Abstract
A pan-virus DNA microarray (Virochip) was used to detect a human metapneumovirus (hMPV) strain associated with a critical respiratory tract infection in an elderly adult with chronic lymphocytic leukemia. This infection had previously eluded diagnosis despite extensive microbiological testing for possible etiologic agents. The patient's hMPV strain did not grow in viral culture, and only one of five specific reverse transcription-PCR assays for hMPV was positive.
CASE REPORT
A 65-year-old male with a history of untreated Rai stage 3 chronic lymphocytic leukemia (CLL) presented in April 2006 with an acute febrile respiratory illness. One week prior, he and his wife had taken a cruise trip to Greece and Turkey. During the trip, the patient, his wife, and several people on the cruise developed “flu-like” symptoms. Additional potential exposures to pathogens included a recent visit by the patient in November to his daughter, who lives on a farm in Indiana, and rodents running near the hot tub at his home in the city of Los Altos, CA. Upon his return to the United States, the patient was seen in an urgent care clinic, presenting with a 3-day history of fever and cough, and was prescribed a 7-day course of doxycycline, which was followed by 3 days of levofloxacin due to persistent symptoms. Despite antibiotic therapy, the patient's symptoms continued to worsen over the next 10 days, and he was admitted to the hospital for further workup.
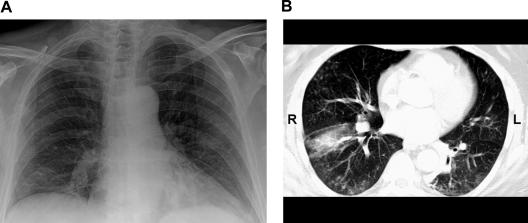
On admission, the patient had fever to 39.7°C, cough, and dyspnea (oxygen saturation, 93% on room air). His leukocyte count was 98,000/mm3, with 48% lymphocytes (patient's baseline for his CLL). A chest X ray and computed tomography scan showed a right-lower-lobe consolidation and bilateral reticulonodular infiltrates (Fig. 1). Gram staining of a sputum sample demonstrated no white blood cells or organisms, and subsequent cultures of the sputum grew only normal respiratory flora. Fluorescent microscopy and culture of the sputum for acid-fast bacilli were negative. Blood cultures demonstrated no growth after 5 days of incubation. Direct fluorescent-antibody (DFA) testing from a nasopharyngeal swab using the D3-DFA respiratory virus kit (Diagnostic Hybrids, Athens, OH) failed to detect respiratory syncytial virus (RSV), adenovirus, influenza A and B viruses, and parainfluenza virus types 1, 2, and 3. The patient was given nasal oxygen and started on ceftriaxone and azithromycin for presumed community-acquired pneumonia.
FIG. 1.
Chest imaging of the patient. A chest X ray taken on admission shows bilateral reticulonodular infiltrates and a right-lower-lobe consolidation (A), while a computed tomography scan (representative slice) (B) reveals in addition a bilateral “tree-in-bud” pattern consistent with a diffuse bronchiolitis.
Despite these interventions, the patient continued to deteriorate clinically and, by hospital day 7, was intubated for progressive respiratory distress. A bronchoscopy with bronchoalveolar lavage (BAL) was performed. The patient's antibiotic coverage was expanded empirically to imipenem, linezolid, liposomal amphotericin, and azithromycin.
Extensive diagnostic testing for an infectious agent was negative. These studies included bacterial, acid-fast bacillus, and fungal cultures on blood, sputum, and BAL fluid, stains and cultures for Pneumocystis jiroveci, urine Legionella antigen, serum Cryptococcus antigen, urine Histoplasma antigen, and buffy coat examination with Wright-Giemsa and Gomori methenamine silver stains, as well as coccidioidomycosis, histoplasmosis, Mycoplasma, and Chlamydia antibody titers. Viral cultures of the BAL fluid and a nasopharyngeal aspirate failed to identify a virus by hemadsorption or cytopathic effect, despite incubation for up to 21 days using five permissive cell lines (two rhesus monkey kidney cell lines, two human fibroblast cell lines, and a continuous A549 human alveolar epithelial cell line). Shell vial cultures for RSV, influenza A virus, and cytomegalovirus were negative. No cytomegalovirus DNA was detected by PCR analysis of BAL fluid or blood. DFA testing of the BAL fluid for RSV, influenza A and B viruses, parainfluenza virus types 1, 2, and 3, and adenovirus was negative. Bone marrow biopsy and flow cytometric analysis of the patient's peripheral blood cells failed to reveal evidence for prolymphocytic or Richter's transformation to acute leukemia; also consistent with this was a normal serum immunoglobulin G (IgG) level of 875 mg/dl.
Informed consent for microarray analysis was obtained under protocols approved by the Stanford University Institutional Review Board (IRB) committee. The patient's BAL fluid at hospital day 7 was flash frozen in liquid nitrogen and stored in a sterile container for analysis using the Virochip, a DNA microarray bearing the most conserved sequences of all known viruses (NCBI GEO, platform GPL3429, series GSE6214) (4, 21). RNA extracted from the specimen was amplified, labeled, and hybridized to the array as previously described (4). Analysis of the microarray hybridization using the E-Predict analysis tool (17) revealed a pattern consistent with infection by human metapneumovirus (hMPV). Surprisingly, initial testing with four clinically validated reverse transcription-PCR (RT-PCR) assays performed on the same BAL specimen using hMPV-specific primers, including one assay in a real-time PCR format, was negative (8, 11, 14). This suggested the possibility that the patient might be harboring a divergent hMPV isolate.
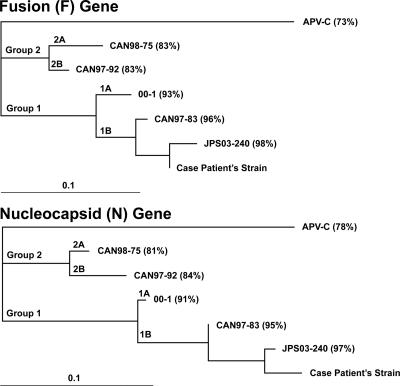
To explore this possibility, the 70-mer oligonucleotide probe on the microarray with the highest hMPV intensity, derived from the nucleoprotein capsid gene of hMPV, was selected. The nucleoprotein capsid gene sequences from five hMPV strains spanning the four different subgroups were downloaded from GenBank and aligned in parallel with the oligonucleotide probe by using ClustalW (16). This multiple sequence alignment was used to design a “spiked” degenerate primer, MPVN-Stf3-f. The “spiked” primer was then employed in conjunction with a random amplicon library derived from the patient's sample to amplify a 450-bp viral fragment that was sequenced and found to share 96% identity with a recently characterized hMPV isolate from Japan (6). Additional sequencing of the complete nucleoprotein and fusion genes of hMPV by primer walking confirmed that the patient's isolate was indeed hMPV, with base mismatches in the primers used to detect hMPV likely responsible for the failure of the previously performed RT-PCR assays (Table 1). However, subsequent testing with an additional pair of primers was used for RT-PCR, and this primer set (MPVN-F1-FAM/MPVN-R1), from Falsey et al. (8), did detect hMPV. Phylogenetic analysis revealed that the patient's hMPV strain was a divergent member of subgroup 1B (Fig. 2). An indirect immunofluorescence assay for hMPV showed a strongly positive hMPV-specific IgG titer of 1:1,028 on hospital day 3 (day 13 after onset of symptoms), with a further eightfold increase in antibody titer over the next 2 weeks.
TABLE 1.
Primers used for detection of hMPVa
| Primer name | Sequence | Target region | Nucleotide position | Expected size of fragment (bp) | Reference |
|---|---|---|---|---|---|
| Published primer pairs | |||||
| MPVF-1f | 5′-CTTTGGACTTAATGACAGATG-3′ | Fusion gene | 3707-3724 | 449 | 14 |
| MPVF-1r | 5′-GTCTTCCTGTGCTAACTTTG-3′ | 4153-4134 | |||
| MPVN-3f | 5′-GAGAAGAGCTGGGTAGAAG-3′ | Nucleoprotein gene | 397-415 | 389 | 14 |
| MPVN-3r | 5′-CAAACAAACTTTCTGCT-3′ | 786-770 | |||
| MPVF-F1-FAM | 5′-GAGCAAATTGAAAATCCCAGACA-3′ | Fusion gene | 3288-3310 | 388 | 8 |
| MPVF-R1-FAM | 5′-GAAAACTGCCGCACAACATTTAG-3′ | 3674-3652 | |||
| MPVN-F1-FAM | 5′-CAACAGGAAGCAAAGCAGAAAG-3′ | Nucleoprotein gene | 757-778 | 174 | 8 |
| MPVN-R1 | 5′-CAGATTCAGGACCCATTTCTC-3′ | 951-931 | |||
| MPV-sense | 5′-CAAGTGTGACATTGCTGACCTGAA-3′ | Fusion gene | 3591-3614 | 118 | 11 |
| MPV-antisense | 5′-ACTGCCGCACAACATTTAGAAA-3′ | 3668-3652 | |||
| MPV-probe | 5′-TGGCYGTYAGCTTCAGTCAATTCAACAGA-3′ | 3617-3645 | |||
| “Spiked” primer | |||||
| MPVN-Stf3-f | 5′-GAAAAAGTRAAYACTRTATCAGAAAC-3′ | Nucleoprotein gene | 1368-1393 |
The patient's bronchoalveolar lavage sample was tested by RT-PCR for hMPV using 5′ clinically validated primer sets. All failed, with the exception of primer set MPVN-F1-FAM/MPVN-R1 from Falsey et al. (8). Base mismatches relative to the patient's hMPV strain in the primer sequences used to detect hMPV are underlined. The nucleotide positions shown are relative to the genomic sequence of hMPV strain 00-1.
FIG. 2.
Phylogenetic analysis of the patient's hMPV strain. Partial sequences of the highly conserved fusion genes (895 bp) and nucleocapsid genes (681 bp) from the patient's hMPV strain, the closest GenBank relative (JPS03-240), representative members from each of the four hMPV subgroups (CAN98-75, CAN97-92, 00-1, and CAN97-83), and avian pneumovirus type C (APV-C) were analyzed. For each sequence, the nucleotide percent identity to the patient's hMPV strain is shown in parentheses.
By the time the diagnosis of hMPV was confirmed, the patient was slowly recovering with aggressive supportive care in the intensive care unit and therefore was not treated with antiviral drugs despite a vigorous discussion of the risks and benefits involved. He received more than 2 weeks of empirical antibiotic treatments for bacterial and fungal pathogens and was discharged, after 3 weeks in the hospital, in stable condition.
The differential diagnosis for an immunocompromised patient presenting with a severe community-acquired respiratory illness is broad. Although many such cases are bacterial in origin, consideration must be given to infection by atypical organisms, including endemic or opportunistic fungi, mycobacteria, and viruses. The exposure history suggested histoplasmosis, coccidioidomycosis, or hantavirus as unlikely but possible atypical etiologic agents. Given the patient's history of CLL with high lymphocyte count, pulmonary leukostasis was also a consideration. However, his clinical picture of fever and bronchiolitis was much more consistent with an infectious process, as was the history of concomitant respiratory illness in several of his fellow travelers.
Community-acquired infections caused by respiratory viruses have been associated with pneumonia and death in immunocompromised individuals (3, 5, 9, 10). In normal adult hosts, viruses such as RSV, influenza virus, rhinovirus, and coronavirus are typically associated with benign, self-limited upper respiratory tract symptoms, such as sore throat, cough, and fever. In immunocompromised patients, however, these same viruses have been associated with life-threatening lower respiratory tract infections, such as bronchiolitis and pneumonia (3, 5, 9). The duration of viral shedding can be prolonged in immunocompromised patients as well, with a mean duration of 16 days (range, 1 to 30 days) in one study of RSV infections in bone marrow transplant recipients (10). Thus, it is not surprising that the case patient's hMPV was still detectable at day 17 after onset of symptoms.
hMPV is a novel RNA virus that was discovered in The Netherlands in 2001 (12, 19). Although thought to be primarily a disease of childhood (with nearly 100% seroprevalence by the age of 5 years), hMPV has recently been linked to severe respiratory infection in elderly and immunocompromised individuals (1, 8, 20). There have been several reported deaths associated with hMPV infection, all in immunocompromised patients (2, 7, 13, 15). The diagnosis of hMPV infection is problematic, as the virus is difficult to isolate from routine cell culture lines (20). RT-PCR examination of respiratory secretions is currently the clinical test of choice to reliably diagnose hMPV infection (20), yet there is currently no consensus on the optimal primer set for hMPV detection. As shown here, the sensitivity of RT-PCR may vary depending on which primer set is selected or which gene is targeted. An RT-PCR assay may also lack sensitivity due to base pair mismatches in the primer sequences. Immunofluorescence is an alternative means to diagnosing hMPV infection, but it has not yet been adopted for clinical use (11, 15).
We have previously described the development of the Virochip, a pan-virus microarray platform that is capable of simultaneous detection of known as well as novel viruses in a single assay (4, 21). The Virochip is currently a research tool, and several issues must be addressed before it can be used on a routine basis for clinical virus detection. These issues include cost, accuracy, reproducibility, and sensitivity/specificity for virus detection in comparison with traditional laboratory tests such as culture, DFA, and specific PCR; research in these areas is ongoing. Nevertheless, in the diagnosis of patients with unexplained critical respiratory illness, the Virochip offers several potential advantages over conventional methods. Unlike PCR, the Virochip assay uses sequences derived from multiple genomic regions, rather than a single conserved region, for detection, and the use of 70-mer oligonucleotides on the array allows better tolerance of sequence mismatches than the shorter sequences employed as primers in RT-PCRs, resulting in improved detection of divergent or novel virus strains (18, 22). The Virochip is also a comprehensive detection platform that does not rely on a priori knowledge of the specific viral pathogen to be detected.
In summary, a pan-virus microarray, the Virochip, was used to detect an hMPV strain associated with a critical respiratory illness that was not detected by viral culture or by the majority of RT-PCR assays tested. Viruses should be considered in the differential diagnosis of critical respiratory illnesses, especially in immunocompromised patients. However, this case illustrates the potential pitfalls of virus-specific PCR for diagnosis, especially given the limited sequences available and inherent sequence diversity for many viruses. It also demonstrates the potential utility of new technologies such as the Virochip in aiding such diagnosis of viral infections in the future.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the fusion gene and nucleocapsid gene of the patient's hMPV strain are EF088284 and EF088285, respectively.
Acknowledgments
This research was supported by the Doris Duke Charitable Foundation (C.Y.C., D.G., and J.L.D.) and the Howard Hughes Medical Institute (D.G. and J.L.D.).
We thank Chao Pan and Thomas Kwok for expert technical assistance. We also thank Amy Kistler, Anatoly Urisman, and Patrick Tang for helpful discussions and comments.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Boivin, G., G. De Serres, M. E. Hamelin, S. Cote, M. Argouin, G. Tremblay, R. Maranda-Aubut, C. Sauvageau, M. Ouakki, N. Boulianne, and C. Couture. 2007. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long-term care facility. Clin. Infect. Dis. 44:1152-1158. [DOI] [PubMed] [Google Scholar]
- 2.Cane, P. A., B. G. van den Hoogen, S. Chakrabarti, C. D. Fegan, and A. D. Osterhaus. 2003. Human metapneumovirus in a haematopoietic stem cell transplant recipient with fatal lower respiratory tract disease. Bone Marrow Transplant. 31:309-310. [DOI] [PubMed] [Google Scholar]
- 3.Chemaly, R. F., S. Ghosh, G. P. Bodey, N. Rohatgi, A. Safdar, M. J. Keating, R. E. Champlin, E. A. Aguilera, J. J. Tarrand, and I. I. Raad. 2006. Respiratory viral infections in adults with hematologic malignancies and human stem cell transplantation recipients: a retrospective study at a major cancer center. Medicine (Baltimore) 85:278-287. [DOI] [PubMed] [Google Scholar]
- 4.Chiu, C. Y., S. Rouskin, A. Koshy, A. Urisman, K. Fischer, S. Yagi, D. Schnurr, P. B. Eckburg, L. S. Tompkins, B. G. Blackburn, J. D. Merker, B. K. Patterson, D. Ganem, and J. L. DeRisi. 2006. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin. Infect. Dis. 43:e71-e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couch, R. B., J. A. Englund, and E. Whimbey. 1997. Respiratory viral infections in immunocompetent and immunocompromised persons. Am. J. Med. 102:2-9. (Discussion, 102:25-26.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebihara, T., R. Endo, H. Kikuta, N. Ishiguro, H. Ishiko, M. Hara, Y. Takahashi, and K. Kobayashi. 2004. Human metapneumovirus infection in Japanese children. J. Clin. Microbiol. 42:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Englund, J. A., M. Boeckh, J. Kuypers, W. G. Nichols, R. C. Hackman, R. A. Morrow, D. N. Fredricks, and L. Corey. 2006. Brief communication: fatal human metapneumovirus infection in stem-cell transplant recipients. Ann. Intern. Med. 144:344-349. [DOI] [PubMed] [Google Scholar]
- 8.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh, S., R. Champlin, R. Couch, J. Englund, I. Raad, S. Malik, M. Luna, and E. Whimbey. 1999. Rhinovirus infections in myelosuppressed adult blood and marrow transplant recipients. Clin. Infect. Dis. 29:528-532. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, S., R. E. Champlin, J. Englund, S. A. Giralt, K. Rolston, I. Raad, K. Jacobson, J. Neumann, C. Ippoliti, S. Mallik, and E. Whimbey. 2000. Respiratory syncytial virus upper respiratory tract illnesses in adult blood and marrow transplant recipients: combination therapy with aerosolized ribavirin and intravenous immunoglobulin. Bone Marrow Transplant. 25:751-755. [DOI] [PubMed] [Google Scholar]
- 11.Landry, M. L., D. Ferguson, S. Cohen, T. C. Peret, and D. D. Erdman. 2005. Detection of human metapneumovirus in clinical samples by immunofluorescence staining of shell vial centrifugation cultures prepared from three different cell lines. J. Clin. Microbiol. 43:1950-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McIntosh, K., and A. J. McAdam. 2004. Human metapneumovirus—an important new respiratory virus. N. Engl. J. Med. 350:431-433. [DOI] [PubMed] [Google Scholar]
- 13.O'Gorman, C., E. McHenry, and P. V. Coyle. 2006. Human metapneumovirus in adults: a short case series. Eur. J. Clin. Microbiol. Infect. Dis. 25:190-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peret, T. C., G. Boivin, Y. Li, M. Couillard, C. Humphrey, A. D. Osterhaus, D. D. Erdman, and L. J. Anderson. 2002. Characterization of human metapneumoviruses isolated from patients in North America. J. Infect. Dis. 185:1660-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumino, K. C., E. Agapov, R. A. Pierce, E. P. Trulock, J. D. Pfeifer, J. H. Ritter, M. Gaudreault-Keener, G. A. Storch, and M. J. Holtzman. 2005. Detection of severe human metapneumovirus infection by real-time polymerase chain reaction and histopathological assessment. J. Infect. Dis. 192:1052-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urisman, A., K. F. Fischer, C. Y. Chiu, A. L. Kistler, S. Beck, D. Wang, and J. L. DeRisi. 2005. E-Predict: a computational strategy for species identification based on observed DNA microarray hybridization patterns. Genome Biol. 6:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. Derisi. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathogens 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Hoogen, B. G., D. M. Osterhaus, and R. A. Fouchier. 2004. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr. Infect. Dis. J. 23:S25-S32. [DOI] [PubMed] [Google Scholar]
- 21.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, D., A. Urisman, Y. T. Liu, M. Springer, T. G. Ksiazek, D. D. Erdman, E. R. Mardis, M. Hickenbotham, V. Magrini, J. Eldred, J. P. Latreille, R. K. Wilson, D. Ganem, and J. L. DeRisi. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 1:E2. [DOI] [PMC free article] [PubMed] [Google Scholar]