Abstract
In screening amplified poly(A) mRNA from hippocampal dendrites and growth cones in culture to determine candidates for local translation, we found that select transcription factor mRNAs were present. We hypothesized that synthesis of transcription factor proteins within dendrites would provide a direct signaling pathway between the distal dendrite and the nucleus resulting in modulation of gene expression important for neuronal differentiation. To evaluate this possibility, radiolabeled amplified antisense RNA was used to probe slot blots of transcription factor cDNAs as well as arrayed blots of zinc finger transcription factors. The mRNAs encoding the cAMP response element binding protein (CREB), zif 268, and one putative transcription factor were detected. We expanded upon these results showing that CREB protein is present in dendrites, that translation of CREB mRNA in isolated dendrites is feasible and that CREB protein found in dendrites can interact with the cis-acting cyclic AMP reponse element DNA sequence by using an in situ Southwestern assay. Further, CREB protein in dendrites is not transported to this site from the cell body because fluorescently tagged CREB microperfused into the soma did not diffuse into the dendrites. In addition, CREB protein microperfused into dendrites was rapidly transported to the nucleus, its likely site of bioactivity. Lastly, by using the isolated dendrite system we show that phosphorylation of Ser-133 on CREB protein can occur in isolated dendrites independent of the nucleus. These data provide a regulatory pathway in which transcription factors synthesized and posttranslationally modified in dendrites directly alter gene expression bypassing the integration of signal transduction pathways that converge on the nucleus.
Many extracellular signals modulate neuronal gene transcription via protein cascade signaling and directly affect dendritic outgrowth and synaptogenesis. However, the presence of mRNAs in dendrites (1–3) and dendritic growth cones (4) of hippocampal neurons suggests that some proteins necessary for neuronal development may be synthesized within dendrites and their growth cones. The population of dendritic and growth cone mRNAs includes cytoskeletal elements, neurotrophic factor receptors, synthetic enzymes, and certain neurotransmitter subunits and the relative abundance of many of these mRNAs in cultured hippocampal neurons is developmentally regulated.
It follows that regulation of local translation of dendritic mRNAs, especially in response to neurotrophic factors, is likely important during dendritic arborization. Previously translation of transfected mRNA within dendrites and dendritic growth cones proved that local protein synthesis can occur in vivo (4) and supports the hypothesis that proteins are synthesized from mRNAs in dendrites (5–9). Synthesis of proteins within nascent dendrites during development would provide a system to modulate dynamic phases of cell growth such as dendritic arborization, growth cone pathfinding, and synaptogenesis in response to activity-dependent and trophic signals. In previous studies, zif 268 mRNA (a transcription factor) was unexpectedly detected in several dendrites (K. Miyashiro and J.E., unpublished observations). In light of this data and the dynamic changes in gene expression that coincide with dendritic arborization and synapse formation, we hypothesized that select transcription factor mRNAs are localized in dendrites and dendritic growth cones. Further, we would expect the transcription factor proteins corresponding to the dendritically localized mRNAs to be synthesized, posttranslationally modified, and transported to the nucleus to modulate gene transcription in response to trophic or other cues. Such a signaling pathway would imprint the nuclear transcriptional response directly based upon the local environment of the dendrite. Such “nuclear imprinting” would allow signals acting on tips of growing dendrites to control gene transcription via direct production and modification of transcription factors rather than through integrated signal produced by the averaging of activated signal transduction cascades converging on transcription factors confined to the nucleus.
METHODS
Hippocampal Cultures.
Primary dissociated neuronal cultures were generated from embryonic rat hippocampi on day 18 (10). Dendrites (from 48- and 72-hr cultures) were distinguished from axons on the basis of tapering caliber, nascent branch pattern, immunoreactivity for microtubule-associated protein 2 and absence of GAP 43 antigenicity (4). Growth cones were identified by phase contrast microscopy. Individual dendritic growth cones were transected at the attachment point by using glass microelectrodes filled with reagents necessary for cDNA synthesis (3, 4). Transected growth cones were gently aspirated into the electrodes where cDNA synthesis then proceeded for 90 min at 40°C. Cognate cell bodies were separately aspirated for comparison.
Antisense RNA (aRNA) Amplification and Reverse Northern Blotting.
aRNA synthesis was as described (3, 4, 11). The success rate for aRNA amplification whether the mRNA source is a cell body or process is ≈75%. Failure to amplify is likely due to the introduction of RNAse into the samples. Reverse Northern blotting involved linearizing 1 μg of each cDNA with a restriction enzyme followed by application to nylon membrane as described (3, 4). With appropriate exposure times the relative hybridization signal is quantifiable (4, 12). The length of cDNA clones varies but because each contains the 3′-UTR of the corresponding mRNA this has no influence on detection of signal. pBluescript plasmid cDNA (pBS) serves as a negative control (no pBS mRNA in cells) hence light hybridization to pBS seen in Fig. 1A is the background hybridization observed for that particular batch of aRNA.
Figure 1.
CREB mRNA presence in growth cones and dendrites. (A) Representative reverse Northern blot probed with 32P-CTP radiolabeled aRNA from a single dendritic growth cone. Slot a1, microtubule-associated protein 2; a2, CREB δ; a3, calmodulin-dependent kinase; b1, c-fos; b2, c-jun; b3, C82 (a C2H2 zinc finger cDNA); c1, zif268; c2, orthodenticle 1; c3, pBS. (B) Reverse Northern blot probed with 32P-CTP radiolabeled aRNA from a single cell body. The arrangement of cDNAs is as described for A. (C) Localization of CREB mRNA by using digoxigenin-labeled CREB cRNA as a probe in cultured rat hippocampal cells by using in situ hybridization. The neuronal soma and the CREB positive processes exhibit a purple color because of NTB staining of the dig-labeled cRNA that is hybridized to CREB mRNA. A single CREB mRNA containing process is indicated by the arrow. The inset shows background staining when in situ hybridization is performed with no cRNA probe added to fixed cells. Bar = 15 μM. (D) Localization of CREB mRNA by using digoxigenin-labeled CREB cRNA as a probe in neurons from normal human temporal neocortex by using in situ hybridization. Apical dendrite of a layer V pyramidal cell (arrow). Several adjacent cell bodies contain abundant CREB mRNA within the soma (out of focal plane). Inset depicts competition control on an adjacent tissue section (arrow). Positive in situ hybridization in dendrites was observed in human brain neocortex from four individuals. The competition control shows the in situ hybridization signal produced when unlabeled antisense cRNA is added to prehybridization mix at a concentration of 50× the amount of labeled cRNA to be used in the hybridization step. Between prehybridization and hybridization the unhybridized unlabeled cRNA is washed away. Bar = 10 μM.
Hybridization intensity of cell bodies or growth cone aRNA-cDNA hybrids was quantified by scanning densitometry of the autoradiographs by using imagequant 3.3 software. Hybridization levels were expressed as a percentage of calmodulin-dependent kinase hybridization, because this mRNA has been previously identified in dendritic growth cones (4), and levels of cyclic AMP reponse element binding protein (CREB) were compared between cell bodies and growth cones.
In Situ Hybridization.
CREB cRNA was made by using digoxigenin-labeled UTP by using the RNA labeling mix of Boehringer Mannheim. In situ hybridization was as described (13). The hybridized digoxigenin-labeled cRNA was detected by using an anti-digoxigenin-alkaline phosphatase conjugated antibody (Boehringer Mannheim). 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indoyl phosphate was used as the enzymatic stain to detect the antibody-dig complex. Competition hybridization experiments with high molar excess of unlabeled antisense cRNA probe added to the prehybridization solution served as a specificity control to block hybridization by the digoxigenin-labeled cRNA. Also, digoxigenin-labeled antisense RNA controls (a control for nonspecific hybridization of the probe based upon G-C content of the probe) were performed and showed no hybridization.
Immunohistochemistry.
Hippocampal neurons were immunolabeled with rabbit polyclonal antibodies to nonphosphorylated CREB, Ser-133 phospho-CREB (Ab244), or the leucine zipper-containing region of CREB (ZE244) in 0.1 M Tris/5% horse serum overnight at 4°C. Antibody labeling was performed via the avidin-biotin method (ABC Vectastain, Vector Laboratories) and visualized with 3,3′-diaminobenzidine. Immunoreactive cell bodies or dendrites were counted by using a low power objective field (20×) to establish the approximate frequency of immunolabeling. Control experiments included exclusion of the primary antibody and preimmunoabsorption of the antibody (Ab244) with CREB peptide.
In Situ Southwestern Assay.
The cyclic AMP response element (CRE) octamer (5′-TGACGTCA-3′; 100 ng) was radiolabeled with T4 polynucleotide kinase and 32P-γATP. The labeled octamer was then ligated with 1 mg unlabeled oligonucleotide. Hippocampal neurons fixed in 4% paraformaldehyde for 2–3 min were incubated for 20 min in 0.1 M Tris/0.1% Triton-X, then overnight at room temperature in DNA binding buffer (250 mM Hepes (pH 7.4)/5× Denhardt’s solution/200 mg/ml sheared single-stranded salmon sperm DNA/0.01 mM MgCl2/0.03 mM KCl). The various probes were added to fresh binding buffer, and applied to cells for 4 hr at room temperature. For competition experiments, cells were exposed to 5 mg unlabeled ligated CRE concatamer for 4 hr prior to addition of the CRE probe, and then washed in binding buffer. The coverslips were dipped in Kodak NT2 photographic emulsion and exposed for 72 hr at 4°C. Slides were developed in Kodak D19 developer at 14°C, rinsed in dH2O, and viewed under darkfield microscopy. Critical features of this methodology include fixation time (fixation for >5 min results in no signal because of too much cross-linking while <2 min fixation results in no signal because of subsequent washing away of protein and probe from the coverslip), type of fixative (paraformaldehyde works better than ethanol), specific activity of the probe (≈108 works well with higher specific activities producing high backgrounds) and use of appropriate washing conditions (the coverslips must be washed extensively to remove nonspecific associated CREs).
Control experiments to determine specificity of the in situ Southwestern assay assay included probing the cells with unligated 32P-CRE oligonucleotide, preincubation of the tissue section with excess (>5 mg) unlabeled ligated CRE to compete for binding of the 32P-CRE concatamer, and probing with nonsense palindrome probe (inverted CRE sequence, 5′-CAGTACTG-3′).
Single Dendrite Transfection.
Capped CREB-myc mRNA synthesized in vitro by using the Ambion Megascript kit, was complexed with the cationic lipid 1,3-Di-oleoyloxy-2-(6-carboxy-spermyl)-propylamid (Boehringer Mannheim) and carrier tRNA (5 μg) in buffered saline (20 mM Hepes/150 mM NaCl) at room temperature for 20 min prior to use. Removal of the cell soma, dendritic transfection and myc-immunohistochemistry were as previously described (4).
Several control experiments were as described previously including treatment of the cells with 30 μM cyclohexamide to block protein synthesis or 2 units of RNAse A to destroy the reporter CREB-myc mRNA.
Fluorescent Tagging and Microperfusion of Protein.
CREB protein (10 mg in 10 ml sodium bicarbonate 0.2M, pH 8.3) was mixed with 2 ml of Oregon Green 514 carboxylic acid, succinimidyl ester (10 mg/ml in dimethyl sulfoxide; no. O6139, Molecular Probes) and stirred for 60 min at room temperature. Two milliliters of freshly prepared 1.5 M hydroxylamine (pH 8.5) were added and the solution incubated for 30 min. Addition of 10 ml of a nonspecific peptide bound to silica beads was followed by incubation at room temperature for 30 min. After centrifugation at 12,000 × g for 30 sec the liquid phase was removed and drop dialyzed at room temperature in the dark for 2 hr against intracellular patch pipette solution (145 mM K gluconate/8 mM KCl/10 mM K-Hepes, pH 7.2/3 mM MgATP, osmolality 287 mosmol/kg).
Hippocampal neurons were visualized with a Zeiss Axioskop by using a 63×, 0.9 numerical aperture water immersion infinity-corrected objective. Cells were voltage clamped employing the whole-cell patch clamp technique (14) by patching on the soma (1.2–3 Mohms) or the dendrites (12–18 Mohms). The extracellular solution contained 150 mM NaCl, 2.5 mM CaCl2, 1.5 mM MgCl2 and 10 mM Na-Hepes (pH 7.4), osmolality 295 mosmol/kg. The intracellular patch pipette solution was supplemented with Oregon Green 514 and ogCREB or ogBSA. Promptly after establishing the whole-cell recording the free fluorophore diffused from the patch pipette into the cytosol and equilibrated (≈2 min). An initial image of the fluorescence emitted from the cell was taken at this time by using a CCD imaging camera (1- or 5-sec exposures; EEV chip, Princeton Instruments) employing epifluorescence microscopy with 485 and 530 nm DF band pass excitation and emission filters (full half-width maximums of 22 and 30 nm respectively), and a 505 nm long pass 45° dichroic filter. Subsequent images were taken for up to 70 min provided the holding current was less than −100 pA at a holding potential of −60 mV. ΔF/F images were calculated after correcting for background fluorescence, by subtracting the initial image from subsequent ones, and dividing the resulting difference by the initial image. The ΔF/F images, therefore, provide a measure of the slower diffusion/transport of the fluorescently tagged proteins and are independent of such variables as differences in optical pathlength or nonuniformity of incident light.
RESULTS
32P-CTP radiolabeled aRNA was used to screen reverse Northern blots containing candidate transcription factor full-length cDNAs including the δ isoform of CREB, c-fos, c-jun, zif268, orthodenticle 1, brain factor 1/brain factor 2, and hairy enhancer of split 1 all of which are important in neuronal development. The α subunit of Ca2+/calmodulin-dependent kinase and microtubule-associated protein 2 cDNAs, previously identified in dendrites and dendritic growth cones (1–4), were detected and served as positive controls. CREB and zif 268 mRNAs were detected in growth cones but c-fos, c-jun, orthodenticle 1, hairy enhancer of split 1, brain factor 1, and brain factor 2 mRNAs were at or below pBS background (Fig. 1A). Whereas, all candidate genes were detected in the cognate cell bodies that is similar to data reported in previous localization studies in embryonic neurons by using in situ hybridization (Fig. 1B; see also refs. 15, 16). The pBS background is lower in the illustrated cell body sample than in the process. pBS backgrounds vary with aRNA preparation and should be subtracted from each specific probe hybridization to eliminate the background contribution to hybridization. The aRNA procedure and expression profiling have been shown to exhibit a linear and quantifiable analysis of mRNA levels (4, 12). Comparison and expression of the relative levels of CREB as a percentage of calmodulin-dependent kinase mRNA from processes isolated from over 30 cells shows that CREB mRNA is present in processes at ≈20% of the levels of calmodulin-dependent kinase mRNA. To gain a broader view of transcription factor mRNAs in the dendritic domain, arrayed blots of several hundred −C2H2 zinc finger containing cDNAs were probed with radiolabeled aRNA from cell bodies or growth cones. Hybridization to several C2H2 cDNAs probed with cell body derived aRNA (>15 positives) and growth cone derived aRNA (five positives) was observed. Partial sequence of ≈2,400 bases of one of these cDNAs (C82) yielded a mouse zinc finger protein containing 12 zinc finger motifs with partial sequence homology to Krox20 (ref. 17, Fig. 1 A and B). The finding of only a few −C2H2 transcription factor mRNAs and the data from the original reverse Northern analysis (Fig. 1 A and B) in dendrites highlights the selectivity of transcription factor mRNA localization in growth cones. Because of the potential importance of CREB in neuronal plasticity, the dendritic localization in cultured neurons was verified by single cell PCR, DNA sequencing of the PCR product (data not shown), and in situ hybridization. In situ hybridization was performed on primary hippocampal cells by using a digoxigenin-labeled cRNA probe followed by 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indoyl phosphate staining (13). Staining in the dendritic process confirms the subcellular localization of CREB (Fig. 1C). Additionally, to show that CREB mRNA is localized to dendrites in intact tissue and is not an artifact of dispersed cell culturing, as well as to show that dendritic CREB mRNA is present in other species, in situ hybridization was used to localize CREB mRNA in dendrites of human neocortical neurons in brain tissue sections (Fig. 1D). Specificity controls are shown in the insets in Fig. 1 C and D. CREB in situ hybridization was also observed sections of rat and human hippocampus (data not shown).
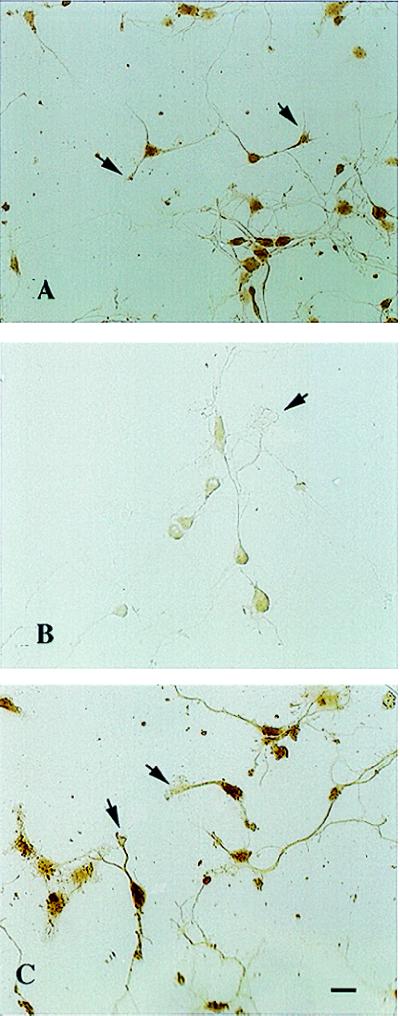
To determine whether CREB protein is present in neuronal processes, cultured hippocampal neurons were stained with polyclonal antibodies against either non- or phosphorylated CREB (Ab244; dilution range of 1:200–1:500) and the leucine zipper domain of CREB (ZE244; dilution range of 1:200–1:500). Ab244 and ZE244 staining was dense in the nucleus and light in somatic, dendritic, and dendritic growth cone cytoplasm (Fig. 2 A and C). Approximately 80% of the cultured neurons exhibited CREB immunoreactivity in dendritic processes. Growth cones with broadened lamellopodia were most heavily stained while many smaller growth cones were not or lightly labeled. No CREB immunoreactivity was seen in morphologically identified axons. Staining was specific for CREB because absorption of Ab244 antibodies with CREB peptide markedly diminished immunolabeling in both nuclei and dendrites (Fig. 2B).
Figure 2.
Immunolabeling of hippocampal dendritic growth cones with CREB antibodies at 48 hr in culture. (A) Intense staining of the nucleus with lighter immunoreactivity in the surrounding cytoplasm extending into the dendrite and growth cone (arrow) by using Ab244. (B) Dramatic reduction of immunoreactivity following prior immunoabsorption of Ab244 with CREB protein. Note absent dendritic growth cone labeling (arrow) and only trace labeling within the nucleus. (C) Similar pattern of immunolabeling in dendrites and dendritic growth cones (arrows) with ZE244. Bar = 20 μM.
CREB in dendrites may exist in either mono- or dimeric form. The majority of cytoplasmic CREB protein exists as nonphosphorylated monomers although a small fraction of cytoplasmic CREB (likely dimeric) is phosphorylated (18). The dimeric form of CREB is predominantly the form that interacts with the CRE, however, CREB monomers can bind the CRE in vitro (19). Thus, because both forms can bind to CRE, we sought to determine whether dendritically localized CREB can bind to CRE concatemers. We checked for binding of the CRE octanucleotide recognized by CREB to the latter’s DNA binding domain/leucine zipper region by using an in situ Southwestern assay. This assay permits the detection of functional transcription factors in paraformaldehyde fixed cells by binding labeled cis-acting elements to the transcription factor. The CRE concatamers bound to CREB protein in the nuclei, dendrites, and dendritic growth cones within neurons (Fig. 3 A–D), corroborating the immunohistochemical identification of CREB in these domains. Indeed, the number of in situ Southwestern assay positive dendrites in a high power field matched the number that were CREB immunopositive. Labeling was dense within the nucleus and more punctate labeling was present within dendrites and dendritic growth cones. Axons showed no binding. Unligated 32P-CRE oligonucleotide did not bind to protein in the cells (Fig. 3E). Moreover, preincubation of the tissue section with excess (>5 mg) unlabeled ligated CRE effectively abolished 32P-CRE concatamer binding. Finally, CRE concatamer binding does not reflect nonspecific interaction of double-stranded DNA with other transcription factors, because a nonsense palindrome probe (inverted CRE sequence, 5′-CAGTACTG-3′) was ineffective (Fig. 3F).
Figure 3.
Binding of radiolabeled CRE to hippocampal neurons (in situ Southwestern assay). (A–C) binding of CRE concatamer to hippocampal neurons at 48 hr in culture (darkfield, 150×). Grains are very dense in nucleus (arrowhead) similar to CREB immunoreactivity but are also detected within the dendrite and dendritic growth cones (arrow). There was no clear binding to axons. (D) Phase contrast view of (C). (E) Unligated CRE exhibited no binding to the neurons. (F) No signal was observed with a ligated inverted CRE palindrome sequence (magnification, ×50).
To demonstrate that CREB mRNA is recognized by dendroplasmic translational machinery, a myc-tagged CREB mRNA was transfected into isolated dendrites (cell bodies removed) by using 1,3-Di-oleoyloxy-2-(6-carboxy-spermyl)-propylamid lipid (Boehringer Mannheim) as the carrier (4). Following application of the lipid:mRNA, dendrites were treated with brain-derived neurotrophic factor or NT3 (100–200 ng/ml) to stimulate protein synthesis (8). The translated fusion protein was detected with anti-myc antibodies (Fig. 4A) while control transfections with lipid encoated pGEM plasmid showed no c-myc immunoreactivity (Fig. 4B). Similarly, treatment with cyclohexamide or RNAse eliminated CREB-myc immunoreactivity. Finally, CREB-myc was not detected in experiments in which brain-derived neurotrophic factor or NT3 were excluded, suggesting that these compounds are important in vivo stimulators of dendritic protein synthesis (4, 9). We conclude that CREB mRNA can be translated locally in individual isolated dendrites and growth cones.
Figure 4.
Local synthesis of CREB protein from CREB mRNA transfected into individual hippocampal dendrites and growth cones. (A) Single transected dendrite (larger arrow) exhibiting c-myc staining adjacent to nonimmunoreactive (nontransfected) cell bodies (smaller arrow). (B) No immunoreactivity is observed in cells transfected with lipid encoated pGEM plasmid mRNA. Bar = 20 μM.
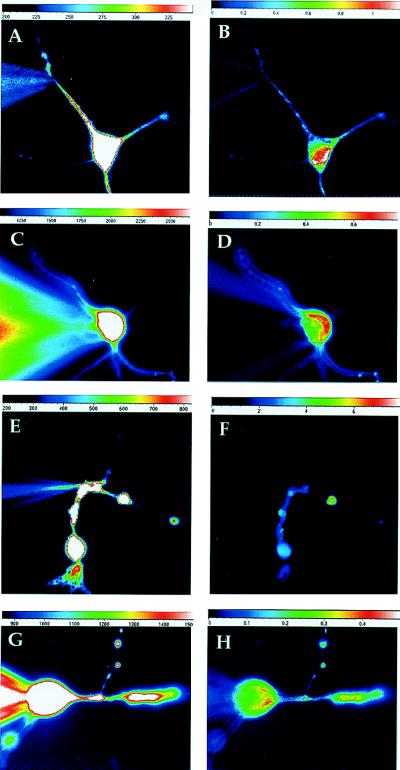
To check for antero- or retrograde transport of dendritic CREB, CREB protein was tagged by linking succinimyl-groups of the fluorescent marker Oregon Green (Molecular Probes) to its lysines (ogCREB). Similarly tagged BSA (ogBSA) served as a diffusion and nonspecific protein control. Both tagged proteins were separately microperfused into the cell body or distal dendrite via a recording electrode under whole cell patch clamp conditions. When perfused into the soma of hippocampal cells, ogCREB did not diffuse into dendrites or axons but concentrated in the nucleus (Fig. 5 C and D). When perfused into dendrites ogCREB moved unidirectionally into the soma and became concentrated in the nucleus (Fig. 5 A and B). In contrast, ogBSA diffused rapidly from the soma or dendrites of neurons throughout the cell (Fig. 5 E–H). These data show that the machinery necessary to transport CREB to the nucleus is present within the dendritic domain. Thus, CREB protein synthesized in dendrites can affect gene expression directly by its selective and rapid transport to the nucleus, likely via interaction of its nuclear localization signal with the translocation machinery (13). These CREB perfusion data also suggest that CREB does not act as a RNA-binding protein to chaperone RNA to the dendrite because microperfused fluorescent CREB does not move from the cell soma to the dendrites that would be expected for a RNA-transport protein.
Figure 5.
Perfusion of ogCREB and ogBSA into dendrite or soma of hippocampal neurons. Because of the limited time the baseline images of ogCREB (A and C) and ogBSA (E and G) show the fluorescence resulting primarily from diffusion of free fluorofluor throughout the cell. The ΔF/F images are a measure of diffusion of ogCREB (B and D) or ogBSA (F and H) and is independent of the pathlength. (A) Image of the cell 300 sec after starting ogCREB and free fluorophore diffusion in the dendrite. Light emitted by the free fluorophore could be easily detected in the soma and numerous dendritic processes. (B) The ΔF/F image taken 21 min later with the baseline image subtracted, showed considerable increase in the fluorescence emitted at the soma, particularly the nucleus, without much change in fluorescence emitted from the dendrites. This difference is due to the larger size and slower diffusion of ogCREB relative to free fluorophore. When ogCREB and free fluorophore are microperfused into the cell body the free fluorophore is easily seen in C, after 135 sec of diffusion in the cell body and the nuclear accumulation of ogCREB can be seen in the ΔF/F image in D, after 867 sec. These same experiments were repeated with ogBSA and free fluorofluor diffusion into the dendrites as shown in E, after 180 sec whole-cell and F, (ΔF/F) after 870 sec. The cell body perfusion of ogBSA is shown in comparison of G, after 211 sec, and (H) (ΔF/F)after 680 sec whole-cell ogBSA.
Transcriptional activation by CREB occurs only if Ser-133 is phosphorylated. To determine whether synaptic modulators could enhance phosphorylation of CREB in dendrites we tested DHPG, a mGluR1 agonist, which had previously been shown to stimulate protein synthesis in synaptodendrosome preparations (20). Application of 50 μM 3,5-dihydroxyphenylglycine for 15 min induced phosphorylation of Ser-133 of CREB in processes severed from their somas (Fig. 6 A and B) as detected with a phospho-Ser-133 CREB specific antibody (ref. 21; Upstate Biotechnology, dilution range of 1:750–1:1000). In Fig. 6A the cell body is left intact close to the severed process (arrow) while in Fig.6B the cell body has been completely removed with the remaining process indicated by a series of arrows. There is little phospho-Ser133 CREB in nonstimulated neuronal cell nucleus or process (Fig. 6C). These data show that Ser-133 phosphorylation can occur independently of the nucleus.
Figure 6.
Phosphorylation of Ser-133-CREB in isolated dendrites. Processes were severed from soma by using a glass micromanipulator. (A and B) The hippocampal cell cultures were bath treated with 50 μM DHPG for 15 min and then the samples were processed for immunohistochemistry by using a CREB phospho-Ser-133-specific antibody and diaminobenzene staining. (A) The cell soma and severed process (arrow) are next to one another. (B) The cell soma has been removed and the remaining process is highlighted by small arrows. (C) Cultures prepared and examined in the same manner as in A and B with the exception that there was no DHPG treatment are shown. The severed process is indicated by the arrow. There is little basal phospho-Ser-133 CREB in the cell nucleus or severed process.
DISCUSSION
Many transcription factors such as CREB, regulate gene expression in response to membrane depolarization, Ca2+ influx, and cAMP-mediated second messenger systems. CREB phosphorylation is tightly coupled to synaptic plasticity and genesis of dendritic spines (22–24) suggesting that cellular events occurring at distance from the nucleus can modulate the activation state of CREB within the nucleus. Because of this, most studies have focused on the regulation of CREB and other transcription factors within the nucleus. The hypothesis that transcription factor mRNAs present within dendrites are locally synthesized into proteins that are retrogradely transported into the nucleus is a view of how changes in gene expression during development may be regulated by distant cellular events. Indeed, the selective transport of microperfused CREB from the dendrite to the nucleus demonstrates that such a pathway exists in vivo and provides a mechanism by which trophic or activity-dependent cues induce local synthesis of transcription factors that are imported to the nucleus and modulate dendritic outgrowth and synaptic connectivity. Dendritic transcription factors may serve to imprint upon the nucleus an image of events occurring in the distal dendrite. Such a nuclear imprinting effect may play an important role in defining activity-dependent states and connectional status of dendritic synapses as dendritic arbors extend into cortex.
Appearance of phospho-Ser-133 CREB in dendrites after mGluR1 stimulation (Fig. 6) provides compelling evidence for how specific stimuli can induce the formation of activated CREB. The possibility that the phosphorylation pattern of CREB differs in different cellular compartments is appealing. The composite of these phosphorylation events likely alters the transcriptional activity of CREB. Unfortunately, no reagents are currently available to assess the state of other putative phosphorylation sites on CREB.
An important consideration, in the concept of “nuclear imprinting” is how the small amount of dendritically synthesized CREB contributes to the total functional nuclear pool. It is difficult to quantitatively compare basal CREB levels or increases in CREB levels in different subregions of neurons because of the large basal levels of CREB protein (Fig. 2) and harvesting of the large number of processes required for Western blot analysis is impractical. However, because basal levels of phospho-CREB are low the DHPG experiment permits us to estimate the relative contribution of dendritic phospho-CREB (dpCREB) to the functional pool of nuclear phospho-CREB (npCREB). Based upon the relative immunoreactivity, 5–10% of the npCREB pool is present in an individual dendrite. Also the dpCREB signal observed in a severed process (Fig. 6A, arrow) is similar to that observed in the process that is still attached to the soma suggesting that much of the phospho-CREB seen in the dendrite is phosphorylated in the dendrite and is not delivered by transport or diffusion from the nucleus to the process. It is reasonable to speculate that when a neuron is stimulated, in vivo, it receives a localized signal, which may differentially stimulate dpCREB production relative to CREB phosphorylation in the nucleus such that there is dpCREB production with little to no stimulation of CREB phosphorylation in the nucleus. This dpCREB, synthesized in the absence of npCREB production, would likely be translocated to the nucleus where it would represent a much larger fraction of the npCREB pool hence this 5–10% estimate (based upon bath application of a receptor agonist) likely underestimates the relative contribution of dpCREB to the nuclear CREB pool.
In toto, these data provide a mechanism by which spatially localized pharmacologic and electrophysiologic signals can effect changes in neuronal gene transcription. The phosphorylation and dephosphorylation of locally synthesized CREB and other transcription factors in the dendrite, which maybe distinct from that in the nucleus, provides a compelling mechanism by which the spatial specificity of Ca2+ entry into the dendrite and mediate regulation of gene expression (25–27).
Acknowledgments
P.B.C. is a Howard Hughes Medical Institute Physician Post-Doctoral Fellow. Partial support for this work came from grants AG9900 and a National Alliances for Research on Schizophrenia and Depression Established Investigator award to J.E. We thank M. Price and M. Dichter for generation of hippocampal neurons, M. Montminy and J. Hoeffler for CREB cDNA, antibodies, and protein, and J.W. Nagle for DNA sequencing. We also thank T. Burcynski, J. Meinkoth, and P. DeWeer for helpful discussions.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: aRNA, amplified antisense RNA; CRE, cyclic AMP response element; CREB, CRE binding protein; pBS, pBluescript plasmid cDNA; DHPG, 3,5-dihydroxyphenylglycine.
References
- 1.Davis L, Banker G, Stewart O. Nature (London) 1987;330:477–480. doi: 10.1038/330477a0. [DOI] [PubMed] [Google Scholar]
- 2.Garner C, Tucker R, Matus A. Nature (London) 1988;336:374–377. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 3.Miyashiro K, Dichter M, Eberwine J. Proc Natl Acad Sci USA. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crino P, Eberwine J. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 5.Eberwine J, Crino P, Dichter M. Neuroscientist. 1995;54:200–210. [Google Scholar]
- 6.Steward O, Banker G. Trends Neurosci. 1992;15:180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- 7.Tiedge H, Brosius J. J Neurosci. 1996;16:7171–7181. doi: 10.1523/JNEUROSCI.16-22-07171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang H, Schuman E. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 9.Weiler I J, Greenough W T. Proc Natl Acad Sci. 1993;90:7168–7171. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchhalter J, Dichter M. Brain Res Bull. 1991;26:333–338. doi: 10.1016/0361-9230(91)90003-3. [DOI] [PubMed] [Google Scholar]
- 11.Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Proc Natl Acad Sci USA. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lockhart D, Dong H, Byrne M, Follettie M, Gallo M, Chee M, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Nat Biotech. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 13.Cao Y, Wilcox K, Martin C, Eberwine J, Dichter M. Proc Natl Acad Sci. 1996;93:9844–9849. doi: 10.1073/pnas.93.18.9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamill O, Marty A, Neher E, Sakmann B, Sigworth F. Pflügers Arch. 1981;391:85–90. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 15.Frantz G, Weimann J, Levin M, McConnell S. J Neurosci. 1994;14:5725–5740. doi: 10.1523/JNEUROSCI.14-10-05725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xuan S, Baptista C, Balas G, Tao W, Soares V, Lai E. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 17.Chavrier P, Janssen-Timmen U, Mattei M, Zerial M, Bravo R, Charnay P. Mol Cell Biol. 1989;9:787–797. doi: 10.1128/mcb.9.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deisseroth K, Biko H, Tsien R. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, Gonzalez G, Biggs W, Montminy M. Nature (London) 1988;334:494–500. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 20.Weiler I J, Irwin S, Klintsova A, Spencer C, Brazelton A, Miyashiro K, Comery T, Patel B, Eberwine J, Greenough W. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginty D, Kornhauser J, Thompson M, Bading H, Mayo K, Takahashi J, Greenberg M. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 22.Waeber G, Habener J. Mol Endocrinol. 1991;5:1431–1438. doi: 10.1210/mend-5-10-1431. [DOI] [PubMed] [Google Scholar]
- 23.Lee J. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 24.Murphy D, Segal M. Proc Natl Acad Sci USA. 1997;94:1482–1487. doi: 10.1073/pnas.94.4.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheng M, McFadden G, Greenberg M. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- 26.Brindle P, Montminy M. Curr Opin Gen Dev. 1992;2:199–204. doi: 10.1016/s0959-437x(05)80274-6. [DOI] [PubMed] [Google Scholar]
- 27.Ginty D. Neuron. 1997;18:183–186. doi: 10.1016/s0896-6273(00)80258-5. [DOI] [PubMed] [Google Scholar]