Abstract
A serotype 1 Streptococcus pneumoniae strain isolated by blood culture from a woman with pneumonia was found to harbor insertion sequence (IS) 1515 in the pneumolysin gene, abolishing pneumolysin expression. To our knowledge, this is the first report of an IS in the pneumolysin gene of S. pneumoniae.
Streptococcus pneumoniae is a bacterial pathogen frequently isolated from young, elderly, and immunocompromised subjects. Pneumococci are divided into 90 serotypes based on their capsular polysaccharide (1). Serotype 1, one of the most prevalent invasive serotypes, is associated with complicated pneumonia, empyema, peritonitis, and salpingitis (4).
Pneumolysin (Ply), a pneumococcal virulence factor (3) classically defined as a pore-forming toxin inhibited by cholesterol, is produced by virtually all clinical isolates of S. pneumoniae (2). The enzyme is cytotoxic for ciliated epithelial cells, and its cytotoxic effects can directly inhibit phagocyte and immune cell function (8). Ply-deficient pneumococci exhibit a reduced capacity for (i) nasopharynx colonization, (ii) lower respiratory tract infection, (iii) intrapulmonary growth, (iv) survival in the bloodstream, and (v) cerebrospinal fluid invasion (7).
The Ply amino acid sequence is generally highly conserved among pneumococci, but some clinical isolates have been shown to bear mutations in the corresponding gene, ply. These mutations are found predominantly in serotypes 1, 7, and 8. A recent study (4) showed that more than 50% of serotype 1 isolates harbored ply mutations. Analysis of these strains by multilocus sequence typing (MLST) revealed that two sequence types (ST), ST227 and ST306, predominated. ST306 harbored mutations at six amino acid positions (Y150H, T1721, K224R, A265S, ΔV270, and ΔK271), whereas ST227 harbored a single mutation (D380N). Ply activity was conserved in ST227 isolates, whereas ST306 isolates were unable to form pores in erythrocyte membranes (4).
Here, we report the insertion of an insertion sequence (IS) into the coding sequence of the ply gene in a type 1 clinical isolate of S. pneumoniae.
The serotype 1 S. pneumoniae strain Lim45, which belongs to our clinical strain collection, was isolated by blood culture from a woman with acute infection of the right upper pulmonary lobe. This isolate was identified using the Gram and optochin tests and subsequently used as positive control in the development of a method for identifying S. pneumoniae species based on the amplification by PCR of an internal fragment of the ply gene. However, in strain Lim45, PCR amplification of genomic DNA with primers Ia (5′-ATTTCTGTAACAGCTACCAACGA-3′) and Ib (5′-GAATTCCCTGTCTTTTCAAAGTC-3′) targeting an internal fragment of ply (9) yielded a 1,220-bp DNA fragment instead of the expected 348-bp product (data not shown). Sequence analysis of this amplified fragment with both strands (Perkin-Elmer Applied Biosystems, Les Ulis, France) revealed the presence of an 871-bp foreign DNA fragment inserted within the ply coding sequence. The insert corresponded to IS1515, which was first described for the capsule-encoding gene cap1E of a serotype 1 S. pneumoniae strain (6). Primers LysI (5′-CCGACTTCTTATCTAGCC-3′) and LysII (5′-TTTGTCGCAAGCATTCTC-3′) were designed to amplify and sequence the entire ply gene of strain Lim45. Sequence analysis of the 2,460-bp amplicon showed that (i) IS1515 was inserted at nucleotide position 430 of the ply coding sequence and was flanked by two direct AAT repeats, indicating duplication of the target sequence, and that (ii) the ply coding sequence of Lim45 harbored nine mutations, only one of which induced an amino acid change, D380N, which was described as characteristic of ST227 (4). Surprisingly, MLST, performed as described elsewhere (4), showed that strain Lim45 belongs to ST228 rather than to ST227. These two ST have two allelic variations, gdh and gki, which differ by two and ten nucleotides, respectively (www.mlst.net). Comparison of the nucleotide sequence of the ply gene harboring IS1515 (2,290 bp) with the 38 ply allele sequences listed in the NCBI databank revealed 100% identity with the sequence of the ply-14 allele (accession number EF413948, unpublished reference).
Multiple copies of IS1515 (from 1 to 13) have been detected in the genome of most type 1 S. pneumoniae strains but not in most other serotypes (6). To determine the number of copies of IS1515 in the genome of strain Lim45, we used Southern analysis of EcoRI-digested chromosomal DNA, using as a probe an internal IS1515-specific fragment obtained by PCR amplification with primers ISI (5′-TATCAGATTATTCAGCACCGA-3′) and ISII (5′-TCACGTCAATAGCCACA-3′), as described elsewhere (5). As one EcoRI cleavage site is present in IS1515 but not in the DNA fragment used as a specific probe, the number of hybridization bands should reflect the IS1515 copy number, which was nine in strain Lim45 (data not shown).
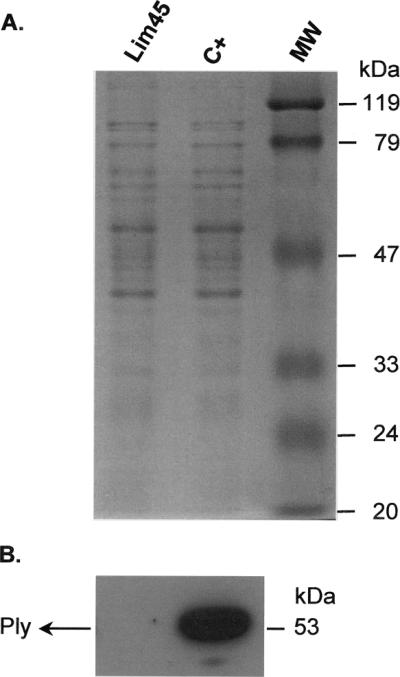
To determine whether IS1515 affected the synthesis of the ply gene product, we used Western blotting analysis of crude bacterial lysates probed with an anti-Ply polyclonal antibody. As shown in Fig. 1, no Ply was detected in strain Lim45, whereas a control strain with no ply insertion sequence was positive. This suggests that IS1515 can abolish the synthesis of the ply gene product in strain Lim45.
FIG. 1.
(A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of total protein from clinical pneumococcal isolate Lim45 (serotype I) and from a control (C+) pneumolysin-positive pneumococcal isolate. (B) Immunoblot analysis of Ply (pneumolysin) in clinical pneumococcal isolate Lim45 (serotype I) and in a control Ply-positive pneumococcal isolate. The membranes were probed with a rabbit antihuman-pneumolysin polyclonal primary antibody (1:5,000) and then incubated with a donkey anti-rabbit horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences Europe GmbH, Vienna, Austria). Immunoreactive bands were detected with an enhanced chemiluminescence kit (Perbio Science Deutschland GmbH, Bonn, Germany). The position of the translational product in the control strain is indicated by Ply (53 kDa). MW, molecular mass.
Kirkham et al. described a serotype 1 strain of S. pneumoniae with a 24-bp insertion in the ply gene, resulting in eight extra amino acids in the Ply protein. The latter insertion considerably reduced Ply expression, as only a faint band was observed by Western blotting; furthermore, the strain was avirulent in mice (4). In our study, strain Lim45 was pathogenic, since it was isolated by blood culture from a woman with acute lobar pneumonia. Although Ply has been reported to be essential for pneumococcal invasive diseases, our findings suggest that virulence factors other than Ply must play an important role in invasive processes. Furthermore, the role of insertion sequences in the population dynamics of S. pneumoniae is largely unknown. The presence of IS1515 in the ply gene supports an important role of mobile insertion sequences in the stability of the pneumococcal genome. Our findings also indicate that insertion sequences may silence the expression of important virulence factors (e.g., capsule, Ply, etc.) under selection pressures encountered in the host.
With the emergence of penicillin- and multidrug-resistant S. pneumoniae strains, the effective means to control pneumococcal disease will rely on preventive strategies, such as vaccination with commercially available vaccines, the 23-valent capsular polysaccharide vaccine (ineffective in children <2 years old), and the 7-valent capsular polysaccharide protein conjugate vaccine. Since protection from these strategies is somewhat limited and varies globally with the pneumococcal serotype prevalence (4), new conjugate vaccines including more serotypes are in development, but their preparations are technically complicated and very expensive (10). Therefore, pneumococcal vaccines based on proteins such as Ply, pneumococcal surface antigen, pneumococcal surface protein A, pneumococcal surface protein C, pneumococcal histidine triad proteins, pneumococcal protective protein A, neuraminidase, and autolysin or iron transport proteins were suggested as interesting alternatives (10). Our findings indicating that Ply does not seem to be fully essential for the invasive process of pneumococcus bring into question whether Ply is a potential candidate for a future vaccine.
Nucleotide sequence accession number.
The nucleotide sequence of the ply gene harboring IS1515 (2.290 bp) has been deposited in the EMBL/GenBank databases under accession number EF490446.
Acknowledgments
We acknowledge use of the pneumococcal MLST database, which is located at Imperial College London and is funded by the Wellcome Trust.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirst, R. A., A. Kadioglu, C. O'Callaghan, and P. W. Andrew. 2004. The role of pneumolysin in pneumococcal pneumonia and meningitis. Clin. Exp. Immunol. 138:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jedrzejas, M. J. 2001. Pneumococcal virulence factors: structure and function. Microbiol. Mol. Biol. Rev. 65:187-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirkham, L. A. S., J. M. C. Jefferies, A. R. Kerr, Y. Jing, S. C. Clarke, A. Smith, and T. J. Mitchell. 2006. Identification of invasive serotype 1 pneumococcal isolates that express nonhemolytic pneumolysin. J. Clin. Microbiol. 44:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mammeri, H., M. Van De Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz, R., R. Lopez, and E. Garcia. 1998. Characterization of IS1515, a functional insertion sequence in Streptococcus pneumoniae. J. Bacteriol. 180:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orihuela, C. J., G. Gao, K. P. Francis, J. Yu, and E. I. Tuomanen. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 190:1661-1669. [DOI] [PubMed] [Google Scholar]
- 8.Rubins, J. B., and E. N. Janoff. 1998. Pneumolysin: a multifunctional pneumococcal virulence factor. J. Lab. Clin. Med. 131:21-27. [DOI] [PubMed] [Google Scholar]
- 9.Salo, P., A. Örtqvist, and M. Leinonen. 1995. Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J. Infect. Dis. 171:479-482. [DOI] [PubMed] [Google Scholar]
- 10.Tai, S. S. 2006. Streptococcus pneumoniae protein vaccine candidates: prospectives, activities and animal studies. Crit. Rev. Microbiol. 32:139-153. [DOI] [PubMed] [Google Scholar]