Abstract
The development of cervical cancer is strongly associated with the presence of persistent high-risk (HR) human papillomavirus (HPV) infection. Recently, the commercially manufactured PCR-based Roche AMPLICOR (AMP) and LINEAR ARRAY (LA) HPV tests have become available for HPV detection. However, knowledge of their clinical performance compared to the U.S. Food and Drug Administration-approved Hybrid Capture 2 (HC2) assay is limited. This study evaluated the concordance between the HC2, AMP, and LA tests in detecting HR-HPV among a cohort of 1,679 women with previous abnormal Pap smear results. Overall, 1,393 specimens (81.3%) generated concordant results for HR-HPV presence or absence by the three assays. The concordance levels were substantial between the HC2 and AMP tests (84.4%, κ = 0.6419) and between the HC2 and LA tests (84.0%, κ = 0.6341) and nearly perfect between the AMP and LA tests (97.8%, κ = 0.9441). HR-HPV prevalence, as detected by the AMP or LA tests, was significantly higher among women with cytological or histological high-grade disease (CIN2 or greater) than that detected by HC2 (P < 0.0001). The AMP and LA tests exhibited greater sensitivity, but lower specificity, than HC2 for detecting HR-HPV among this cohort of women with underlying cervical abnormalities, particularly among subjects with histologically proven high-grade disease. Both PCR-based HPV tests may be valuable in the management of care for women with underlying cervical abnormalities, in predicting treatment success, and in studying the clearance or acquisition of new infections.
Persistent infection with high-risk (HR) human papillomaviruses (HPV) has been demonstrated as the necessary causal factor for developing high-grade cervical intraepithelial neoplasia (CIN) and potential progression to carcinoma of the cervix (1, 2, 21, 33, 47). There are more than 100 recognized HPV genotypes, of which approximately 40 have tropism specifically for the anogenital mucosa. Based on their epidemiological association with high-grade CIN and cervical cancer, anogenital HPV genotypes are divided into HR and low-risk (LR) types (11, 22). Nearly all cases of cervical cancer have been shown to contain HR-HPV genotypes, with declining proportions found among high-grade and low-grade CIN lesions, respectively (1, 7, 22, 47). Most HPV infections, regardless of risk association, are transient and asymptomatic and cleared by host immunity without leading to obvious cellular abnormalities. However, persistent infection with the same HR-type substantially increases the risk for developing high-grade dysplasia (≥CIN2), and ultimately cervical cancer (14, 16, 30, 48).
Considering the substantial association between HR-HPV infection and cervical cancer, it has been proposed that incorporation of highly sensitive HPV detection methods, such as HPV DNA testing, into current screening programs may improve the identification of cervical abnormalities in women. Several studies have shown that HR-HPV testing, either as an independent screening tool or as an adjunct to current Pap smear cytology, could improve the efficacy of population-based screening programs in detecting underlying lesions or their subsequent development, for triage of women with equivocal or minor cytological lesions and, as a follow-up test for women treated for high-grade lesions to predict treatment success or failure (4, 9, 19, 42, 49).
There are a myriad of molecular tests for detecting HPV infection, of which the Hybrid Capture 2 assay (HC2; Digene Corp., Gaithersburg, MD) is the only commercial HPV DNA test currently approved by the U.S. Food and Drug Administration (5, 8, 27, 46). In addition, there are numerous PCR amplification methods in widespread use, although most largely constitute nonstandardized in-house assays. PCR-based systems frequently used for HPV detection include the primer sets GP5+/GP6+ (10, 15) and MY09/11 and their subsequently modified and improved derivatives PGMY09/11 (12, 13) and the SPF10 system (18, 24, 25), which have been coupled with reverse line-dot blot systems for additional HPV genotyping (25, 36, 43, 44, 45). Only recently have such broad-spectrum PCR-based HPV assays become commercially available, in particular, the AMPLICOR HPV (AMP) test and the LINEAR ARRAY HPV genotyping (LA) test (Roche Molecular Systems, Alameda, CA) (20, 32, 44). These are both qualitative HPV tests that utilize primer sets that amplify a pool of HPV types, with AMP capable of detecting up to 13 HR-HPV genotypes (identical to those detected by HC2) and LA capable of detecting and distinguishing up to 37 individual genotypes (including the 13 HR types detectable by the AMP test and HC2). It has been postulated that these PCR-based HPV tests could provide alternative and rapid means for detecting HR-HPV in clinical samples. However, information on their performance relative to the HC2 assay in clinical settings is very limited.
Therefore, the purpose of the present study was to assess the concordance levels between the HC2, AMP, and LA HPV tests in detecting HR-HPV in cervical specimens collected from women being managed and treated for an abnormal Pap smear result and thereby provide insight into their clinical utility.
MATERIALS AND METHODS
Study population and specimen collection.
The study cohort comprised 1,679 women who presented for evaluation of a previously diagnosed abnormal Pap smear result between May 2001 and June 2005 at the Dysplastic Clinic of the Royal Women's Hospital, Melbourne, Australia. Of the study cohort, 97% of the women were referred with abnormal Pap smear results, varying from low-grade to high-grade dysplasia. The remaining women were referred due to postcoital bleeding, an abnormal-looking cervix, or ongoing review of previous abnormality. Prior to undergoing treatment for histologically defined abnormalities, a cervical specimen for ThinPrep cytology, along with simultaneous HR-HPV detection by HC2, was collected from each woman. Specimens were collected by Cervex brush (Rovers Medical Devices B.V., The Netherlands) and rinsed into ThinPrep vials containing PreservCyt fixative solution (Cytyc Corp., Boxborough, MA). Specimens were routinely analyzed by HC2 assay on a fortnightly basis, while testing for HR-HPV by AMP and LA tests was performed in batches. The mean age of women in the present study, at the time of specimen collection, was 29.8 years (standard deviation, 7.9 years; range, 15.5 to 66.2 years; median, 28.2 years).
Cytological and histological analysis.
Cervical specimens, collected on the same day as the treatment, were analyzed for cytology by liquid-based Pap smear and, where available, tissue collected during excision treatment was histologically assessed. All women were examined by colposcopy both prior to and on the day of treatment. Of the women who had a biopsy collected prior to treatment (ca. 89%), not on the same day as the operation, 95% were shown to have histologically confirmed disease (i.e., low-grade dysplasia or worse). The average lag time between this prior histological examination and sample collection for the present study was 76 days (range, 9 to 462 days). Of the 1,679 patients, excision (including loop electrosurgical excision procedure or cone biopsy) was used to treat 899 (53.5%) women, enabling subsequent histological analysis, while the remaining 780 (46.5%) were treated by laser ablation, where no sample was available for histology. The decision on treatment type was independently made by approximately 10 doctors in the early phase of the study. All women were treated by either excision or laser for high-grade dysplasia or persistent low-grade dysplasia. Women with normal histology prior to treatment (ca. 5%) were treated on the basis of persistent abnormal Pap smear results or other symptoms such as postcoital bleeding. A subset of the histology results (n = 148) had a random adjudicated “blinded” pathological review by an independent histologist.
HC2 assay.
All 1,676 cervical specimens collected in PreservCyt, for liquid-based Pap screening, prior to treatment were routinely tested by HC2 for high-risk HPV DNA according to the manufacturer's instructions. according to the manufacturer's protocol, a 4-ml aliquot of PreservCyt specimen was processed, centrifuged, and resuspended into a 150-μl mixture of specimen transport medium and denaturation buffer (Digene). A 75-μl aliquot of this resuspension was utilized in the HC2 assay, an amount equivalent to 2 ml of the original specimen.
DNA isolation.
For AMP and LA HPV testing, cellular and viral DNA was extracted from PreservCyt cervical specimens by using the automated MagNA Pure LC (MP) isolation and purification system (Roche Molecular Systems) with a modified protocol (38). Briefly, 1-ml aliquots of PreservCyt specimens were centrifuged at 13,000 × g for 20 min prior to supernatant discard and resuspension of cell pellets into 200 μl of sterile phosphate-buffered saline, which were extracted by MP using the DNA-I isolation kit into a final volume of 100 μl.
AMP test.
The AMP test involved amplification of a target region within the genomic DNA, hybridization to a microwell plate, and subsequent colorimetric detection. The PCR utilized biotinylated primers specific for a target sequence, approximately 165 bp in size, spanning a variable region within the polymorphic L1 gene of the HPV genome. These primers are specifically targeted to amplify HPV DNA from the same 13 HR anogenital types detectable by HC2 (i.e., types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68). An additional biotinylated primer set simultaneously amplifies a region (∼268 bp) within the human β-globin gene to ascertain specimen adequacy, DNA extraction, and amplification efficiencies (12, 13). PCR was performed in a 100-μl reaction volume according to the manufacturer's recommendations. To maintain an equivalent amount of DNA to the standard AmpliLute protocol (equivalent to approximately 100 μl of original specimen), 10 μl of MP-extracted DNA was used (38). In brief, the AmpliLute protocol extracts 250 μl of specimen by an QIAamp MinElute column and QIAvac 24 Plus vacuum system (QIAGEN, Inc., Valencia, CA) into a final volume of 120 μl, with 50 μl of DNA extract used in the LA of AMP HPV test (equivalent to 104 μl of original specimen). This adjustment was performed to account for the increased volume of PreservCyt sample processed for DNA extraction (1 ml versus conventional 200 μl), thereby maintaining an equivalent proportion of cervical cells to the standard protocol (38). Amplification and detection protocols were performed as described by the manufacturer. Amplification was performed in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). For detection, absorbance readings of greater than 0.2 were classified as positive for either HPV or β-globin presence.
LA HPV genotyping test.
The LA HPV genotyping test included PCR amplification of target DNA followed by hybridization using a reverse line blot system for simultaneous detection of up to 37 anogenital HPV genotypes (i.e., genotypes 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73, 81, 82, 83, 84, IS39, and CP6108) (45). The LA HPV genotyping test amplifies a region (∼450 bp) within the HPV L1 gene using a pool of biotinylated PGMY primers. As with the AMP test, this test incorporates the amplification of a region within the β-globin gene as an internal control. PCR was performed in a reaction volume of 100 μl using 50 μl of MP-extracted DNA (equivalent to 500 μl of original specimen). Five times more DNA was used in the LA test relative to the AMP test to increase the sensitivity of the LA test in concordance with the standard AmpliLute manual extraction method, as previously described (38). Denatured PCR amplicons were hybridized and detected using the recommended LA protocol, with the modification of using an orbital microplate shaker and dry-air incubator at 53°C in place of the shaking water bath (37, 40). The LA HPV genotyping strips were manually interpreted using the HPV reference guide provided and classified as LA positive (LA+; detection of at least 1 of the 13 HR-HPV genotypes) or LA negative (LA−; detection of exclusively LR-HPV genotypes).
Statistical analysis.
Statistical analyses were performed using 2×2 contingency tables (http://www.graphpad.com/quickcalcs/), with two-sided P values calculated by using either the Fisher exact test or the McNemar's test for comparison of paired proportions. All P values of <0.05 were considered statistically significant. Agreement between tests was assessed by Cohen's kappa statistic, with values of 0.00 to 0.20 indicating poor agreement, 0.21 to 0.40 indicating fair agreement, 0.41 to 0.60 indicating moderate agreement, 0.61 to 0.80 indicating substantial agreement, and 0.81 to 1.00 indicating nearly perfect agreement.
RESULTS
A total of 1,679 cervical specimens were analyzed by the HC2, AMP, and LA tests. Three specimens were negative for β-globin and HPV by the two PCR-based tests and removed from subsequent analyses. Collectively, specimen adequacy, as determined by the positive β-globin results, was 99.8%. Of the remaining 1,676 valid specimens, HR-HPV prevalence was as follows: HC2, 64.0%; AMP, 73.3%; and LA, 72.6% (Table 1). Of these 1,676 specimens, 1,393 (81.3%) showed complete concordance for the presence or absence of HR-HPV by the three tests (1,009 positive and 384 negative). The concordance levels between HC2 and AMP and between HC2 and LA were 84.4% (κ = 0.6419) and 84.0% (κ = 0.6341), respectively. Concordance between AMP and LA was 97.8% (κ = 0.9441) (Table 1).
TABLE 1.
Concordance between HC2, AMP, and LA HPV tests in detecting HR-HPV genotypes among 1,676 PreservCyt specimens
| Test resulta | No. of specimens (%) with AMP HPV test resultb
|
Total no. of specimens (%) | ||
|---|---|---|---|---|
| Positive | Negative | |||
| HC2 result | ||||
| Positive | 1,020 | 52 | 1,072 (64.0) | |
| Negative | 209 | 395 | 604 (36.0) | |
| Total | 1,229 (73.3) | 447 (26.7) | 1,676 | |
| No. of specimens (%) with LA-HPV test resultc
|
||||
| Positive
|
Negative
|
|||
| HC2 result | ||||
| Positive | 1,010 | 62 | 1,072 (64.0) | |
| Negative | 206 | 398 | 604 (36.0) | |
| Total | 1,216 (72.6) | 460 (27.4) | 1,676 | |
| No. of specimens (%) with LA-HPV test resultd
|
||||
| Positive
|
Negative
|
|||
| AMP result | ||||
| Positive | 1,204 | 25 | 1,229 (73.3) | |
| Negative | 12 | 435 | 447 (26.7) | |
| Total | 1,216 (72.6) | 460 (27.4) | 1,676 | |
A positive result refers to HR-HPV detection; a negative result reflects the absence of HR-HPV.
Absolute agreement = 84.4%, κ = 0.6419 (95% confidence interval [CI] = 0.6032 to 0.6806; P < 0.0001). Agreement between tests was assessed by Cohen's kappa statistic. P values were calculated by using McNemar's test.
Absolute agreement = 84.0%, κ = 0.6341 (95% CI = 0.5950 to 0.6732; P < 0.0001).
Absolute agreement = 97.8%, κ = 0.9441 (95% CI = 0.9263 to 0.9619; P = 0.0485).
The discordant results between the three HPV tests are summarized in Table 2. When HC2 is compared to AMP, discordant results were observed among 261 (15.6%) specimens, comprising 52 HC2+/AMP− and 209 HC2−/AMP+ results. Of the 209 HC2−/AMP+ specimens, 195 (93.3%) were shown to contain an HR-HPV genotype by LA, with the remaining 14 being either exclusively LR-HPV types (n = 5) or having no detectable HPV type (n = 9). Of the 52 HC2+/AMP− specimens, 51 did not contain any of the 13 HR-HPV genotypes, as identified by LA. The remaining specimen was identified as having the HR-HPV genotype 45 (Table 2). When HC2 was compared to the LA test, 268 (16.0%) discordant results were observed, comprising 62 HC2+/LA− and 206 HC2−/LA+ findings. Of the 206 HC2−/LA+ specimens, 195 (94.7%) were shown to contain HR-HPV by the AMP test. Of the 62 HC2+/LA− specimens, HR-HPV was detected in 11 specimens by the AMP test, with 51 specimens AMP negative. Finally, when we compared the two PCR-based tests, AMP and LA, only 37 (2.2%) discordant results were observed, comprising 25 AMP+/LA− and 12 AMP−/LA+ results. Of the 25 AMP+/LA− specimens, 11 were identified as having HR-HPV by HC2, with 14 HC2 negative. Of the 12 AMP−/LA+ specimens, 1 specimen was HC2 positive and 11 were HC2 negative, the latter comprising HPV genotypes 16, 18, and 68 (one of each) and 45, 52, 58, and 59 (two of each) (Table 2).
TABLE 2.
Summary of discordant HPV results
| HR-HPV resulta | No. of discordant results
|
Total no. of specimens | |||||
|---|---|---|---|---|---|---|---|
| LA
|
AMP
|
HC2
|
|||||
| Positive | Negative | Positive | Negative | Positive | Negative | ||
| HC2+/AMP− | 1b | 51c | 52 | ||||
| HC2−/AMP+ | 195d | 14e | 209 | ||||
| Total | 261 | ||||||
| HC2+/LA− | 11f | 51c | 62 | ||||
| HC2−/LA+ | 195d | 11g | 206 | ||||
| Total | 268 | ||||||
| AMP+/LA− | 11f | 14e | 25 | ||||
| AMP−/LA+ | 1b | 11g | 12 | ||||
| Total | 37 | ||||||
An LA+ result refers to the detection of a HR-HPV genotype; likewise, an LA− result refers to the absence of an HR-HPV genotype.
Specimen shown to have HPV genotype 45 (by LA).
Specimens possibly representing HC2 cross-reactive genotypes. A total of 32 had at least one of the types 53, 66, 67, 70, 71, 73, or CP6108, and 13 had one of the types 6, 11, 26, 40, 42, 83, or 84, totaling 45 of 51 (88.2%) with known cross-reactive types (type 53 [n = 15], type 42 [n = 9], type 66 [n = 6], type 73 [n = 6], type CP6108 [n = 6], and type 6 [n = 5]). The other six specimens were nondetectable types (n = 4) or HPV-54 and HPV-61.
Due to increased sensitivity of the two PCR-based assays.
Nine specimens had nondetectable HPV types, the other five had exclusively LR types: 55 and 62; 6, 55, 72, and 84; 61 and 62; CP6108; or 54.
Possibly due to a combination of lower sensitivity of the LA and HC2 cross-reactive types, since seven contained at least one of the types (53, 66, 67, 70, 71, 73, or CP6108) and a further two contained one of the types (6, 11, 26, 40, 42, 83, or 84), totaling 9 of 11 (81.8%) with known cross-reactive types (66 [n = 6] and CP6108 [n = 2]).
Specimens with HR-HPV types 16 (n = 1), 18 (n = 1), 45 (n = 2), 52 (n = 2), 58 (n = 2), 59 (n = 2), and 68 (n = 1).
Liquid-based cytology results, from cervical smears at time of treatment, were available for 1,560 (93.1%) of women, whereas histological results from tissue obtained during treatment were available for 899 (53.6%) women, since the remainder were treated by laser ablation. All cytological normal results were from women with a previous abnormal Pap smear result. Among the 531 women with “normal” cytology at the time of treatment, 252 (47.5%) were treated by an excision procedure, of whom 69 (27.4%) were diagnosed with histological high-grade dysplasia (≥CIN2). In addition, of these 531 women, 282 had a colposcopy examination at time of treatment, with 251 (89.0%) reported as having minor abnormalities or worse (i.e., ≥CIN1), including 38.3% with ≥CIN2. The correlation of HR-HPV detection with liquid-based cytology and histology results is detailed in Tables 3 and 4, respectively, with a result of ≥CIN2 defined as “disease positive” for calculating clinical sensitivity and specificity. When we compared HR-HPV detection with cytological prediction (n = 1,560), the clinical sensitivities and specificities were determined as follows: HC2, 87.4 and 46.6%; AMP, 95.2 and 36.9%; and LA, 94.8 and 38.0%, respectively (Table 3). When we compared HR-HPV detection to histological diagnoses (n = 899), the clinical sensitivities and specificities were as follows: HC2, 79.0 and 55.7%; AMP, 91.7 and 49.7%; and LA, 91.0 and 50.8%, respectively (Table 4). When we used HR-HPV detection as a predictor of underlying high-grade cervical disease (i.e., ≥CIN2), there was a significant difference in predicting cervical dysplasia, with the AMP or LA HPV test superior to HC2 (P < 0.0001).
TABLE 3.
Relationship between cytological diagnosis and HR-HPV testing
| HR-HPV resulta | Cytological prediction (no. of specimens)b
|
Sensitivity or specificity (95% CI) | Positive or negative predictive values (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Normal | CIN1 | CIN2 | CIN3 | ACISc | Cancerd | ≥CIN2 | NAe | |||
| HC2+ | 161 | 397 | 240 | 209 | 1 | 1 | 451 | 63 | 87.4 (85.8-89.0) | 44.7 (42.2-47.2) |
| HC2− | 370 | 116 | 49 | 15 | 0 | 1f | 65 | 53 | 46.6 (44.1-49.0) | 88.2 (86.6-89.8) |
| AMP+ | 246 | 413 | 265 | 223 | 1 | 2 | 491 | 79 | 95.2 (94.1-96.2) | 42.7 (40.2-45.2) |
| AMP− | 285 | 100 | 24 | 1 | 0 | 0 | 25 | 37 | 36.9 (34.5-39.3) | 93.9 (92.7-95.1) |
| P (HC2 vs AMP) | <0.0001 | 0.2507 | 0.0025 | 0.0004 | 1.0 | 1.0 | <0.0001 | |||
| LA+ | 243 | 404 | 263 | 223 | 1 | 2 | 489 | 80 | 94.8 (93.7-95.9) | 43.0 (40.6-45.5) |
| LA− | 288 | 109 | 26 | 1 | 0 | 0 | 27 | 36 | 38.0 (35.6-40.4) | 93.6 (92.4-94.8) |
| P (HC2 vs LA) | <0.0001 | 0.6508 | 0.0062 | 0.0004 | 1.0 | 1.0 | <0.0001 | |||
| Total | 531 | 513 | 289 | 224 | 1 | 2 | 516 | 116 | ||
An LA+ result refers to the detection of a HR-HPV genotype; likewise, an LA− result refers to the absence of an HR-HPV genotype.
P values, derived as indicated in column 1, were calculated by using the Fisher exact test.
This specimen was also diagnosed as ACIS by histology.
That is, possible malignancy as determined by cytology. One specimen was diagnosed as a malignancy, and the other was diagnosed as ACIS by histology.
NA, not available (including 29 unsatisfactory results).
HC2-negative specimen from a woman with a possible malignancy (cytology) and ACIS (by histology).
TABLE 4.
Relationship between histological diagnosis and HR-HPV testing
| HR-HPV resulta | Histology diagnosis (no. of specimens)b
|
Sensitivity or specificity (95% CI) | Positive or negative predictive values (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Normal | CIN1 | CIN2 | CIN3 | ACISc | SCCd | ≥CIN2 | NAe | |||
| HC2+ | 59 | 103 | 147 | 260 | 5 | 9 | 421 | 489 | 79.0 (76.3-81.7) | 72.2 (69.3-75.1) |
| HC2− | 111 | 93 | 57 | 51 | 4f | 0 | 112 | 288 | 55.7 (52.5-59.0) | 64.6 (61.4-67.7) |
| AMP+ | 66 | 118 | 176 | 295 | 9 | 9 | 489 | 556 | 91.7 (89.9-93.5) | 72.7 (69.3-75.1) |
| AMP− | 104 | 78 | 28 | 16 | 0 | 0 | 44 | 221 | 49.7 (46.5-53.0) | 80.5 (77.9-83.1) |
| P (HC2 vs AMP) | 0.4998 | 0.1538 | 0.0006 | <0.0001 | 0.0824 | 1.0 | <0.0001 | |||
| LA+ | 66 | 114 | 176 | 291 | 9 | 9 | 485 | 551 | 91.0 (89.1-92.9) | 72.9 (70.0-75.8) |
| LA− | 104 | 82 | 28 | 20 | 0 | 0 | 48 | 226 | 50.8 (47.6-54.1) | 79.5 (76.8-82.1) |
| P (HC2 vs LA) | 0.4998 | 0.3096 | 0.0006 | <0.0001 | 0.0824 | 1.0 | <0.0001 | |||
| Total | 170 | 196 | 204 | 311 | 9 | 9 | 533 | 777 | ||
An LA+ result refers to the detection of a HR-HPV genotype; likewise, an LA− result refers to the absence of an HR-HPV genotype.
P values, derived as indicated in column 1, were calculated by using the Fisher exact test.
This specimen was also diagnosed as ACIS by histology.
SCC, squamous cell carcinoma.
NA, not available.
Four HC2-negative specimens were from women with ACIS (by histology).
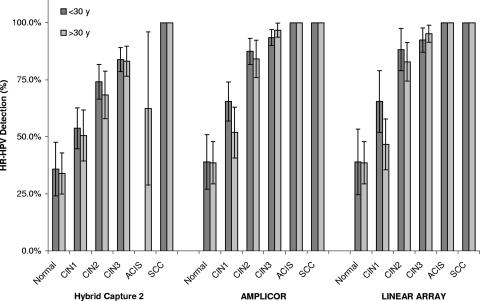
Overall, HR-HPV prevalence increased in parallel with increasing cytological and histological severity regardless of age stratification (Fig. 1 and 2) with the exception of an HC2-negative result from a possible malignancy (as predicted by cytology), diagnosed as adenocarcinoma in situ (ACIS) by histology, and four HC2-negative results among nine women with histologically diagnosed ACIS. Of the 1,560 cytological results, a significantly higher proportion of specimens tested positive for HR-HPV by either the AMP or the LA test than by HC2 in women with a normal Pap smear result, as well as in women diagnosed with CIN2 or CIN3 (together with high-grade abnormality, i.e., ≥CIN2) (Table 3). HC2 and either PCR-based test performed equally in women with a CIN1 Pap smear result, as well as in the detection of the ACIS specimen. Of the 899 histological results, a significantly higher proportion of specimens tested positive for HR-HPV by either PCR method in women with diagnosed CIN2 or CIN3, than by HC2 (Table 4). All three tests performed similarly among the normal and CIN1 groups, as well as in detecting the nine squamous cell carcinoma cases. There were no significant differences between the AMP and LA tests among all cytological and histological results. When subjects were stratified by age (<30 years or ≥ 30 years), there was no difference in HR-HPV prevalence as determined by HC2, AMP, or LA among women with cytologically predicted CIN1 (data not shown), However, when the same age stratification was applied to women with “normal” cytology, HR-HPV prevalence was significantly lower as determined by HC2 than as determined by either AMP or LA (P = 0.0043). Among women either normal or with CIN1 cytology, HR-HPV prevalence was significantly lower in women ≥30 years old than those <30 years old when tested by HC2 (P = 0.0444 and P < 0.0001), AMP (P = 0.0047 and P = 0.0007), and LA (P = 0.0081 and P < 0.0001) (Fig. 1). This was not the case among age-stratified women examined histologically, with the only significant difference being among women (<30 years old or ≥30 years old) with CIN1 and tested by LA (P = 0.0117) (Fig. 2).
FIG. 1.
Relationship between cytological prediction and HR-HPV testing by the HC2, AMP, and LA HPV tests. The prevalence of HR-HPV in women stratified by age (<30 years old and ≥30 years old) is graphed against increasing cytological severity.
FIG. 2.
Relationship between histological diagnosis and HR-HPV testing by the HC2, AMP, and LA HPV tests. HR-HPV prevalence in women stratified by age (<30 years old and ≥30 years old) is graphed against increasing histological severity.
DISCUSSION
In this study, the concordance levels of three commercially available HPV detection kits—the U.S. Food and Drug Administration-approved HC2 assay and the PCR-based AMP and LA tests—were evaluated for the detection of 13 recognized HR-HPV genotypes in a highly referred cohort of 1,679 women with previously reported abnormal Pap smear results. Overall, agreement levels varied from 84.0% (HC2 versus LA, κ = 0.6341), to 84.4% (HC2 versus AMP, κ = 0.6419), to 97.8% (AMP versus LA, κ = 0.9441), indicating substantial correlation between HC2 and either PCR-based test and nearly perfect correlation between both Roche HPV tests. Both PCR-based tests identified a significantly higher prevalence of HR-HPV than HC2: 72.6 and 73.3% versus 64.0%, respectively (P < 0.0001). This finding was not unexpected due to the higher analytical sensitivity of PCR-based methods over signal amplification assays, such as HC2. The high β-globin positivity (99.8%) further confirms the high efficiency of the MP extraction system as a means of specimen processing prior to HPV testing by either PCR-based assay, as reported previously (38).
Given that both Roche HPV tests have only recently become commercially available, there are few population-based studies comparing their performance in HR-HPV detection. Two recent studies reported comparative analyses between the HC2 and the AMP tests in their ability to detect HR-HPV genotypes among cervical specimens, with similar levels of concordance identified between tests (29, 32). The first study analyzed a cohort of 167 women presenting for either screening, evaluation of an abnormal Pap smear result, or follow-up posttreatment, for the presence of 13 HR-HPV genotypes, with concordance between HC2 and AMP reported at 83% (32). The second study prospectively evaluated 312 cervical scrape specimens for the same 13 HR-HPV types. Concordance levels between the HC2 and AMP tests were 85.9% (29). A very similar level of agreement between HC2 and AMP was identified in the present study at 84.4%, based on a significantly larger study cohort and thus with higher levels of confidence. A comparable concordance level was found when HC2 was compared to the LA test at 84.0%.
The positivity rates, varying from 64.0% (HC2) to 72.6 to 73.3% (LA and AMP, respectively), of HR-HPV among this cohort of women with a previous abnormal Pap result were similar to the rate of 65.9% detected by AMP among a cohort of 270 women being managed for an abnormal Pap smear result (20). The higher rates of HR-HPV detected by AMP in the present study, compared to that reported earlier (20), may be attributable to the difference in mean age of the cohorts (29.8 years versus 35.0 years), since HPV prevalence has been shown to decline with age (3). As would be expected, the AMP positivity rates of HR-HPV among screening cohorts are considerably lower (20, 29, 32, 44). It should be reiterated that the present study population is a highly referred group, having previously reported abnormal Pap smear results, with an inflated disease prevalence and a consequent selection bias. Therefore, the current clinical performance of these HPV assays must be viewed in this light and should not be inappropriately extrapolated to more general screening populations where these tests will be most likely used.
The levels of discordance between HPV tests varied from 15.6 to 16.0% between HC2 and PCR-based tests to only 2.2% between the AMP and LA tests among the 1,676 valid specimens. The majority of discordance comprised specimens testing as HC2−/PCR+, a finding that can be attributed to the increased sensitivity of PCR-based testing. Of the HC2+/AMP− discrepancies, 51 of 52 specimens were confirmed as containing no HR-HPV by LA, with 1 specimen containing HPV-45. LA genotyping demonstrated 47 of 51 of these specimens contained at least 1 LR-HPV genotype, with 4 of 51 remaining uncharacterized. A number of recent studies have demonstrated a level of internal cross-reactivity with specific LR-HPV genotypes when the HC2 HR-HPV assay is used (6, 26, 28, 41). Interestingly, all LR-HPV genotypes detected among the 51 HC2+/AMP−/LA− specimens have been described as potential HC2 cross-reactive types. Notably, 32 contained at least one of the types (types 53, 66, 67, 71, 73, and CP6108), described as cross-reactive (26), with an additional 13 containing the cross-reactive types (types 6, 11, 26, 40, 42, 83, and 84), also described previously (41). The remaining two HC2+/AMP−/LA− specimens were composed of single HPV infections (HPV-54 or HPV-61), described as cross-reactive (6, 28). The predominant cross-reactive genotypes identified among the HC2+/PCR− specimens included HPV-53 (n = 15), HPV-42 (n = 9), HPV-66 (n = 6), HPV-73 (n = 6), and HPV-CP6108 (n = 6). HPV-53 and HPV-66 are the most widely identified cross-reactive genotypes (6, 26, 28, 41), which have been suggested as potential HR types through epidemiological association with cervical cancer (22, 23, 39), and may be considered clinically important.
To evaluate the ability of the HC2 assay and the AMP and LA tests to detect HR-HPV genotypes in this highly referred cohort of women with previously identified abnormal Pap smear results, histological diagnosis or cytological prediction were used as standards against which the three HPV tests were assessed. Despite the substantial levels of concordance observed between tests, the AMP and LA tests generated significantly higher rates of HR-HPV positivity than HC2 among women with normal Pap smear results (including smears classified as ≤CIN1). It should be emphasized that the cytologically normal women at the time of treatment were initially enrolled in the present study cohort due to a previous abnormal Pap smear result. Therefore, it is highly likely that a considerable proportion of these cytologically normal women harbor underlying residual disease, possibly missed due to the lower sensitivity of cytology. This is supported by the fact that 27.4% of the available 252 histological samples (with normal cytology) were diagnosed with ≥CIN2 and that 38.2% of 282 women with normal cytology had a colposcopy examination prior to treatment reported as ≥CIN2. In women reported as having normal or CIN1 cytology, the prevalence of HR-HPV was significantly higher among women under 30 years of age than in women ≥30 years old, using either the AMP or the LA test. In a recent study it was reported that despite being cytologically “normal” a single HR-HPV positive result can still be considered substantially predictive of future high-grade CIN development (17). However, it should be noted that HR-HPV prevalence in the present study was not statistically different among women with “normal histology” when HC2 was compared to either the AMP or the LA test. Given the higher sensitivity of histology over cytology, the use of histology as the standard should be considered a more accurate method for comparing the clinical value of these HPV tests.
It has been well established that HPV testing has far greater sensitivity for HPV detection than conventional cytology, although at a cost of reduced specificity. The negative predictive values of HR-HPV tests (i.e., the proportion of women testing negative who are correctly diagnosed as being not at risk) is generally very high, approaching 100% for lesions classified as ≥CIN3, far greater than that of cytology (31, 35, 49). In the present study, the negative predictive values were high (higher when HR-HPV was compared to cytology than to histology). However, it should be noted that the values calculated here were based on the diagnosis of ≥CIN2, thereby lowering the predictive values. When we based these values on ≥CIN3, the values were indeed near 100% (particularly compared to cytology). In the present study, compared to cytology, 1 of 227 (0.4%) ≥CIN3 samples was classified HR-HPV negative by AMP and LA (7.0% by HC2); compared to histology, this value varied from 16 of 329 (4.8%) for AMP to 20 of 329 (6.1%) for LA, among ≥CIN3 samples (15.5% by HC2).
Of importance to the statistical analysis of this data set, particularly the low specificity levels, is the fact that the study cohort was composed of women who were referred due to an initial abnormal Pap smear result after routine screening and, therefore, likely to have been HPV positive. All women were referred for a colposcopy examination, and many lesions could have been biopsied, resulting in the initiation of an immune response and subsequent clearance of the lesion. However, it has been reported that the clearance of HPV genotypes occurs later than the regression of cervical abnormalities (34). Therefore, despite the initial cervical abnormalities that initiated the referral, specimens collected some time later for DNA testing, cytology, and/or histology (range, 9 to 462 days; median, 65 days) may have been obtained subsequent to disease regression, although still harboring detectable HR-HPV DNA. It would be reasonable to expect that in a primary screening setting, the specificity of both PCR-based HPV tests would be substantially greater.
In conclusion, the commercially available AMP and LA HPV tests are relatively easy to use and provide a rapid and standardized method for detecting and genotyping HPV, respectively. In the current setting, both PCR-based tests exhibited greater sensitivity for detecting HR-HPV, particularly among women with high-grade disease (≥CIN2), and yet lower specificity than HC2. Both the AMP and the LA tests should be considered for clinical use in the detection of HR-HPV, in particular for a test of cure, given their greater sensitivity and, in conjunction with the LA test, could be used to predict treatment success or failure, reinfection, and/or new infections. Finally, the high sensitivity of the Roche HPV tests should be considered of significant value as an epidemiological tool for prevalence studies as well as for pre- and postprophylactic vaccine intervention for analyses of HPV infections.
Acknowledgments
Roche Molecular Systems supplied the AMP and LA HPV amplification and detection kits required for this study.
Footnotes
Published ahead of print on 9 May 2007.
REFERENCES
- 1.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, and K. V. Shah. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 2.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burk, R. D., P. Kelly, J. Feldman, J. Bromberg, S. H. Vermund, J. A. DeHovitz, and S. H. Landesman. 1996. Declining prevalence of cervicovaginal human papillomavirus infection with age is independent of other risk factors. Sex Transm. Dis. 23:333-341. [DOI] [PubMed] [Google Scholar]
- 4.Carozzi, F. M., M. Confortini, S. Cecchini, S. Bisanzi, M. P. Cariaggi, G. Pontenani, M. R. Raspollini, C. Sani, M. Zappa, and S. Ciatto. 2005. Triage with human papillomavirus testing of women with cytologic abnormalities prompting referral for colposcopy assessment. Cancer 105:2-7. [DOI] [PubMed] [Google Scholar]
- 5.Castle, P. E., A. T. Lorincz, I. Mielzynska-Lohnas, D. R. Scott, A. G. Glass, M. E. Sherman, J. E. Schussler, and M. Schiffman. 2002. Results of human papillomavirus DNA testing with the hybrid capture 2 assay are reproducible. J. Clin. Microbiol. 40:1088-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castle, P. E., M. Schiffman, R. D. Burk, S. Wacholder, A. Hildesheim, R. Herrero, M. C. Bratti, M. E. Sherman, and A. Lorincz. 2002. Restricted cross-reactivity of hybrid capture 2 with nononcogenic human papillomavirus types. Cancer Epidemiol. Biomarkers Prev. 11:1394-1399. [PubMed] [Google Scholar]
- 7.Clifford, G. M., J. S. Smith, M. Plummer, N. Munoz, and S. Franceschi. 2003. Human papillomavirus types in invasive cervical cancer worldwide: a meta-analysis. Br. J. Cancer 88:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox, J. T., A. T. Lorincz, M. H. Schiffman, M. E. Sherman, A. Cullen, and R. J. Kurman. 1995. Human papillomavirus testing by hybrid capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. Am. J. Obstet. Gynecol. 172:946-954. [DOI] [PubMed] [Google Scholar]
- 9.Cuzick, J., A. Szarewski, G. Terry, L. Ho, A. Hanby, P. Maddox, M. Anderson, G. Kocjan, S. T. Steele, and J. Guillebaud. 1995. Human papillomavirus testing in primary cervical screening. Lancet 345:1533-1536. [DOI] [PubMed] [Google Scholar]
- 10.de Roda Husman, A. M., J. M. Walboomers, A. J. van den Brule, C. J. Meijer, and P. J. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76:1057-1062. [DOI] [PubMed] [Google Scholar]
- 11.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 324:17-27. [DOI] [PubMed] [Google Scholar]
- 12.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlee, A. Hildesheim, M. H. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, G. Y., R. D. Burk, S. Klein, A. S. Kadish, C. J. Chang, P. Palan, J. Basu, R. Tachezy, R. Lewis, and S. Romney. 1995. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J. Natl. Cancer Inst. 87:1365-1371. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs, M. V., P. J. Snijders, A. J. van den Brule, T. J. Helmerhorst, C. J. Meijer, and J. M. Walboomers. 1997. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kjaer, S. K., A. J. van den Brule, G. Paull, E. I. Svare, M. E. Sherman, B. L. Thomsen, M. Suntum, J. E. Bock, P. A. Poll, and C. J. Meijer. 2002. Type specific persistence of high risk human papillomavirus (HPV) as indicator of high grade cervical squamous intraepithelial lesions in young women: population based prospective follow up study. BMJ 325:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kjaer, S., E. Hogdall, K. Frederiksen, C. Munk, A. van den Brule, E. Svare, C. Meijer, A. Lorincz, and T. Iftner. 2006. The absolute risk of cervical abnormalities in high-risk human papillomavirus-positive, cytologically normal women over a 10-year period. Cancer Res. 66:10630-10636. [DOI] [PubMed] [Google Scholar]
- 18.Kleter, B., L. J. van Doorn, J. ter Schegget, L. Schrauwen, K. van Krimpen, M. Burger, B. ter Harmsel, and W. Quint. 1998. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am. J. Pathol. 153:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorincz, A. T., and R. M. Richart. 2003. Human papillomavirus DNA testing as an adjunct to cytology in cervical screening programs. Arch. Pathol. Lab. Med. 127:959-968. [DOI] [PubMed] [Google Scholar]
- 20.Monsonego, J., J. M. Bohbot, G. Pollini, C. Krawec, C. Vincent, I. Merignargues, F. Haroun, P. Sednaoui, L. Monfort, R. Dachez, and K. Syrjanen. 2005. Performance of the Roche AMPLICOR human papillomavirus (HPV) test in prediction of cervical intraepithelial neoplasia (CIN) in women with abnormal PAP smear. Gynecol. Oncol. 99:160-168. [DOI] [PubMed] [Google Scholar]
- 21.Munoz, N., and F. X. Bosch. 1996. The causal link between HPV and cervical cancer and its implications for prevention of cervical cancer. Bull. Pan. Am. Health Organ. 30:362-377. [PubMed] [Google Scholar]
- 22.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, C. J. Meijer, et al. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 23.Munoz, N., X. Castellsague, A. B. de Gonzalez, and L. Gissmann. 2006. HPV in the etiology of human cancer. Vaccine 24-S3:S1-S10. [DOI] [PubMed] [Google Scholar]
- 24.Perrons, C., B. Kleter, R. Jelley, H. Jalal, W. Quint, and R. Tedder. 2002. Detection and genotyping of human papillomavirus DNA by SPF10 and MY09/11 primers in cervical cells taken from women attending a colposcopy clinic. J. Med. Virol. 67:246-252. [DOI] [PubMed] [Google Scholar]
- 25.Perrons, C., R. Jelley, B. Kleter, W. Quint, and N. Brink. 2005. Detection of persistent high-risk human papillomavirus infections with hybrid capture II and SPF10/LiPA. J. Clin. Virol. 32:278-285. [DOI] [PubMed] [Google Scholar]
- 26.Peyton, C. L., M. Schiffman, A. T. Lorincz, W. C. Hunt, I. Mielzynska, C. Bratti, S. Eaton, A. Hildesheim, L. A. Morera, A. C. Rodriguez, R. Herrero, M. E. Sherman, and C. M. Wheeler. 1998. Comparison of PCR- and hybrid capture-based human papillomavirus detection systems using multiple cervical specimen collection strategies. J. Clin. Microbiol. 36:3248-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poljak, M., A. Brencic, K. Seme, A. Vince, and I. J. Marin. 1999. Comparative evaluation of first- and second-generation Digene hybrid capture assays for detection of human papillomaviruses associated with high or intermediate risk for cervical cancer. J. Clin. Microbiol. 37:796-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poljak, M., I. J. Marin, K. Seme, and A. Vince. 2002. Hybrid Capture II HPV Test detects at least 15 human papillomavirus genotypes not included in its current high-risk probe cocktail. J. Clin. Virol. 25-S. 3:S89-S97. [DOI] [PubMed] [Google Scholar]
- 29.Poljak, M., K. Fujs, K. Seme, B. J. Kocjan, and E. Vrtacnik-Bokal. 2005. Retrospective and prospective evaluation of the Amplicor HPV test for detection of 13 high-risk human papillomavirus genotypes on 862 clinical samples. Acta Dermatovenerol. Alp. Panonica Adriat. 14:147-152. [PubMed] [Google Scholar]
- 30.Remmink, A. J., J. M. Walboomers, T. J. Helmerhorst, F. J. Voorhorst, L. Rozendaal, E. K. Risse, C. J. Meijer, and P. Kenemans. 1995. The presence of persistent high-risk HPV genotypes in dysplastic cervical lesions is associated with progressive disease: natural history up to 36 months. Int. J. Cancer 61:306-311. [DOI] [PubMed] [Google Scholar]
- 31.Rozendaal, L., J. Westerga, J. C. van der Linden, J. M. Walboomers, F. J. Voorhorst, E. K. Risse, M. E. Boon, and C. J. Meijer. 2000. PCR based high risk HPV testing is superior to neural network based screening for predicting incident CIN III in women with normal cytology and borderline changes. J. Clin. Pathol. 53:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandri, M. T., P. Lentati, E. Benini, P. Dell'Orto, L. Zorzino, F. M. Carozzi, P. Maisonneuve, R. Passerini, M. Salvatici, C. Casadio, S. Boveri, and M. Sideri. 2006. Comparison of the Digene HC2 assay and the Roche AMPLICOR human papillomavirus (HPV) test for detection of high-risk HPV genotypes in cervical samples. J. Clin. Microbiol. 44:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiffman, M. H., H. M. Bauer, R. N. Hoover, A. G. Glass, D. M. Cadell, B. B. Rush, D. R. Scott, M. E. Sherman, R. J. Kurman, and S. Wacholder. 1993. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 85:958-964. [DOI] [PubMed] [Google Scholar]
- 34.Schiffman, M., C. M. Wheeler, P. E. Castle, et al. 2002. Human papillomavirus DNA remains detectable longer than related cervical cytologic abnormalities. J. Infect. Dis. 186:1169-1172. [DOI] [PubMed] [Google Scholar]
- 35.Snijders, P. J., A. J. van den Brule, and C. J. Meijer. 2003. The clinical relevance of human papillomavirus testing: relationship between analytical and clinical sensitivity. J. Pathol. 201:1-6. [DOI] [PubMed] [Google Scholar]
- 36.Soderlund-Strand, A., P. Rymark, P. Andersson, J. Dillner, and L. Dillner. 2005. Comparison between the Hybrid Capture II test and a PCR-based human papillomavirus detection method for diagnosis and posttreatment follow-up of cervical intraepithelial neoplasia. J. Clin. Microbiol. 43:3260-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens, M. P., S. M. Garland, and S. N. Tabrizi. 2006. Human papillomavirus genotyping using a modified linear array detection protocol. J. Virol. Methods 135:124-126. [DOI] [PubMed] [Google Scholar]
- 38.Stevens, M. P., E. Rudland, S. M. Garland, and S. N. Tabrizi. 2006. Assessment of MagNA Pure LC extraction system for detection of human papillomavirus (HPV) DNA in PreservCyt samples by the Roche AMPLICOR and LINEAR ARRAY HPV tests. J. Clin. Microbiol. 44:2428-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens, M. P., S. N. Tabrizi, M. A. Quinn, and S. M. Garland. 2006. Human papillomavirus genotype prevalence in cervical biopsies from women diagnosed with cervical intraepithelial neoplasia or cervical cancer in Melbourne, Australia. Int. J. Gynecol. Cancer 16:1017-1024. [DOI] [PubMed] [Google Scholar]
- 40.Tabrizi, S. N., M. Stevens, S. Chen, E. Rudland, J. R. Kornegay, and S. M. Garland. 2005. Evaluation of a modified reverse line blot assay for detection and typing of human papillomavirus. Am. J. Clin. Pathol. 123:896-899. [DOI] [PubMed] [Google Scholar]
- 41.Terry, G., L. Ho, P. Londesborough, J. Cuzick, I. Mielzynska-Lohnas, and A. Lorincz. 2001. Detection of high-risk HPV types by the hybrid capture 2 test. J. Med. Virol. 65:155-162. [PubMed] [Google Scholar]
- 42.The Atypical Low-Grade Squamous Intraepithelial Lesions Triage Study Group. 2000. Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. J. Natl. Cancer Inst. 92:397-402. [DOI] [PubMed] [Google Scholar]
- 43.van Doorn, L. J., A. Molijn, B. Kleter, W. Quint, and B. Colau. 2006. Highly effective detection of human papillomavirus 16 and 18 DNA by a testing algorithm combining broad-spectrum and type-specific PCR. J. Clin. Microbiol. 44:3292-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Ham, M. A., J. M. Bakkers, G. K. Harbers, W. G. Quint, L. F. Massuger, and W. J. Melchers. 2005. Comparison of two commercial assays for detection of human papillomavirus (HPV) in cervical scrape specimens: validation of the Roche AMPLICOR HPV test as a means to screen for HPV genotypes associated with a higher risk of cervical disorders. J. Clin. Microbiol. 43:2662-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Hamont, D., M. A. van Ham, J. M. Bakkers, L. F. Massuger, and W. J. Melchers. 2006. Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the Roche linear array HPV genotyping test. J. Clin. Microbiol. 44:3122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vince, A., N. Kutela, J. Iscic-Bes, V. Harni, M. Ivanisevic, Z. Sonicki, Z. Culig, and M. Poljak. 2002. Clinical utility of molecular detection of human papillomavirus in cervical samples by hybrid capture technology. J. Clin. Virol. 25(S.3):S109-S112. [DOI] [PubMed] [Google Scholar]
- 47.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 48.Wallin, K. L., F. Wiklund, T. Angstrom, F. Bergman, U. Stendahl, G. Wadell, G. Hallmans, and J. Dillner. 1999. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N. Engl. J. Med. 341:1633-1638. [DOI] [PubMed] [Google Scholar]
- 49.Zielinski, G. D., P. J. Snijders, L. Rozendaal, F. J. Voorhorst, A. P. Runsink, F. A. de Schipper, and C. J. Meijer. 2001. High-risk HPV testing in women with borderline and mild dyskaryosis: long-term follow-up data and clinical relevance. J. Pathol. 195:300-306. [DOI] [PubMed] [Google Scholar]