Abstract
Aichi virus is a new member of the family Picornaviridae, genus Kobuvirus, and is associated with human gastroenteritis. This study detected Aichi virus in 28 of 912 fecal specimens which were negative for rotavirus, adenovirus, norovirus, sapovirus, and astrovirus and were collected in Japan, Bangladesh, Thailand, and Vietnam during 2002 to 2005.
Aichi virus, a small round virus about 30 nm in diameter, was first recognized in 1989 as the cause of oyster-associated nonbacterial gastroenteritis in humans (8-10). The virus was classified into a new genus named Kobuvirus of the family Picornaviridae (11), which contains nine genera, Aphthovirus, Cardiovirus, Enterovirus, Erbovirus, Hepatovirus, Kobuvirus (which includes Aichi virus and bovine kobuvirus), Parechovirus, Rhinovirus, and Teschovirus. The complete Aichi virus genome was determined in 1998 and proved to be a single-stranded positive-sense RNA molecule with 8,251 bases, excluding a poly(A) tail; it contains a large open reading frame with 7,302 nucleotides that encodes a potential polyprotein precursor of 2,433 amino acids (11). In 2000, a reverse transcription (RT)-PCR method for the detection of Aichi virus was developed and a genetic analysis was performed with the 519-base RNA sequences at the putative junction between the C terminus of 3C and the N terminus of 3D. As a result, Aichi virus isolates have been divided into groups 1 (genotype A) and 2 (genotype B) (12).
Studies on Aichi virus were subsequently performed, and it was also detected in Brazil and Germany (2-7). However, there has been limited knowledge about the epidemiology of Aichi virus infection in Asian countries other than Japan and Pakistan. This study was performed to determine the prevalence of Aichi virus in Bangladesh, Thailand, Vietnam, and also in Japan and to provide a better understanding of the epidemiology and genetic relationships between the Aichi virus strains in the present study and the strains previously reported.
A total of 912 stored, extracted RNA samples from fecal specimens known to be negative for rotavirus, adenovirus, norovirus, sapovirus, and astrovirus by RT-PCR that were collected from patients with acute gastroenteritis in Japan (215 samples collected from July 2002 to June 2003), Bangladesh (405 samples collected from October 2004 to September 2005), Thailand (107 samples collected from March 2002 to December 2004), and Vietnam (185 samples collected from October 2002 to September 2003) were used in this study. First, RT was preformed by random priming. Then, to detect Aichi virus, a nested PCR was conducted with the primer sets for amplifying the 3CD junction region as described by Yamashita et al. (12). The first PCR was conducted with primers 6261 (5′-ACACTCCCACCTCCCGCCAGTA-3′) and 6779 (5′-GGAAGAGCTGGGTGTCAAGA) to amplify a 519-bp region between the C terminus of 3C and the N terminus of 3D. Next, a nested PCR was performed with the primer pair C94b-246k (C94b, 5′-GACTTCCCCGGAGTCGTCGTCT-3′; 246k, 5′-GACATCCGGTTGACGTTGAC-3′) to amplify a 223-bp segment within the 3C-3D junction region.
The products from the first PCR were then purified and sequenced in both directions. Phylogenetic analysis was performed by the neighbor-joining method within the MEGA 3.1 analytical package with a bootstrap of 1,000 replicates (1).
Of the 912 samples tested, 28 (3.1%) were positive for Aichi virus. Of these, 14 samples were collected from Japan, 10 were from Bangladesh, 3 were from Vietnam, and 1 was from Thailand. As a result, the prevalence rates of Aichi virus detected in each country were 6.5%, 2.5%, 1.6%, and 0.9%, respectively. Twenty Aichi virus strains were successfully sequenced, including 11 Japanese strains, 5 Bangladeshi strains, 3 Vietnamese strains, and 1 Thai strain.
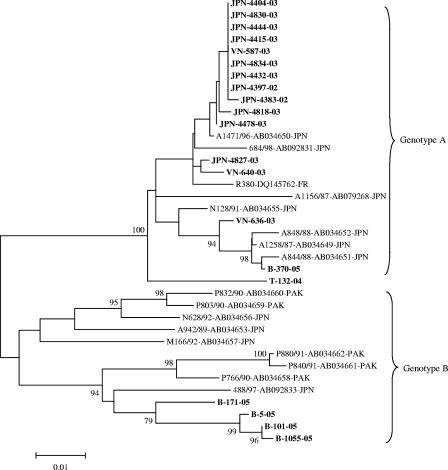
The phylogenetic tree constructed from the 519-nucleotide sequences showed that 11 Japanese strains, 3 Vietnamese strains, 1 Thai strain, and 1 Bangladeshi strain were clustered into the same branch with Japanese reference strains A844/88, A848/88, 684/98, and N128/91 and belonged to genotype A. The four other Bangladeshi strains clustered into genotype B, together with Pakistani strains P832/90 and P840/91 (Fig. 1).
FIG. 1.
Phylogenetic tree constructed from the 519-nucleotide sequences between the C terminus of 3C and the N terminus of 3D. Bootstrap values of greater than 70% are shown at the branch nodes. The Aichi virus strains in this study are in boldface. Abbreviations of locations: JPN, Japan; B, Bangladesh; VN, Vietnam; T, Thailand; FR, France; PAK, Pakistan. Aichi virus strains M166/92, N128/91, and N628/92 were isolated from Japanese travelers returning from Malaysia (M) and Indonesia (N).
From previous studies, Aichi virus was known as one of the agents which cause acute gastroenteritis in human, especially associated with oysters (9, 10). In this study, Aichi virus was found in fecal specimens from diarrheal infants and children that were known to be negative for other common causative agents such as rotavirus, norovirus, adenovirus, astrovirus, and sapovirus. Thus, taken together with previous studies, these findings demonstrated clearly that Aichi virus was related to diarrhea in infants and young children in several Asian countries.
In this study, Aichi virus was detected in Asian countries other than Japan and Pakistan. This is the first finding of Aichi virus in fecal specimens from Bangladesh, Thailand, and Vietnam. These results also provide useful information for the epidemiological understanding of Aichi virus in Japan. Since Aichi virus is a new virus discovered recently, further genetic studies of it are definitely necessary.
Nucleotide sequence accession numbers.
The nucleotide sequences of the strains studied here were assigned accession numbers EF079149 to EF079162 and EF466010 to EF466015.
Acknowledgments
We are grateful to Teruo Yamashita for kindly providing Aichi virus reference strains.
This study was supported by grants-in-aid from the Ministry of Education and Sciences and the Ministry of Heath, Labor and Welfare, Japan, and by the Asian Development Bank.
Footnotes
Published ahead of print on 23 May 2007.
REFERENCES
- 1.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 2.Nagashima, S., J. Sasaki, and K. Taniguchi. 2005. The 5′-terminal region of the Aichi virus genome encodes cis-acting replication elements required for positive- and negative-strand RNA synthesis. J. Virol. 79:6918-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagashima, S., J. Sasaki, and K. Taniguchi. 2003. Functional analysis of the stem-loop structures at the 5′ end of the Aichi virus genome. Virology 313:56-65. [DOI] [PubMed] [Google Scholar]
- 4.Oh, D. Y., P. A. Silva, B. Hauroeder, S. Diedrich, D. D. Cardoso, and E. Schreier. 2006. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch. Virol. 151:1199-1206. [DOI] [PubMed] [Google Scholar]
- 5.Sasaki, J., Y. Kusuhara, Y. Maeno, N. Kobayashi, T. Yamashita, K. Sakae, N. Takeda, and K. Taniguchi. 2001. Construction of an infectious cDNA clone of Aichi virus (a new member of the family Picornaviridae) and mutational analysis of a stem-loop structure at the 5′ end of the genome. J. Virol. 75:8021-8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki, J., S. Nagashima, and K. Taniguchi. 2003. Aichi virus leader protein is involved in viral RNA replication and encapsidation. J. Virol. 77:10799-10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasaki, J., and K. Taniguchi. 2003. The 5′-end sequence of the genome of Aichi virus, a picornavirus, contains an element critical for viral RNA encapsidation. J. Virol. 77:3542-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita, T., S. Kobayashi, K. Sakae, S. Nakata, S. Chiba, Y. Ishihara, and S. Isomura. 1991. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 164:954-957. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita, T., K. Sakae, Y. Ishihara, S. Isomura, and E. Utagawa. 1993. Prevalence of newly isolated, cytopathic small round virus (Aichi strain) in Japan. J. Clin. Microbiol. 31:2938-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita, T., K. Sakae, S. Kobayashi, Y. Ishihara, T. Miyake, A. Mubina, and S. Isomura. 1995. Isolation of cytopathic small round virus (Aichi virus) from Pakistani children and Japanese travelers from Southeast Asia. Microbiol. Immunol. 39:433-435. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita, T., K. Sakae, H. Tsuzuki, Y. Suzuki, N. Ishikawa, N. Takeda, T. Miyamura, and S. Yamazaki. 1998. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J. Virol. 72:8408-8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita, T., M. Sugiyama, H. Tsuzuki, K. Sakae, Y. Suzuki, and Y. Miyazaki. 2000. Application of a reverse transcription-PCR for identification and differentiation of Aichi virus, a new member of the picornavirus family associated with gastroenteritis in humans. J. Clin. Microbiol. 38:2955-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]