exists. In contrast, the sequence variation both on the nucleotide and amino acid level is considerably greater for parvovirus B19. Thus, it may be speculated that hBoV has entered the human population more recently than parvovirus B19. Alternatively, the diverse degrees of variation may be due to different replication strategies.
In conclusion, we have successfully established and used a real-time PCR assay for the detection of hBoV DNA. Application of this assay for quantitative analysis has shown that hBoV DNA can be found in NPA samples at very high titers. In addition to the respiratory tract, hBoV was also found in stool samples. However, the clinical relevance of hBoV infections remains uncertain both for the respiratory and for the gastrointestinal tract.
Acknowledgments
We thank the technicians of the viral diagnostic lab for skillful and dedicated assistance and Kirsty McPherson for helpful comments on the manuscript.
Footnotes
Published ahead of print on 2 May 2007.
REFERENCES
- 1.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, J. C., K. K. Singh, S. A. Spector, and M. H. Sawyer. 2006. Human bocavirus: prevalence and clinical spectrum at a children's hospital. Clin. Infect. Dis. 43:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastien, N., K. Brandt, K. Dust, D. Ward, and Y. Li. 2006. Human bocavirus infection, Canada. Emerg. Infect. Dis. 12:848-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binn, L. N., E. C. Lazar, G. A. Eddy, and M. Kajima. 1970. Recovery and characterization of a minute virus of canines. Infect. Immun. 1:503-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, E. H., H. J. Lee, S. J. Kim, B. W. Eun, N. H. Kim, J. A. Lee, J. H. Lee, E. K. Song, S. H. Kim, J. Y. Park, and J. Y. Sung. 2006. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000-2005. Clin. Infect. Dis. 43:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, J. Y., T. H. Han, C. K. Kim, and S. W. Kim. 2006. Bocavirus infection in hospitalized children, South Korea. Emerg. Infect. Dis. 12:1254-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouchier, R. A., G. F. Rimmelzwaan, T. Kuiken, and A. D. Osterhaus. 2005. Newer respiratory virus infections: human metapneumovirus, avian influenza virus, and human coronaviruses. Curr. Opin. Infect. Dis. 18:141-146. [DOI] [PubMed] [Google Scholar]
- 8.Foulongne, V., Y. Olejnik, V. Perez, S. Elaerts, M. Rodiere, and M. Segondy. 2006. Human bocavirus in French children. Emerg. Infect. Dis. 12:1251-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foulongne, V., M. Rodiere, and M. Segondy. 2006. Human bocavirus in children. Emerg. Infect. Dis. 12:862-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan, N. M., W. Dove, A. F. Abu-Zeid, H. E. Shamoon, S. A. Abd-Eldayem, and C. A. Hart. 2006. Human bocavirus infection among children, Jordan. Emerg. Infect. Dis. 12:1418-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kesebir, D., M. Vazquez, C. Weibel, E. D. Shapiro, D. Ferguson, M. L. Landry, and J. S. Kahn. 2006. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J. Infect. Dis. 194:1276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 13.Lu, X., M. Chittaganpitch, S. J. Olsen, I. M. Mackay, T. P. Sloots, A. M. Fry, and D. D. Erdman. 2006. Real-time PCR assays for detection of bocavirus in human specimens. J. Clin. Microbiol. 44:3231-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, X., R. Endo, N. Ishiguro, T. Ebihara, H. Ishiko, T. Ariga, and H. Kikuta. 2006. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J. Clin. Microbiol. 44:1132-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manning, A., V. Russell, K. Eastick, G. H. Leadbetter, N. Hallam, K. Templeton, and P. Simmonds. 2006. Epidemiological profile and clinical associations of human bocavirus and other human parvoviruses. J. Infect. Dis. 194:1283-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohayem, J., S. Berger, T. Juretzek, O. Herchenroder, M. Mogel, M. Poppe, J. Henker, and A. Rethwilm. 2004. A simple and rapid single-step multiplex RT-PCR to detect norovirus, astrovirus, and adenovirus in clinical stool samples. J. Virol. Methods 118:49-59. [DOI] [PubMed] [Google Scholar]
- 17.Simon, A., P. Groneck, B. Kupfer, R. Kaiser, G. Plum, R. L. Tillmann, A. Muller, and O. Schildgen. 2007. Detection of bocavirus DNA in nasopharyngeal aspirates of a child with bronchiolitis. J. Infect. 54:e125-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sloots, T. P., P. McErlean, D. J. Speicher, K. E. Arden, M. D. Nissen, and I. M. Mackay. 2006. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J. Clin. Virol. 35:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smuts, H., and D. Hardie. 2006. Human bocavirus in hospitalized children, South Africa. Emerg. Infect. Dis. 12:1457-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storz, J., J. J. Leary, J. H. Carlson, and R. C. Bates. 1978. Parvoviruses associated with diarrhea in calves. J. Am. Vet. Med. Assoc. 173:624-627. [PubMed] [Google Scholar]
- 21.Tattersall, P., M. Bergoin, M. E. Bloom, K. E. Brown, R. M. Linden, N. Muzyczka, C. R. Parrish, and P. Tijsses. 2005. Family Parvoviridae, p. 353-369. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom.
- 22.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2005. Croup is associated with the novel coronavirus NL63. PLoS Med. 2:e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissbrich, B., F. Harms, and V. ter Meulen. 1995. Detection of enterovirus RNA and herpesvirus DNA in CSF by multiplex PCR. QIAGEN News 2:4-5. [Google Scholar]
- 24.Weissbrich, B., F. Neske, J. Schubert, F. Tollmann, K. Blath, K. Blessing, and H. W. Kreth. 2006. Frequent detection of Bocavirus DNA in German children with respiratory tract infections. BMC Infect. Dis. 6:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]