Abstract
Shiga toxin 2 (Stx2) from Shiga toxin-producing Escherichia coli (STEC) was subtyped by a method involving partial sequencing of the stxAB2 operon. Of 255 strains from the Danish STEC cohort, all 20 cases of hemolytic-uremic syndrome were associated with subtype Stx2 (11 cases), subtype Stx2c (1 case), or the two combined (8 cases).
Infections caused by Shiga toxin-producing Escherichia coli (STEC), alternatively known as verocytotoxin-producing E. coli, account for the most severe symptoms among those caused by diarrheagenic E. coli, sometimes leading to bloody diarrhea (BD) or hemolytic-uremic syndrome (HUS). STEC is defined by the ability to produce Shiga toxins (Stx or verocytotoxin) 1 and 2, which are two sequentially and antigenetically distinct toxin groups transcribed from the stxAB1 and stxAB2 operons, respectively. Besides the stx gene(s), STEC strains often carry the gene encoding the adherence factor intimin (eae), which is an outer membrane protein (14). It has previously been shown that there exists an increased risk for developing HUS when both stx2 and eae are present in the infecting strain (4, 7, 11). Additionally, a number of studies have documented that subtypes stx2 and stx2c are more often associated with HUS than the other stx2 subtypes (5, 6, 8, 15), but stx2d (9, 13, 16)- and stx2e (11)-containing strains have also been isolated from humans with HUS. PCR-restriction fragment length polymorphism has often been the preferred tool for subtyping stx2 variants (2, 3, 5, 6, 8, 10, 17-19, 22). These methods are, however, vulnerable to single-nucleotide changes and are difficult to interpret if the strain contains more than one subtype or if the fragments generated are small or of similar sizes. For a new and improved subtyping method for the Stx2 toxin, we applied partial sequencing of the most variable part of the stxAB2 operon. By applying this method to the stx2-positive isolates from the Danish National STEC cohort from 1997 to 2003, we hoped to clarify the epidemiology of human stx2 subtypes and to correlate specific subtypes with clinical manifestations of BD and HUS.
(The results of this study were presented in part at the 14th European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], Prague, Czech Republic, May 2004 [poster no. P972], and at the 2nd Met-Vet-Net Meeting, Qawra, Malta, May 2006 [poster no. HMI&ME23]).
All clinical STEC strains included in this study were characterized at the Unit of Gastrointestinal Infections, Statens Serum Institut (SSI), Denmark. About 90% of the strains were isolated at the SSI from individuals suffering from diarrhea, using DNA dot blot hybridization with broad-range polynucleotide DNA probes derived from plasmids pNTP705 for stx1 (23), pDEP28 for stx2 (21), and pCVD434 for eae (12). The remaining STEC strains originated either from patients with diarrhea who had stool cultures analyzed for the most common STEC O groups (at regional clinical laboratories) or from asymptomatic carriers analyzed because of close contact to STEC-infected individuals. The data set consists of 44 different O groups and is presumably not biased toward certain O groups; there were no general outbreaks within the study period. Retrieval of patient information concerning the illness was performed throughout the study period by patient interviews as described previously (7). STEC strains were grown on agar plates under standard conditions and prepared for PCR in the following way. One colony was transferred to 100 μl 10% Chelex 100 (Bio-Rad, Hercules) in 10 mM Tris-HCl, 1 mM EDTA, pH 8, boiled for 5 min, and centrifuged briefly. The supernatant was used directly for PCR. Each template was subjected to PCR with the following primers: F4, 5′-GGCACTGTCTGAAACTGCTCCTGT (matched all subtypes except stx2f); R1, 5′-ATTAAACTGCACTTCAGCAAATCC (matched all subtypes except stx2e and stx2f); F4-f, 5′-CGCTGTCTGAGGCATCTCCGCT (matched stx2f); and R1-e/f, 5′-TAAACTTCACCTGGGCAAAGCC (matched stx2e and stx2f). PCRs were carried out in a total reaction volume of 25 μl containing 1× PCR buffer [50 mM Tris-HCl, 10 mM KCl, 5 mM (NH4)2SO4, pH 8.3], 2.6 mM MgCl2, 260 μM of each deoxynucleoside triphosphate, 1.25 U Taq polymerase (FastStart, Roche Diagnostics GmbH, Mannheim, Germany), 0.05 μM of each of the four primers, and 5 μl template. The thermocycler conditions were as follows: 95°C for 2 min, followed by 35 cycles of 94°C for 50 s, 58°C for 40 s, and 72°C for 50 s, and finally 72°C for 3 min. The completed PCR products were purified by a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and sequenced at MWG-BIOTECH AG, Ebersberg, Germany. If the first sequencing attempt using primer F4 or R1 failed, amplicons were sequenced with primer F4-f or R1-e/f. Sequencing with forward and reverse PCR primers generated a 491-bp internal high-quality sequence on the approximately 620-bp PCR product. The nucleotide sequence was translated into 159 amino acids, covering 95 residues of the C-terminal part of subunit A and 64 residues of the N-terminal part of subunit B (Fig. 1). By analyzing the positions of double peaks in the sequence chromatogram, different subtypes present in the same strain could be identified.
FIG. 1.
A total of 255 STEC strains were subjected to partial sequencing of the stxAB2 operon, and the coding regions were translated. The figure shows an alignment (ClustalW) of the resulting 159 amino acids (95 amino acids of the C-terminal part of subunit A [positions 225 to 319] and 64 amino acids of the N-terminal part of subunit B [positions 1 to 64]). The toxin variants are named by subtype, O group, and strain name. With respect to the 159 residues, the sequences for the following toxin variants are registered under the respective GenBank accession numbers: Stx2-O48-94C, Z37725; Stx2-O157-EDL933, X07865; Stx2c-O157-C394-03, DQ235774; Stx2c-O157-FLY16, AB015057; Stx2d-O73-C165-02, DQ059012; Stx2d-O8-C466-01B, DQ235775; Stx2d3-O157-7279, X61283; Stx2e-O101-E-D43, X81417; Stx2e-ONT-26725-97, AJ567998; Stx2g-Out-S-8, AB048227; Stx2c-O111-PH, L11078; Stx2c-O118-EH250, AF043627.
All observed chromatograms with two different toxins corresponded to the superimposition of two known sequences; if previously unknown sequences are present in a strain harboring two different toxin variants, further confirmatory analyses may be required. Construction of alignments and phylogenetic trees was performed by DNAStar software (DNAStar Inc., Madison, WI).
This partial sequencing of the stxAB2 operon offers an easy and specific subtyping method for the Stx2 toxins. Based on the 159 amino acids, 12 different variants were identified in the Danish cohort (Fig. 1 and 2), named by subtype (nomenclature according to reference 20), O group, and strain name. Nine of these were already present in GenBank, and among these, two were for the first time found in humans (Stx2c-O157-FLY16 and Stx2g-Out-S-8).
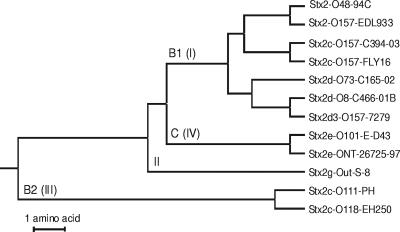
FIG. 2.
Phylogenetic tree (unweighted-pair group method using average linkages) based on the alignment shown in Fig. 1 of the 159 amino acids partially covering the stxAB2 operon of the 12 toxin variants from this study. Roman numbers (I, II, III, and IV) and capital letters (B1, B2, and C) refer to previous groupings introduced in references 1 and 2, respectively. The same clustering was observed when the entire sequence of 408 joined A- and B-subunit stx2 holotoxin sequences in GenBank was analyzed.
From 1 January 1997 to 28 October 2003, the Danish National STEC cohort contained 272 strains positive for the stx2 gene. Of these, 17 could not be recovered from the archives or did not grow and the resulting 255 strains (of which 39 contained two different toxins) were subtyped by the present method (Table 1). Fifty-two patients (20%) reported being infected during foreign travel. Clinical data concerning HUS were available for 241 patients, and among these patients, 20 (8.3%) developed HUS. Clinical data concerning BD were available for 199 patients, of which 90 (45%) had BD. Eighteen of the 20 HUS cases, but only 40% of all cases, were from children 7 years of age or below. All 20 HUS cases were associated with subtype Stx2 and/or Stx2c, i.e., 19 HUS cases were associated with subtype Stx2 (Stx2-O157-EDL933 or Stx2-O48-94C) either alone (11 cases) or in combination with variant Stx2c-O157-FLY16 (8 cases). One strain associated with HUS was found to contain variant Stx2c-O157-FLY16 alone. Subtypes Stx2 (Stx2-O157-EDL933 and Stx2-O48-94C) and Stx2c (Stx2c-O157-FLY16) accounted for the majority of cases (83%) associated with BD. O157 was by far the most common O group, accounting for 109 (42.7%) of 255 Stx2-positive strains, and was associated with 12 HUS cases. All O157 strains from patients with HUS were stx1 negative and eae positive. To calculate adjusted odds ratios (ORs), multivariate logistic regression analysis was performed using SAS version 9.1 (Cary, NC). The crude and adjusted ORs for the association between HUS and subtypes Stx2 and Stx2c and age are shown in Table 2. eae was strongly associated with HUS but could not be included in the model, since this gene was identified in isolates from all 20 HUS cases. As expected (7), there was no association between HUS and individual O groups, including O group O157 (crude OR, 2.18; 95% confidence interval [CI], 0.86 to 5.56), in this data set of exclusively stx2-positive isolates. Subtype Stx2 was strongly associated with HUS in both the crude and the adjusted analysis, while subtype Stx2c was associated only in the adjusted analysis. Comparable results were found when the analysis was restricted to the 63% eae-positive strains (not shown). Our results do not allow conclusions about a causal mechanism but strongly suggest that the stx2 and the stx2c genes, as opposed to other stx2 genes, are associated with development of HUS. Because stx2c (i.e., toxin type Stx2c-O157-FLY16) was present at the same time as subtype stx2 in eight out of nine isolates from HUS patients, it is not clear from our data if stx2c can contribute to the development of HUS on its own or merely assists subtype stx2. It may also be speculated that the one strain associated with HUS containing stx2c alone has lost a bacteriophage carrying an stx2 gene.
TABLE 1.
Stx2 subtype distribution among 255 strains from the Danish STEC cohort, listed with the clinical outcomes for HUS and BD
| Toxin variant(s)a | Stx2 subtype(s) | No. (%) of strains
|
No. (%) of patients with indicated disease
|
||
|---|---|---|---|---|---|
| Total | Positive for eae | HUSb | BDc | ||
| Stx2-O157-EDL933 | 2 | 38 (14.9) | 38 (23.8) | 4 (20.0) | 19 (21.1) |
| Stx2-O48-94C | 2 | 32 (12.5) | 27 (16.9) | 7 (35) | 10 (11.1) |
| Stx2c-O157-FLY16 | 2c | 54 (21.2) | 49 (30.6) | 1 (5) | 24 (26.7) |
| Stx2c-O157-C394-03 | 2c | 16 (6.3) | 16 (10) | 6 (6.6) | |
| Stx2d-O8-C466-01B | 2d | 2 (0.8) | 1 (0.6) | 1 (1.1) | |
| Stx2d-O73-C165-02 | 2d | 1 (0.4) | 1 (1.1) | ||
| Stx2d3-O157-7279 | 2d | 4 (1.6) | |||
| Stx2c-O118-EH250 | 2c | 29 (11.4) | 2 (2.3) | ||
| Stx2c-O111-PH | 2c | 34 (13.3) | 5 (5.5) | ||
| Stx2e-O101-E-D43 | 2e | 1 (0.4) | |||
| Stx2e-ONT-26725-97 | 2e | 3 (1.2) | 1 (0.6) | ||
| Stx2g-Out-S-8 | 2g | 2 (0.8) | 1 (0.6) | ||
| Stx2-O157-EDL933 + Stx2c-O157-FLY16 | 2 + 2c | 26 (10.2) | 26 (16.3) | 7 (35) | 20 (22.2) |
| Stx2-O48-94C + Stx2c-O157-FLY16 | 2 + 2c | 1 (0.4) | 1 (0.6) | 1 (5) | |
| Stx2-O157-EDL933 + Stx2c-O118-EH250 | 2 + 2c | 1 (0.4) | 1 (1.1) | ||
| Stx2-O48-94C + Stx2d3-O157-7279 | 2 + 2d | 1 (0.4) | |||
| Stx2c-O111-PH + Stx2c-O118-EH250 | 2c + 2c | 7 (2.7) | |||
| Stx2-O48-94C + Stx2d-O8-466-01B | 2 + 2d | 2 (0.8) | |||
| Stx2c-O157-FLY16 + Stx2d-O8-C466-01B | 2c + 2d | 1 (0.4) | 1 (1.1) | ||
| Total | 255 | 160 | 20 | 90 | |
The first 12 rows comprise the 216 strains that contained one toxin variant, and the remaining rows comprise the 39 strains that contained two different toxin variants. ONT, O nontypeable; Out, O untypeable.
Information concerning HUS was available for 241 of the 255 patient strains; of these patients, 20 had HUS.
Information concerning BD was available for 199 of the 255 patient strains; of these patients, 90 had BD.
TABLE 2.
Crude and adjusted odds ratios for development of HUS
| Determinant and value | No. of patients
|
OR (95% CI)a for indicated analysis
|
||
|---|---|---|---|---|
| Total (n = 241) | With HUS (n = 20) | Crude | Adjusted | |
| Subtype Stx2b | ||||
| No | 145 | 1 | 1c | 1 |
| Yes | 96 | 19 | 35.5 (4.67-270) | 43.1 (4.9-380) |
| Subtype Stx2cd | ||||
| No | 147 | 11 | 1 | 1 |
| Yes | 94 | 9 | 0.34e (0.15-1.0) | 3.29 (1.04-10.5) |
| Age (yrs) | ||||
| >7 | 144 | 2 | 1 | 1 |
| ≤7 | 97 | 18 | 16.2 (3.66-71.5) | 10.4 (2.71-50.0) |
| eae | ||||
| No | 88 | 0 | 1 | 1 |
| Yes | 153 | 20 | NDf | ND |
ND, not defined.
Presence of Stx2-O157-EDL933 or Stx2-O48-94C.
Reference value.
Presence of Stx2c-O157-FLY16, Stx2c-O157-C394-93, Stx2c-O118-EH250, or Stx2c-O111-PH.
The OR was 1.31 (95% CI, 0.52 to 3.29) when only subtypes Stx2c-O157-FLY16 and Stx2c-O157-C394-93 were included.
P < 0.001 (Fisher exact test).
Subtype Stx2c appeared to consist of two separate groups of toxins. The amino acid sequences of toxin variants Stx2c-O118-EH250 and Stx2c-O111-PH were divergent from those of the two other Stx2c variants. Furthermore, they were found only in eae-negative strains and never in strains from HUS cases. We therefore would suggest the introduction of a distinct nomenclature for these subtypes, which we propose to designate subtype stx2b.
Nucleotide sequence accession numbers.
Three sequences, Stx2d-O73-C165-02, Stx2c-O157-C394-03, and Stx2d-O8-C466-01B, were new. They were subjected to full gene sequencing and deposited in GenBank under accession numbers DQ059012, DQ235774, and DQ235775, respectively.
Acknowledgments
We thank Joan Nevermann Jensen for technical assistance and Karen A. Krogfelt for critical revision of the manuscript.
Footnotes
Published ahead of print on 19 April 2007.
REFERENCES
- 1.Asakura, H., S. Makino, H. Kobori, M. Watarai, T. Shirahata, T. Ikeda, and K. Takeshi. 2001. Phylogenetic diversity and similarity of active sites of Shiga toxin (stx) in Shiga toxin-producing Escherichia coli (STEC) isolates from humans and animals. Epidemiol. Infect. 127:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. [DOI] [PubMed] [Google Scholar]
- 3.Bertin, Y., K. Boukhors, N. Pradel, V. Livrelli, and C. Martin. 2001. Stx2 subtyping of Shiga toxin-producing Escherichia coli isolated from cattle in France: detection of a new Stx2 subtype and correlation with additional virulence factors. J. Clin. Microbiol. 39:3060-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caprioli, A., I. Luzzi, A. Gianviti, H. Russmann, and H. Karch. 1995. Pheno-genotyping of verotoxin 2 (VT2)-producing Escherichia coli causing haemorrhagic colitis and haemolytic uraemic syndrome by direct analysis of patients’ stools. J. Med. Microbiol. 43:348-353. [DOI] [PubMed] [Google Scholar]
- 6.Eklund, M., K. Leino, and A. Siitonen. 2002. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J. Clin. Microbiol. 40:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ethelberg, S., K. E. Olsen, F. Scheutz, C. Jensen, P. Schiellerup, J. Engberg, A. M. Petersen, B. Olesen, P. Gerner-Smidt, and K. Molbak. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 10:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 9.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 10.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins, C., G. A. Willshaw, J. Evans, T. Cheasty, H. Chart, D. J. Shaw, G. Dougan, G. Frankel, and H. R. Smith. 2003. Subtyping of virulence genes in verocytotoxin-producing Escherichia coli (VTEC) other than serogroup O157 associated with disease in the United Kingdom. J. Med. Microbiol. 52:941-947. [DOI] [PubMed] [Google Scholar]
- 12.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindgren, S. W., A. R. Melton, and A. D. O'Brien. 1993. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect. Immun. 61:3832-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piérard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pradel, N., K. Boukhors, Y. Bertin, C. Forestier, C. Martin, and V. Livrelli. 2001. Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients, cattle, and food samples in central France. Appl. Environ. Microbiol. 67:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roldgaard, B. B., F. Scheutz, J. Boel, S. Aabo, A. C. Schultz, T. Cheasty, E. M. Nielsen, K. E. Olsen, and B. B. Christensen. 2004. VTEC O157 subtypes associated with the most severe clinical symptoms in humans constitute a minor part of VTEC O157 isolates from Danish cattle. Int. J. Med. Microbiol. 294:255-259. [DOI] [PubMed] [Google Scholar]
- 18.Russmann, H., E. Kothe, H. Schmidt, S. Franke, D. Harmsen, A. Caprioli, and H. Karch. 1995. Genotyping of Shiga-like toxin genes in non-O157 Escherichia coli strains associated with haemolytic uraemic syndrome. J. Med. Microbiol. 42:404-410. [DOI] [PubMed] [Google Scholar]
- 19.Russmann, H., H. Schmidt, J. Heesemann, A. Caprioli, and H. Karch. 1994. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J. Med. Microbiol. 40:338-343. [DOI] [PubMed] [Google Scholar]
- 20.Scheutz, F., and N. A. Strockbine. 2005. Escherichia, p. 607-624. In G. M. Garrity, D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 21.Thomas, A., H. R. Smith, G. A. Willshaw, and B. Rowe. 1991. Nonradioactively labelled polynucleotide and oligonucleotide DNA probes, for selectively detecting Escherichia coli strains producing Vero cytotoxins VT1, VT2 and VT2 variant. Mol. Cell. Probes 5:129-135. [DOI] [PubMed] [Google Scholar]
- 22.Tyler, S. D., W. M. Johnson, H. Lior, G. Wang, and K. R. Rozee. 1991. Identification of verotoxin type 2 variant B subunit genes in Escherichia coli by the polymerase chain reaction and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 29:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willshaw, G. A., H. R. Smith, S. M. Scotland, and B. Rowe. 1985. Cloning of genes determining the production of vero cytotoxin by Escherichia coli. J. Gen. Microbiol. 131:3047-3053. [DOI] [PubMed] [Google Scholar]