Abstract
The discovery of Helicobacter hepaticus and its role in hepatitis, hepatocellular carcinoma, typhlocolitis, and lower-bowel carcinoma in murine colonies was followed by the isolation and characterization of other Helicobacter spp. involved in enterohepatic disease. Colonization of mouse colonies with members of the family Helicobacteriaceae has become an increasing concern for the research community. From 2001 to 2005, shipments of selected gift mice from other institutions and mice received from specified commercial vendors were screened for Helicobacter spp. by culture of cecal tissue. The identities of the isolates were confirmed by genus-specific PCR, followed by species-specific PCR and restriction fragment length polymorphism analysis. Sequencing of the 16S rRNA gene was performed if the species identity was not apparent. The survey included 79 mice from 34 sources: 2 commercial sources and 16 research sources from the United States and 1 commercial source and 15 research sources from Canada, Europe, or Asia. Helicobacter spp. were cultured from the ceca of 62 of 79 mice. No Helicobacter spp. were found in mice from advertised Helicobacter-free production areas from two U.S. vendors. Multiple Helicobacter spp. were found in mice from one vendor's acknowledged Helicobacter-infected production area. The European commercial vendor had mice infected with novel Helicobacter sp. strain MIT 96-1001. Of the U.S. academic institutions, 6 of 16 (37%) had mice infected with Helicobacter hepaticus; but monoinfection with H. bilis, H. mastomyrinus, H. rodentium, and MIT 96-1001 was also encountered, as were mice infected simultaneously with two Helicobacter spp. Non-U.S. academic institutions had mice that were either monoinfected with H. hepaticus, monoinfected with seven other Helicobacter spp., or infected with a combination of Helicobacter spp. This survey indicates that 30 of 34 (88%) commercial and academic institutions in Canada, Europe, Asia, Australia, and the United States have mouse colonies infected with Helicobacter spp. Mice from 20 of the 34 institutions (59%) were most commonly colonized with H. hepaticus alone or in combination with other Helicobacter spp. These results indicate that a broad range of Helicobacter spp. infect mouse research colonies. The potential impact of these organisms on in vivo experiments continues to be an important issue for mice being used for biomedical research.
Naturally acquired Helicobacter infections have been reported in all commonly used laboratory rodent species (32). Several of the most frequently isolated species cause disease in selected strains of infected mice. Helicobacter hepaticus was first isolated from A/JCr mice in a long-term carcinogenesis study, in which the control animals developed a high incidence of hepatic tumors and hepatitis (5, 31). H. hepaticus is now known to cause hepatitis, liver tumors, cholesterol gallstones, inflammatory bowel disease, and colon cancer in susceptible strains of mice (2, 7, 16, 18, 32). The negative impact of natural infection with H. hepaticus was further documented in 12 National Toxicological Program studies (31). Nine of the 12 studies were confounded by H. hepaticus-induced hepatitis and hepatocellular carcinoma in control mice.
Other Helicobacter spp. are also known to cause disease in laboratory mice. Naturally acquired H. typhlonius causes typhlocolitis in immunocompromised mice (6, 11). H. muridarum may be associated with gastritis (22), whereas H. bilis is associated with moderate hepatitis in aged inbred and outbred mice (10, 12) and typhlocolitis and lower-bowel cancer in mice with intestinal barrier defects (17). Furthermore, H. bilis was reported to confound a study performed to determine the effect of chronic oral supplementation with creatinine in which both control and experimental outbred Swiss mice developed hepatitis (8). Maurer et al. (18) reported that C57L mice infected with H. bilis or coinfected with H. hepaticus and H. rodentium and fed a lithogenic diet developed cholesterol gallstones at an 80% prevalence by 8 weeks, whereas approximately 10% of the uninfected controls developed cholesterol gallstones. H. rodentium may also play a pathogenic role with other Helicobacter spp. and elicit diarrhea and typhlocolitis in immunocompromised mice (19, 28). In an experimental model, Helicobacter sp. strain MIT 96-1001 caused inflammatory bowel disease and cholangiohepatitis in SCID and immunocompetent A/J mice (29). Given the proven potential for Helicobacter spp. to confound research utilizing laboratory mice, we undertook a survey of mice from commercial vendors and academic research facilities to determine the prevalence of Helicobacter spp. in mice used in biomedical research.
MATERIALS AND METHODS
Animals and tissue collection.
A total of 79 mice were assessed for colonization with Helicobacter spp.. The majority of the mice (69/79) in this survey were genetically manipulated. They were sent to MIT principal investigators for research purposes from 16 research institutions in the United States and 16 research and commercial institutions in Canada, Europe, Australia, and Asia. In addition, 10 animals were purchased from two U.S. commercial vendors. The mice were euthanized with CO2, and the cecum was collected from each mouse and stored in brucella broth containing 20% glycerol at −70°C until it was submitted for culture.
Histology.
Representative sections of all the hepatic lobes, gallbladder, stomach, and ileocecal junction were fixed in 10% formalin, embedded in paraffin, and routinely stained with hematoxylin-eosin. A board-certified veterinary pathologist (P.N.) examined all tissue sections for lesions. Because, Helicobacter spp. typically cause inflammation within the stomach, liver, and ileocecocolic junction, special emphasis was placed on these organs. Briefly, the stomach, liver, and ileocecocolic junction were scored for inflammation or any other abnormal finding by using previously described criteria (2).
Bacterial isolation and characterization.
Cecal tissue was homogenized in 1 ml of phosphate-buffered saline, and aliquots were placed on CVA (cefoperazone, vancomycin, and amphotericin B) plates or TVP (trimethoprim, vancomycin, and polymyxin B) plates and filtered through a 0.45-μm-pore-size filter onto Trypticase soy agar plates with 5% sheep blood (all from Remel Laboratories, Lenexus, KS). In-house-prepared selective medium plates were also used and contained the following: blood agar base (Oxoid; Remel), 5% horse blood (Quad Five, Ryegate, Montana), 50 μg amphotericin B/ml, 100 μg vancomycin/ml, 3.3 μg polymyxin B/ml, 200 μg bacitracin/ml, and 10.7 μg nalidixic acid/ml (all from Sigma Chemical Company, St. Louis, MO). After incubation under microaerobic conditions (culture vessels evacuated to 25 in. of mercury and filled with N2-CO2-H2 at 80:10:10) at 37°C, suspect colonies were identified as Helicobacter on the basis of colony morphology, biochemical reaction (assessed for the enzymes catalase, oxidase, and urease), phase microscopy, Gram staining, and Helicobacter genus-specific PCR.
DNA extraction.
For PCR of genomic DNA, isolates were grown on blood agar plates, harvested, and washed once with phosphate-buffered saline; and the High Pure PCR template preparation kit (Roche Molecular Biochemicals, Indianapolis, Indiana) was used for DNA extraction according to the manufacturer's specifications.
Genus-specific PCR.
Helicobacter genus-specific primers that amplify a 1.2-kb product on the 16S rRNA gene were used as described previously (4).
RFLP and species-specific PCR.
The 1.2-kb product from the genus-specific PCR was analyzed by restriction fragment length polymorphism (RFLP) analysis by the method previously published by Shen et al. (26). The resulting gel patterns were compared to the RFLP patterns of known Helicobacter spp. Those patterns resembling the patterns for H. hepaticus, H. bilis, H. typhlonius, and H. rodentium were subjected to species-specific PCR (Table 1) for species confirmation. The entire 16S rRNA gene from isolates that were negative by the species-specific PCR and isolates with RFLP patterns other than those for H. hepaticus, H. bilis, or H. rodentium was sequenced.
TABLE 1.
Helicobacter species-specific primer sets
| Organism | Primer sequence (5′ to 3′) | Product size (bp) | Reference |
|---|---|---|---|
| H. typhlonius | Forward: AGGGACTCTTAAATATGCTCCTAGAGT | 122 | 3 |
| Reverse: ATTCATCGTGTTTGAATGCGTCAA | |||
| H. rodentium | Forward: GTCCTTAGTTGCTAACTATT | 166 | 27 |
| Reverse: AGATTTGCTCCATTTCACAA | |||
| H. bilis | Forward: AGAACTGCATTTGAAACTACTTT | 638 | 10 |
| Reverse: GGTATTGCATCTCTTTGTATGT | |||
| H. hepaticus | Forward: GCATTTGAAACTGTTACTCTG | 417 | 25 |
| Reverse: CTGTTTTCAAGCTCCCC |
Sequence analysis.
Amplification of the 16S rRNA cistrons, 16S rRNA gene sequencing, and analysis of the 16S rRNA data were performed as described elsewhere (21). For alignment, the 16S rRNA gene sequences were entered into RNA, a program designed for the analysis of 16S rRNA. The database contains more than 600 sequences for Helicobacter, Wolinella, Arcobacter, and Campylobacter strains and >2,000 sequences for other bacteria.
Nucleotide sequence accession numbers.
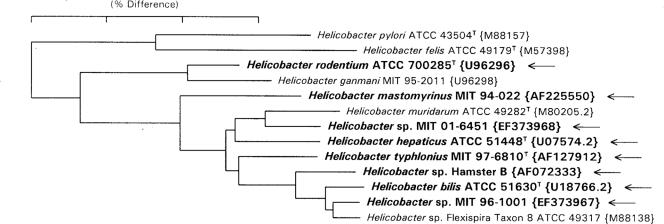
The GenBank accession numbers for the strains examined in this study are included in Fig. 1.
FIG. 1.
Neighbor-joining phylogenetic tree of Helicobacter species recovered from mice. The GenBank accession number of each strain is given in braces. H. pylori and H. felis are included as an outgroup. The species identified in the current survey are shown in boldface and are marked with arrows.
RESULTS
Culture.
Of 79 mice, 62 (78%) were positive by culture for Helicobacter spp. as assessed by biochemical reaction, phase microscopy, Gram staining, and Helicobacter genus-specific PCR. Helicobacter-infected mice came from 30 of the 34 institutions surveyed (Tables 2 and 3). Thus, the overall prevalence of Helicobacter spp. in mice from the institutions in this survey was 88%.
TABLE 2.
Helicobacter species isolated from commercial and research mouse colonies in Europe, Asia, Australia, and Canadaa
| Source | No. of mice tested/no. of Helicobacter spp. culture positive | WT/GEM: genotype (background) | Age (mo) | Helicobacter species identified | No. of isolates identified by:
|
|
|---|---|---|---|---|---|---|
| PCR | Sequencing | |||||
| Commercial company 1 (Switzerland) | 2/2 | GEM (B6/G9) | 12+ | Helicobacter sp. strain MIT 96-1001 | 2 | |
| Research institutions | ||||||
| Institution 1 (Australia) | 2/0 | WT (C57BL/6) | 13+ | Negative | NA | NA |
| Institution 2 (Canada) | 2/2 | GEM: Myr AKT | 9 | H. hepaticus | 2 | NA |
| Institution 3 (Canada) | 2/2 | GEM: Lnk−/− (C57BL/6) | 17 | Mixed culture | NA | 1 |
| Institution 4 (Canada) | 1/1 | WT (C57BL/6) | 8+ | H. hepaticus mixed culture | 1 | |
| Institution 5 (England) | 2/2 | GEM: R6/1 (CBA/C57BL/6 F1) | 6+ | H. hepaticus | 2 | |
| Institution 6 (France) | 1/0 | GEM: Apc Flox (C57BL/6) | 7+ | Negative | NA | NA |
| Institution 7 (France) | 2/1 | GEM: PO cre (C57BL/6) | 10+ | Helicobacter sp. strain Hamster B (NCBI accession no. AF072333) | 1 | |
| Institution 7 (France) | 2/0 | GEM: IGF-IR−/+ | 9+ | Negative | NA | NA |
| Institution 8 (Germany) | 2/2 | GEM: D1 cre | 9+ | H. rodentium | 2 | |
| Institution 9 (Japan) | 2/2 | GEM: Lefty1+/− (C57BL/6) | 5+ | Helicobacter sp. strain MIT 01-6451 | 1 | |
| Institution 10 (Japan) | 2/2 | GEM: WGA | 7+ | Helicobacter sp. strain MIT 01-6451 | 2 | |
| Institution 11 (The Netherlands) | 2/2 | GEM: GFAP-cre (FVB) | 8 | H. hepaticus | 2 | |
| Institution 11 (The Netherlands) | 1/0 | GEM: p53-flox/Rb flox | 7+ | Negative | NA | NA |
| Institution 12 (Netherlands) | 1/1 | GEM: rad23B+/− (C57BL/6) | 12+ | H. hepaticus and H. typhlonius | 2 | |
| Institution 13 (The Netherlands) | 2/2 | GEM: cdx1−/− (C57BL/6) | 8+ | H. typhlonius | 2 | |
| Institution 14 (Scotland) | 3/3 | GEM: Alpha V Flox | 9 | H. hepaticus | 3 | |
| Institution 14 (Scotland) | 2/2 | GEM: Intavflox-cre | 20+ | H. mastomyrinus | 2 | |
| Institution 15 (Sweden) | 2/2 | GEM: Cre 151 | 6 | H. typhlonius | 1 | |
| Total | 35/28 | 16 | 9 | |||
WT, wild type; GEM, genetically engineered mice; NA, not applicable.
TABLE 3.
Helicobacter species isolated from commercial and research mouse colonies in the United Statesa
| Source | No. mice tested/no. of Helicobacter spp. culture positive | WT/GEM: genotype (background) | Age or characteristic | Helicobacter species identified | No. of isolates identified by:
|
|
|---|---|---|---|---|---|---|
| PCR | Sequencing | |||||
| Commercial | ||||||
| Company 1 | 4/0 | WT (AJ) | Various | Negative | NA | |
| Company 2 | 2/2 | WT (BALB/c) | Retired breeders | H. hepaticus, H. bilis, and H. rodentium | 6 | |
| Company 2 | 2/2 | WT (AJ) | Retired breeders | H. hepaticus, H. bilis, and H. rodentium | 6 | |
| Company 2 | 2/0 | WT (AJ) | Retired breeders | Negative | NA | |
| Research institutions | ||||||
| Institution 1 (California) | 1/1 | GEM: HIF 1A (129) | 7 mo+ | H. hepaticus | 1 | |
| Institution 2 (California) | 1/1 | GEM: HYPOE (C57BL/6) | 11 mo+ | H. hepaticus | 1 | |
| Institution 3 (Maryland) | 2/0 | GEM: c-myc (C57BL/6/CBA/J) | 11 mo+ | Negative | NA | |
| Institution 4 (Massachusetts) | 2/2 | GEM: Smo−/− (C57BL/6) | 7 mo | H. hepaticus | 2 | |
| Institution 4 (Massachusetts) | 2/2 | GEM: ICOS KO (129) | 15 wk | H. bilis | 2 | |
| Institution 5 (Massachusetts) | 2/2 | GEM: Fibrogen−/−; VWF−/− | 11 mo | H. hepaticus | 2 | |
| Institution 6 (Massachusetts) | 2/2 | WT (C57BL/6) | 7 mo+ | H. hepaticus | 2 | |
| Institution 7 (Massachusetts) | 2/2 | GEM: CARR | 8 mo+ | H. hepaticus | 2 | |
| Institution 8 (Massachusetts) | 2/2 | GEM: TLR2 (C57BL/6) | 9 wk | H. mastomyrinus | 1 | |
| Institution 8 (Massachusetts) | 2/2 | WT (B6/129) | 6 wk | H. mastomyrinus | 1 | |
| Institution 8 (Massachusetts) | 1/1 | GEM: INS-GASGgly (FVB) | H rodentium | 1 | ||
| Institution 8 (Massachusetts) | 1/0 | GEM: TFF2 (C57BL/6) | 7 mo+ | Negative | NA | |
| Institution 8 (Massachusetts) | 1/0 | GEM: INS-GAS (C57BL/6) | Negative | NA | ||
| Institution 9 (New York) | 1/1 | GEM: DYT (C57BL/6) | 9 mo+ | Helicobacter sp. MIT 96-1001 | 1 | |
| Institution 9 (New York) | 1/1 | GEM: DYT (C57BL/6) | 9 mo+ | H. hepaticus and H. typhlonius | 2 | |
| Institution 10 (New York) | 2/2 | WT (FVB) | 11 mo+ | H. hepaticus and H. typhlonius | 4 | |
| Institution 11 (New York) | 1/1 | GEM: E2F1+/− | 7 mo+ | H. hepaticus and H. typhlonius | 2 | |
| Institution 12 (Ohio) | 2/2 | GEM (C57BL/6-AJ) | 8 mo+ | H. hepaticus | 2 | |
| Institution 13 (Ohio) | 2/2 | GEM: Pdx-cre | 12 mo+ | H. hepaticus and H. typhlonius | 4 | |
| Institution 14 (Tennessee) | 2/2 | GEM: p48-cre | 10 mo+ | H. hepaticus and H. typhlonius | 4 | |
| Institution 15 (Texas) | 1/1 | GEM: mdm2 (129) | 7 mo+ | H. hepaticus and H. typhlonius | 2 | |
| Institution16 (Texas) | 1/1 | GEM: 2-MHCGSK-3BS9A | 12 mo+ | H. hepaticus and H. typhlonius | 2 | 1 |
| Total | 44/34 | 47 | 4 | |||
WT, wild type; GEM, genetically engineered mice; NA, not applicable.
No Helicobacter spp. were found in mice from advertised Helicobacter-free production areas from two U.S. vendors. Multiple Helicobacter spp. were found in mice from one vendor's acknowledged Helicobacter-infected production area. The European vendor had mice culture positive for Helicobacter sp. strain MIT 96-1001. Shipments of mice from 15 of 16 U.S. research institutions (94%) had mice that were positive by culture for Helicobacter spp. Shipments from 13 of 15 non-U.S. research institutions (87%) had mice that were positive by culture for Helicobacter spp.
RFLP analysis followed by species-specific PCR.
Of the 62 Helicobacter-positive cultures, 46 were assessed to be pure cultures. Digestion of the 1.2-kb PCR product with AluI and HhaI showed that 21 were H. hepaticus, 2 were H. bilis, and 3 were H. rodentium, according to their RFLP patterns. The identities of these 26 isolates were further confirmed by species-specific PCR. Fifteen of the remaining 20 isolates were submitted for sequence analysis of the 16S RNA gene (Tables 1 and 2). The remaining five isolates were not submitted for DNA sequence analysis because, based on their RFLP patterns and morphological characteristics, they were identical to an isolate obtained from a cage mate in the same shipment which was submitted for sequence analysis.
Analysis of mixed cultures.
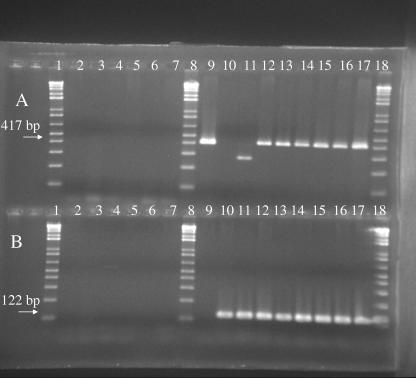
Pure cultures of individual species can be difficult to obtain due to the spreading nature of Helicobacter growth on agar. Mixed RFLP patterns and examination of bacteria by phase microscopy and Gram staining indicated that 16/62 (26%) cultures contained two or more Helicobacter spp. A pure culture of H. hepaticus and H. typhlonius from one set of coinfected mice was obtained only from U.S. institution 16. The remainder of the mixed cultures were assessed by using the species-specific primers listed in Table 1. Figure 2 shows the results of the species-specific PCR for the mixed cultures and for other known murine Helicobacter spp. One shipment from a U.S. commercial vendor had mice that were positive by PCR with primers specific for H. hepaticus, H. bilis, and H. rodentium. Six U.S. research institutions and one European research institution had mice that were positive by PCR with primers specific for H. hepaticus and H. typhlonius. Only H. hepaticus could be identified with species-specific primers and DNA from a mixed culture from mice from institution 4 (Canada). The mixed Helicobacter spp. obtained from institution 3 (Canada) could not be identified with species-specific primers.
FIG. 2.
Results of species-specific PCR for (A) H. hepaticus and (B) H. typhlonius in mixed cultures. Lanes: 1, 1-kb plus ladder; 2, Hamster B (NCBI accession no. AF072333); 3, H. bilisT; 4, H. mastomyrinusT; 5, MIT-1001; 6, H. rodentiumT; 7, novel MIT 01-6451; 8, ladder; 9, H. hepaticusT; 10, H. typhloniusT; 11, H. typhlonius (U.S. institution 16); 12 to 17, mixed cultures from European institution 12 and U.S. institutions 9 to 11 and 13 to 15, respectively; 18, ladder. PCR of these DNA samples with primers specific for H. bilis and H. rodentium gave negative results.
Sequencing of pure cultures.
Of the 20 isolates not identified with the species-specific primers, 14 isolates were analyzed by complete sequencing of the 16S rRNA gene (Tables 2 and 3). Sequence analysis identified H. mastomyrinus (four isolates), H. typhlonius (two isolates), MIT 96-1001 (three isolates), Hamster B (NCBI accession no. AF072333) (one isolate), and novel Helicobacter species strain MIT 01-6451 (three isolates). One DNA sample from a culture from mice from institution 3 (Canada) could not be identified because it contained DNA from more than one organism. Figure 1 shows a phylogenetic tree of the Helicobacter species recovered from the mice. The tree includes H. pylori and H. felis as outgroups. The Helicobacter species recovered from the mice examined in this study are shown in boldface.
Distribution of Helicobacter species.
Overall, this survey found that 2/34 institutions (6%) had mice colonized with at least three Helicobacter spp., 10/34 (29%) had mice colonized with two Helicobacter spp., and 15/34 (47%) had mice colonized with a single Helicobacter species. Institutions in the United States were more likely to have mice colonized with two or more Helicobacter spp., including H. hepaticus with H. rodentium and H. bilis (1/3 commercial institutions), MIT 96-1001 with H. hepaticus and H. typhlonius (institution 9), and H. mastomyrinus with H. rodentium (institution 8). H. hepaticus and H. typhlonius were identified in mice from 7 of 16 research institutions. Outside the United States, one research institution had mice colonized with H. mastomyrinus and H. hepaticus (institution 14) and another had mice colonized with H. hepaticus with H. typhlonius (institution 12).
Among the mice from the 34 institutions surveyed, H. hepaticus, alone or in combination with other Helicobacter spp., was the organism most predominantly found (59%), followed by H. typhlonius (26%) and MIT 96-1001, H. rodentium, and H. mastomyrinus (6% each). One novel Helicobacter species was also cultured from mice from two Asian institutions. Mice in the United States were more likely to be colonized with H. hepaticus. Fifteen of the 18 institutions had mouse colonies infected with H. hepaticus alone or with other species (90%). The prevalence of H. typhlonius in mice from the 18 institutions was also high in the United States (39%). Mice from the 16 non-U.S. institutions were the most likely to be colonized with H. hepticus alone or with other species (37%). However, mice from non-U.S. institutions were also monocolonized with several other Helicobacter spp.
Mice obtained from 11 European institutions were monocolonized with diverse Helicobacter spp., including H. hepaticus, H. typhlonius, MIT 96-1001, H. rodentium, and H. mastomyrinus. Mice from both Asian institutions were colonized with the same novel Helicobacter sp. (strain MIT 01-6451). Shipments from three Canadian institutions had mice that were colonized with H. hepaticus or were cocolonized with more than one Helicobacter species. Mouse colonies in the United States were colonized with six different species (including H. hepaticus, H. typhlonius, MIT 96-1001, H. rodentium, H. mastomyrinus, and H. bilis). H. ganmani was not found in mice from any of the institutions screened. H. bilis was not found in mice from the European institutions.
Histopathology.
No pathology consistent with Helicobacter infection or the age of the mouse was noted in any of the mice. Of those mice whose background strain could be determined, most were of the C57BL/6 background, a strain of mouse known to be resistant to the pathology caused by enterohepatic Helicobacter spp. (16, 31).
DISCUSSION
This study found an 88% prevalence of Helicobacter spp. in mouse colonies from commercial and research institutions around the world. The finding that H. hepaticus is the predominant species in the United States confirms the findings of Shames et al. (25), who also used bacterial culture and PCR to identify H. hepaticus colonization in murine colonies. In a 1996 survey of major commercial vendors in the United States, 28 different strains from a total of 160 mice from four major U.S. vendors were surveyed by bacterial culture. All mice from 2 outbred strains from one vendor were colonized with H. hepaticus, and 9 of 13 inbred mouse strains from another vendor were also infected with H. hepaticus (25). In 1998, Riley et al. (23) used fecal PCR and RFLP analysis to identify H. hepaticus, H. muridarum, and H. bilis, the most common rodent helicobacters known at that time. They reported that of 508 mice, 10.4% were colonized with H. hepaticus, 17.1% were colonized with H. bilis, and <1% had H. muridarum colonization. Our laboratory has reported that one vendor had mice colonized with H. bilis, and a U.S. research institution that also had H. bilis in its colony obtained mice from that vendor (8). Thus, the distribution of Helicobacter species in the research population is, not surprisingly, related to the source of the mice.
In Japan, Goto et al. (13) surveyed 820 mice from 47 colonies in universities, breeding companies, pharmaceutical companies, and national research institutions obtained in 1997 and 1998. Using fecal reverse transcription-PCR, they found that the following strains were present in the colonies: H. hepaticus (25.5% of the colonies); H. rodentium (38.3%); H. hepaticus and H. rodentium (5.7% each); and H. typhlonius-like, H. bilis, and H. westmeadii-like (2.1% each). H. westmeadii was later identified as H. cinaedi (30). A novel Helicobacter was found in both of the two institutions in Japan that we surveyed. Thus, other Helicobacter species are present in Japanese institutions, in addition to those that Goto et al. (13) reported in their paper.
Using fecal PCR-denaturing gradient gel electrophoresis, Grehan et al. surveyed eight mice from three suppliers in Australia (14). They reported that four of eight mice were colonized with H. ganmani, two of eight had H. bilis, three of eight had H. hepaticus, and one of eight had two unique isolates, in addition to H. ganmani. One mouse was negative for Helicobacter DNA. The research institution that we surveyed in Australia had mice that were negative by culture for all helicobacters. We did not use anaerobic conditions for culture and did not isolate H. ganmani from any mice. While it has been reported that H. ganmani grows only anaerobically (24), we have found that the strain obtained from CCUG grows both anaerobically and microaerobically in our laboratory. This may be due to our use of culture in jars evacuated to 25 in. of mercury and then filled with an 80:10:10 mixture of nitrogen-carbon dioxide-hydrogen rather than anaerobic jars with gas-generating packs, as described by Robertson et al. (24). Also, the Helicobacter species present in these mice and the number of mice infected may be underestimated since PCR of DNA directly isolated from fecal samples is more sensitive than culture (9, 20, 25). However, the use of culture techniques allowed the characterization of the Helicobacter spp. isolated and the identification of novel Helicobacter spp.
Nilsson et al. (20) also used PCR-denaturing gradient gel electrophoresis but added pyrosequencing to detect Helicobacter spp. in 15- to 26-week-old mice from four different animal facilities in Sweden. One animal facility had mice negative for Helicobacter. The other vivaria had mice infected with multiple Helicobacter spp., including H. bilis, H. hepaticus, H. typhlonius, H. ganmani, and H. rodentium. The one research institution that we surveyed in Sweden had mice infected with H. typhlonius.
In Germany, Bohr et al. (1) reported on the prevalence and spread of enterohepatic Helicobacter spp. in mice from a specific-pathogen-free animal facility. Eighty-five percent of the mice were colonized, some with multiple Helicobacter species. Five different species were identified by PCR: H. ganmani, H. hepaticus, H. typhlonius, and isolates resembling Hamster B and MIT-5357. The one German institution in this study had mice colonized with H. rodentium.
Other countries in this study that have not previously published information on the prevalence of helicobacters in murine research colonies included Switzerland, England, The Netherlands, Scotland, France, and Canada. All were found to have colonies infected with Helicobacter spp.
The lack of pathology consistent with Helicobacter infection in this study was expected, since most mice in this study were of a C57BL/6 background, and such mice are known to be resistant to enteric and hepatic diseases (16, 31). Nilsson et al. (20) reported that the only pathology seen in their study of 42 young adult mice was hepatitis associated with H. hepaticus-monoinfected C57BL/6 ApoE−/− mice and BALB/cA mice infected with multiple helicobacters (H. hepaticus, H. bilis, H. rodentium, and H. typhlonius). Goto et al. (13) surveyed 820 mice 9 weeks of age or older and found that the only pathology observed was in 5/174 (2.8%) mice colonized with H. rodentium and H. rodentium-like strains. They reported multiple white foci on the livers and suggested that H. rodentium may be associated with hepatitis.
In summary, more than 10 years have elapsed since the original isolation of H. hepaticus, and even though it may confound the results of experiments with infected mice, this Helicobacter is still prevalent in mouse colonies throughout the world. In addition, other Helicobacter spp. known to cause disease in mice are also widespread (8-11, 13-15). Several vendors do maintain mice in Helicobacter-free breeding areas. Requesting animals from these areas, importing them into Helicobacter-free rooms, and implementing strict animal husbandry procedures are ways to control infection. For existing lines of mice, rederivation and housing under Helicobacter-free conditions remain the most reliable options (32).
Acknowledgments
We thank Vivian Ng, Kristen Clapp, Elizabeth Groff, Jennifer Cline, and Noel Radwanski for necropsy and collection of tissue from the mice and Kathleen Cormier and Erin Stefonovich for histology.
This research was supported in part by grants from the National Institutes of Health: P30-ES02109 (to J.G.F.), R01 CA67529 (to J.G.F.), and DE 016937 (to F.E.D.).
Footnotes
Published ahead of print on 16 May 2007.
REFERENCES
- 1.Bohr, U. R., M. Selgrad, C. Ochmann, S. Backert, W. Konig, A. Fenske, T. Wex, and P. Malfertheiner. 2006. Prevalence and spread of enterohepatic Helicobacter species in mice reared in a specific-pathogen-free animal facility. J. Clin. Microbiol. 44:738-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erdman, S. E., T. Poutahidis, M. Tomczak, A. B. Rogers, K. Cormier, B. Plank, B. H. Horwitz, and J. G. Fox. 2003. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am. J. Pathol. 162:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng, S., K. Ku, E. Hodzic, E. Lorenzana, K. Freet, and S. W. Barthold. 2005. Differential detection of five mouse-infecting Helicobacter species by multiplex PCR. Clin. Diagn. Lab. Immunol. 12:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox, J. G., F. E. Dewhirst, Z. Shen, Y. Feng, N. S. Taylor, B. J. Paster, R. L. Ericson, C. N. Lau, P. Correa, J. C. Araya, and I. Roa. 1998. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114:755-763. [DOI] [PubMed] [Google Scholar]
- 5.Fox, J. G., F. E. Dewhirst, J. G. Tully, B. J. Paster, L. Yan, N. S. Taylor, M. J. Collins, Jr., P. L. Gorelick, and J. M. Ward. 1994. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 32:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox, J. G., P. L. Gorelick, M. C. Kullberg, Z. Ge, F. E. Dewhirst, and J. M. Ward. 1999. A novel urease-negative Helicobacter species associated with colitis and typhlitis in interleukin-10-deficient mice. Infect. Immun. 67:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox, J. G., X. Li, L. Yan, R. J. Cahill, R. Hurley, R. Lewis, and J. C. Murphy. 1996. Chronic proliferative hepatitis in A/JCr mice associated with persistent Helicobacter hepaticus infection: a model of helicobacter-induced carcinogenesis. Infect. Immun. 64:1548-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, J. G., A. B. Rogers, M. T. Whary, N. S. Taylor, S. Xu, Y. Feng, and S. Keys. 2004. Helicobacter bilis-associated hepatitis in outbred mice. Comp. Med. 54:571-577. [PubMed] [Google Scholar]
- 9.Fox, J. G., L. Yan, B. Shames, J. Campbell, J. C. Murphy, and X. Li. 1996. Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect. Immun. 64:3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox, J. G., L. L. Yan, F. E. Dewhirst, B. J. Paster, B. Shames, J. C. Murphy, A. Hayward, J. C. Belcher, and E. N. Mendes. 1995. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J. Clin. Microbiol. 33:445-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin, C. L., P. L. Gorelick, L. K. Riley, F. E. Dewhirst, R. S. Livingston, J. M. Ward, C. S. Beckwith, and J. G. Fox. 2001. Helicobacter typhlonius sp. nov., a novel murine urease-negative Helicobacter species. J. Clin. Microbiol. 39:3920-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin, C. L., L. K. Riley, R. S. Livingston, C. S. Beckwith, C. L. Besch-Williford, and R. R. Hook, Jr. 1998. Enterohepatic lesions in SCID mice infected with Helicobacter bilis. Lab. Anim. Sci. 48:334-339. [PubMed] [Google Scholar]
- 13.Goto, K., H. Ohashi, A. Takakura, and T. Itoh. 2000. Current status of Helicobacter contamination of laboratory mice, rats, gerbils, and house musk shrews in Japan. Curr. Microbiol. 41:161-166. [DOI] [PubMed] [Google Scholar]
- 14.Grehan, M., G. Tamotia, B. Robertson, and H. Mitchell. 2002. Detection of Helicobacter colonization of the murine lower bowel by genus-specific PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 68:5164-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hailey, J. R., J. K. Haseman, J. R. Bucher, A. E. Radovsky, D. E. Malarkey, R. T. Miller, A. Nyska, and R. R. Maronpot. 1998. Impact of Helicobacter hepaticus infection in B6C3F1 mice from twelve National Toxicology Program two-year carcinogenesis studies. Toxicol. Pathol. 26:602-611. [DOI] [PubMed] [Google Scholar]
- 16.Ihrig, M., M. D. Schrenzel, and J. G. Fox. 1999. Differential susceptibility to hepatic inflammation and proliferation in AXB recombinant inbred mice chronically infected with Helicobacter hepaticus. Am. J. Pathol 155:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maggio-Price, L., D. Shows, K. Waggie, A. Burich, W. Zeng, S. Escobar, P. Morrissey, and J. L. Viney. 2002. Helicobacter bilis infection accelerates and H. hepaticus infection delays the development of colitis in multiple drug resistance-deficient (mdr1a−/−) mice. Am. J. Pathol. 160:739-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurer, K. J., M. M. Ihrig, A. B. Rogers, V. Ng, G. Bouchard, M. R. Leonard, M. C. Carey, and J. G. Fox. 2005. Identification of cholelithogenic enterohepatic Helicobacter species and their role in murine cholesterol gallstone formation. Gastroenterology 128:1023-1033. [DOI] [PubMed] [Google Scholar]
- 19.Myles, M. H., R. S. Livingston, and C. L. Franklin. 2004. Pathogenicity of Helicobacter rodentium in A/JCr and SCID mice. Comp. Med. 54:549-557. [PubMed] [Google Scholar]
- 20.Nilsson, H. O., I. S. Ouis, U. Stenram, A. Ljungh, A. P. Moran, T. Wadstrom, and W. A. Al-Soud. 2004. High prevalence of Helicobacter species detected in laboratory mouse strains by multiplex PCR-denaturing gradient gel electrophoresis and pyrosequencing. J. Clin. Microbiol. 42:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paster, B. J., and F. Dewhirst. 1988. Phylogeny of Campylobacter, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 22.Queiroz, D. M., C. Contigli, R. S. Coimbra, A. M. Nogueira, E. N. Mendes, G. A. Rocha, and S. B. Moura. 1992. Spiral bacterium associated with gastric, ileal and caecal mucosa of mice. Lab. Anim. 26:288-294. [DOI] [PubMed] [Google Scholar]
- 23.Riley, L. K., C. L. Franklin, R. R. Hook, Jr., and C. Besch-Williford. 1996. Identification of murine helicobacters by PCR and restriction enzyme analyses. J. Clin. Microbiol. 34:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson, B. R., J. L. O'Rourke, P. Vandamme, S. L. On, and A. Lee. 2001. Helicobacter ganmani sp. nov., a urease-negative anaerobe isolated from the intestines of laboratory mice. Int. J. Syst. Evol. Microbiol. 51:1881-1889. [DOI] [PubMed] [Google Scholar]
- 25.Shames, B., J. G. Fox, F. Dewhirst, L. Yan, Z. Shen, and N. S. Taylor. 1995. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J. Clin. Microbiol. 33:2968-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen, Z., Y. Feng, and J. G. Fox. 2000. Identification of enterohepatic Helicobacter species by restriction fragment-length polymorphism analysis of the 16S rRNA gene. Helicobacter 5:121-128. [DOI] [PubMed] [Google Scholar]
- 27.Shen, Z., J. G. Fox, F. E. Dewhirst, B. J. Paster, C. J. Foltz, L. Yan, B. Shames, and L. Perry. 1997. Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int. J. Syst. Bacteriol. 47:627-634. [DOI] [PubMed] [Google Scholar]
- 28.Shomer, N. H., C. A. Dangler, R. P. Marini, and J. G. Fox. 1998. Helicobacter bilis/Helicobacter rodentium co-infection associated with diarrhea in a colony of SCID mice. Lab. Anim. Sci. 48:455-459. [PubMed] [Google Scholar]
- 29.Shomer, N. H., C. A. Dangler, M. D. Schrenzel, M. T. Whary, S. Xu, Y. Feng, B. J. Paster, F. E. Dewhirst, and J. G. Fox. 2001. Cholangiohepatitis and inflammatory bowel disease induced by a novel urease-negative Helicobacter species in A/J and Tac:ICR:HascidfRF mice. Exp. Biol. Med. (Maywood) 226:420-428. [DOI] [PubMed] [Google Scholar]
- 30.Vandamme, P., C. S. Harrington, K. Jalava, and S. L. On. 2000. Misidentifying helicobacters: the Helicobacter cinaedi example. J. Clin. Microbiol. 38:2261-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward, J. M., J. G. Fox, M. R. Anver, D. C. Haines, C. V. George, M. J. Collins, Jr., P. L. Gorelick, K. Nagashima, M. A. Gonda, R. V. Gilden, et al. 1994. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J. Natl. Cancer Inst. 86:1222-1227. [DOI] [PubMed] [Google Scholar]
- 32.Whary, M. T., and J. G. Fox. 2004. Natural and experimental Helicobacter infections. Comp. Med. 54:128-158. [PubMed] [Google Scholar]