Abstract
Viral load testing for cytomegalovirus (CMV) has become the standard for the diagnosis of infection and monitoring of therapy at many transplant centers. However, no viral load test has been approved by the FDA. Therefore, many laboratories rely on laboratory-developed assays. This study evaluated the performance characteristics of two real-time PCR tests developed using the artus CMV analyte-specific reagents (ASRs). One version is distributed by Abbott Molecular and the other by QIAGEN. For plasma specimens, the Abbott test had a limit of detection of 2.3 log10 copies/ml and a linear range up to at least 6.0 log10 copies/ml. Comparison of plasma viral loads using the Abbott test and the Roche Amplicor Monitor test showed a mean difference of −0.012 log10 copies/ml. In addition, the Abbott test viral loads correlated with the Digene Hybrid Capture assay ratios. Viral loads obtained from plasma specimens tested by the Abbott and QIAGEN tests were in very close agreement (mean difference, 0.144 log10 copies/ml). When the QIAGEN test was evaluated with the QIAGEN, MagNA Pure, and easyMAG extraction methods, the viral loads for all three methods were within 0.370 log10 copies/ml. Thus, there is good agreement between viral loads obtained by the different tests using the same extraction method or by the same test using different extraction methods. The availability of real-time PCR ASRs provides additional reagents that can be used for CMV viral load testing.
Cytomegalovirus (CMV) remains an important pathogen for immunocompromised individuals. In many centers, viral load assays are the cornerstone for the diagnosis and monitoring of patients at risk for CMV disease. The clinical utility of CMV viral load testing is supported by a considerable body of literature (7, 9, 11, 12, 14, 15). These assays can be used to determine when to initiate preemptive therapy, to monitor the response to therapy, to determine the duration of therapy, and to assess patients at risk of developing relapsing infection (6, 9, 10, 12). For example, a study of liver transplant recipients showed that a viral load of 2,000 to 5,000 copies/ml in plasma, determined by the Amplicor Monitor CMV test (Roche Diagnostics, Indianapolis, IN), is predictive of the development of active CMV disease (10). Another study showed that when one is monitoring active disease for response to therapy, failure to clear CMV DNA from plasma after a course of ganciclovir therapy increases the risk of relapsing CMV infection (14). Failure of the plasma viral load to decline after several weeks of therapy has been associated with the development of ganciclovir resistance (4). In addition, the half-life of CMV DNA in plasma is longer for patients who relapse than for those who do not (11). These studies all support the use of close monitoring of viral loads to identify those at high risk of relapse, thus allowing the intensification of therapy early in the treatment course.
In spite of the widespread use of CMV viral load measurements, no assays for the quantification of CMV nucleic acid have been approved by the U.S. Food and Drug Administration (FDA). Laboratories that perform CMV viral load testing either modify the Hybrid Capture CMV DNA test (Digene, Gaithersburg, MD) to allow for quantification, use the Amplicor CMV Monitor assay (Roche Diagnostics, Indianapolis, IN), or rely on laboratory-developed PCR tests. Based on the recent proficiency testing results (2006 ID A survey) of the College of American Pathologists, the majority of participating sites rely on laboratory-developed tests. Though these laboratory-developed tests can work very well for any given center, test performance may differ between different laboratories, and without an international standard, viral loads obtained using different tests are difficult to compare and interpret. This lack of agreement poses a significant problem for patients who may be monitored at more than one medical center.
Recently, several analyte-specific reagents (ASRs) for CMV DNA have become available. Currently, limited data are available regarding the performance of laboratory tests developed using these CMV ASRs. This study evaluated the performance characteristics of two tests developed in our laboratory using the artus CMV ASR (Hamburg, Germany), one version of which is distributed by Abbott Molecular (Des Plaines, IL) and another version by QIAGEN (Valencia, CA). Using clinical specimens, viral loads obtained by laboratory-developed tests using the Abbott CMV ASRs (referred to below as the “Abbott test”) or the QIAGEN CMV ASRs (referred to below as the “QIAGEN test”) were compared with those obtained by the Hybrid Capture and Amplicor Monitor tests. The goal was to determine if there is adequate agreement in viral loads among the different tests, which could be used to establish the relevance of the currently published literature on the clinical utility of the Amplicor and Hybrid Capture assays for the these real-time PCR tests. In addition, the impact of extraction methods on viral loads was assessed using the QIAamp DNA minikit (QIAGEN, Valencia, CA), MagNA Pure (Roche Diagnostics), and easyMAG (bioMerieux, Durham, NC) methods for DNA nucleic acid extraction.
MATERIALS AND METHODS
Standard material and samples.
A plasma sample with a high viral load (BBI, West Bridgewater, MA) was used to make serial dilutions in CMV-seronegative human plasma to concentrations of 1.4 to 6.0 log10 copies/ml. The concentration of the BBI material was determined by the manufacturer using the Amplicor Monitor test. Specimens were aliquoted and stored at −70°C until testing.
For the cell-based standard, human foreskin fibroblasts (HFFs) were inoculated with CMV strain AD169 at a multiplicity of infection of 0.03 (6). At 6 h postinfection, the HFFs were harvested by trypsinization, washed, and counted. Dilutions of the HFFs were made with uninfected buffy coat cells to concentrations as high as 105 HFFs per 106 total cells. Aliquots of 200 μl containing 106 total cells were stored at −70°C until testing. For the agreement studies, plasma samples were collected at the Emory Medical Laboratories following an institutional review board-approved protocol and stored at −70°C. In addition, whole-blood specimens submitted to the Clinical Microbiology Laboratory at Rush University Medical Center for the Hybrid Capture assay were collected under an institutional review board-approved protocol. These whole-blood specimens were stored at 4°C for as long as 72 h before separation of the plasma. Plasma specimens were held at −70°C up to 7 days before testing.
Nucleic acid extraction.
For the CMV Monitor assay, nucleic acid was extracted using the MagNA Pure total-nucleic-acid kit as previously described (8). A 200-μl volume of plasma was extracted and eluted in 100 μl, and 50 μl of the eluate was added to the master mix. For the ASRs, a variety of nucleic acid extraction methods were used as described below. Nucleic acid was extracted from 200 μl of sample using the QIAamp DNA minikit (QIAGEN, Valencia, CA) Blood and Body Fluid protocol with the following adjustments: for each specimen, 2 μl of carrier RNA (1 mg/ml) and 5 μl of the internal control were added to 200 μl of buffer AL. The specimen was eluted in 50 μl. The MagNA Pure LC (Roche Diagnostics, Indianapolis, IN) was used to isolate nucleic acid from plasma using the total-nucleic-acid kit. For each specimen, 200 μl of plasma and 5 μl of the internal control were added to 300 μl of lysis buffer. The sample was eluted in 50 μl. For nucleic acid extraction using the NucliSens easyMAG, 200 μl of the sample plus 5.5 μl of the internal control were added to 2,000 μl of lysis buffer. Then 100 μl of magnetic silica was added, and the entire specimen was loaded onto the easyMAG instrument. The sample was eluted in 55 μl.
CMV DNA assays.
The Hybrid Capture system (version 2.0) assay (Digene Corporation, Gaithersburg, MD) was performed according to the manufacturer's protocol for the qualitative assay, which has been approved by the FDA. The Amplicor CMV Monitor test (Monitor; Roche Diagnostics) was performed according to the manufacturer's recommendations except for the nucleic acid extraction, which was performed as described above. The Amplicor CMV Monitor test targets a 365-bp region of the polymerase gene. The linear range of the assay is 300 to 50,000 log10 copies/ml. The Abbott and QIAGEN reagents use the same primers, probes, internal control, and standards, and both target the same 105-bp region of the major immediate-early antigen. For the Abbott test, the working master mix was prepared by adding 598 μl of Enzyme Mix E and 130 μl of Magnesium Mix E into the CMV primer/probe mix tube. After 20 μl of sample eluate from the DNA extraction was added to 30 μl of the working master mix, amplification and detection were performed using the ABI Prism 7000 system (Applied Biosystems Inc., Foster City, CA) with the following parameters: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 55°C for 1 min. The results were quantified using CMV DNA at predetermined concentrations provided by the manufacturer. For the QIAGEN test, 25 μl of TM PCR ASR master mix was added to 5 μl MG-SOL to which 20 μl of each sample eluate was added for a total volume of 50 μl. The QIAGEN quantitation standards were provided by the manufacturer, and the master mix for the standards was prepared as described above, except for the addition of 2 μl of the internal control to the master mix. The cycling parameters were identical to those used for the Abbott test.
Study design.
A detailed analytical evaluation was performed using the Abbott test. Viral loads obtained by the Abbott test and the Amplicor CMV Monitor test or the semiquantitative Hybrid Capture assay were compared. Since the Abbott and QIAGEN tests use the same reagents, the analytical validation was not repeated using the QIAGEN test. The different extraction methods were compared using the QIAGEN test.
Statistical analysis.
Data were log10 transformed prior to analysis. Descriptive statistics and regression line equations were calculated with the analysis tool pack of Microsoft Excel 2000 (Microsoft Corp., Redmond, WA). Agreement between viral loads was assessed by the method of Bland and Altman (2).
RESULTS
Performance characteristics of the Abbott test.
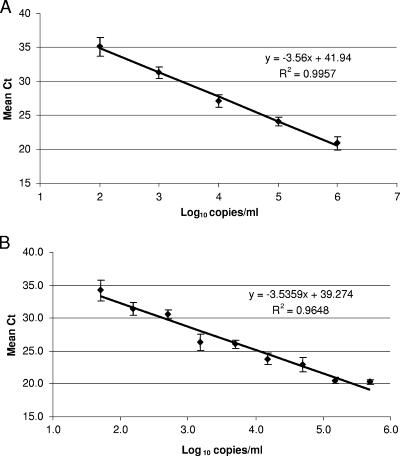
The linear range of the Abbott test was determined by testing aliquots of CMV DNA ranging in concentration from 2.0 to 6.0 log10 copies/ml. Data shown in Fig. 1A are means (±standard deviations [SD]) for samples tested in triplicate in two separate runs. Multiple replicates of samples ranging in concentration from 1.4 to 3.0 log10 copies/ml were tested to determine the limit of detection (Table 1). The assay was linear from 2.0 log10 copies/ml to 6.0 log10 copies/ml (Fig. 1). We did not have a high-titer specimen to measure the linear range beyond 6.0 log10 copies/ml. When a larger number of replicates were tested to assess the limit of detection, the concentration at which CMV DNA was detected in 95% of the replicates was 2.3 log10 copies/ml (200 copies/ml). The reproducibility of the assay (Table 2) varied through the linear range of the assay; the assay has the greatest precision at higher viral loads and is least precise with viral loads at or below 2.3 log10 copies/ml.
FIG. 1.
(A) Linear range of the Abbott test using plasma samples. Data are means (±SD) for samples tested in triplicate in two separate runs (n = 6). (B) Linear range of the Abbott test using cell-based standards. Data are means (±SD) for five to seven replicates tested in a single run.
TABLE 1.
Limit of sensitivity of the Abbott test using plasma samples
| Log10 copies/ml | No. of positive samples/no. tested | % Positive |
|---|---|---|
| 3.0 | 16/16 | 100 |
| 2.3 | 19/20 | 95 |
| 2.0 | 18/20 | 90 |
| 1.7 | 9/13 | 69 |
| 1.4 | 7/20 | 35 |
TABLE 2.
Reproducibility of the Abbott test using plasma samples
| Nominal concn (log10 copies/ml) | No. of samples tested | Mean viral load (log10 copies/ml) | SD (log10 copies/ml) | % CVa |
|---|---|---|---|---|
| 6.0 | 6 | 5.88 | 0.19 | 3.2 |
| 5.0 | 16 | 4.79 | 0.24 | 5.0 |
| 4.0 | 16 | 3.91 | 0.21 | 5.3 |
| 3.0 | 16 | 2.76 | 0.24 | 8.7 |
| 2.3b | 19 | 1.93 | 0.45 | 23.3 |
| 2.0b | 18 | 1.90 | 0.46 | 24.2 |
CV, coefficient of variation.
Only positive samples were included.
The linear range, limit of detection, and reproducibility of the Abbott test were also assessed using a cell-based standard, which was designed to mimic a white blood cell sample. This sample type was tested because whole-blood samples are used for clinical testing in many laboratories. The test was linear from 1.7 log10 copies/ml to 5.2 log10 copies/ml, with a limit of detection of 1.7 log10 copies/ml (Fig. 1B). As seen with the plasma standard, the test was less reproducible near the limit of detection. The coefficient of variation ranged from 1.7% at 5.7 log10 copies/ml to 34.1% at 1.7 log10 copies/ml (data not shown).
Agreement between the Abbott test and the Amplicor CMV Monitor assay.
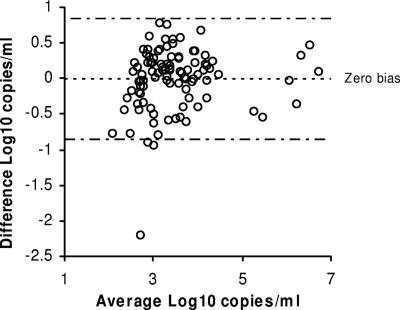
A total of 101 clinical specimens that had detectable viral loads by the Amplicor CMV Monitor assay were tested by the Abbott test (Fig. 2). The one specimen that was positive by the Monitor assay and negative by the Abbott test had a viral load of 2.99 log10 copies/ml in the Monitor assay. For the 100 samples that were positive by both tests, the population mean (SD) was 3.51 (0.89) log10 copies/ml for the Monitor assay and 3.50 (0.95) log10 copies/ml for the Abbott test. Based on the agreement analysis, the mean difference between the two tests was −0.012 log10 copies/ml (95% limits of agreement, −0.869 to 0.845 log10 copies/ml) (Fig. 2). No bias was observed for viral loads obtained with the two tests.
FIG. 2.
Agreement plot for 100 plasma samples with detectable viral loads by the Abbott and Monitor tests. Dotted line, mean difference for the samples; dashed lines, 95% limits of agreement.
Comparison of the Abbott test and the Hybrid Capture assay.
Fifty specimens were tested by both the Abbott test and the Digene Hybrid Capture assay, using plasma and whole-blood specimens, respectively. Twenty-six samples were negative by both tests. Ten specimens were negative by the Hybrid Capture assay and positive by the Abbott test, and all of these had low viral loads, ranging from 1.54 to 2.96 log10 copies/ml, by the Abbott test. Three samples were equivocal by the Hybrid Capture assay and positive by the Abbott test, with viral loads of 2.10, 3.45, and 3.83 log10 copies/ml by the Abbott test (Table 3). Eleven samples were positive by both assays. It was not possible to compare the viral loads for these 11 specimens, since the qualitative version of the Hybrid Capture assay was performed. However, the Hybrid Capture assay ratio did appear to correlate with the Abbott test viral load (Table 3). The lowest Abbott test viral load with a positive Hybrid Capture result was 3.73 log10 copies/ml. Specimens with viral loads between 4 log10 copies/ml and 5 log10 copies/ml had a mean ratio of 18 by the Hybrid Capture assay, while those specimens with viral loads greater than 5.0 log10 copies/ml by the Abbott test had a mean ratio of 182.
TABLE 3.
Comparison of Abbott test viral loads and Hybrid Capture assay ratiosa
| Hybrid Capture ratio | Abbott test viral load (log10 copies/ml) |
|---|---|
| Equivocal | 2.10 |
| Equivocal | 3.45 |
| Equivocal | 3.83 |
| 1 | 3.73 |
| 4 | 4.56 |
| 6 | 4.69 |
| 9 | 4.67 |
| 31 | 5.23 |
| 54 | 4.83 |
| 117 | 6.55 |
| 131 | 5.34 |
| 182 | 5.37 |
| 216 | 5.45 |
| 416 | 6.0 |
The Abbott test used plasma specimens, and the Hybrid Capture assay used whole-blood specimens.
Comparison of ASRs and extraction methods.
Forty-seven plasma samples were tested by the Abbott and QIAGEN tests using the QIAGEN extraction method. Viral loads were detectable in all samples by both tests. The mean viral load (SD) obtained using the Abbott test was 3.89 (1.02) log10 copies/ml, compared to 4.04 (1.01) log10 copies/ml by the QIAGEN test. The mean difference in viral loads between the two tests was 0.144 log10 copies/ml, and the 95% limits of agreement were −0.222 to 0.511 log10 copies/ml.
To assess the impacts of different extraction methods on viral loads, plasma specimens were tested by the QIAGEN test using the QIAGEN, MagNA Pure, and easyMAG extraction methods. Of the 47 specimens used to evaluate the two tests (see above), 34 specimens had adequate volume remaining to assess the different extraction methods. The mean viral loads and SD for the three methods are shown in Table 4. In summary, the differences in viral load obtained by the different tests using the same extraction method or by the same test (QIAGEN test) using different extraction methods are all within 0.370 log10 copies/ml or within 2.3-fold.
TABLE 4.
Comparison of mean viral loads and SD obtained by the QIAGEN test using different extraction methods
| Extraction method | Mean viral load (log10 copies/ml) | SD (log10 copies/ml) |
|---|---|---|
| QIAGEN | 4.09 | 1.05 |
| MAgNA Pure | 3.86 | 0.93 |
| easyMAG | 4.23 | 1.04 |
DISCUSSION
The clinical utility of CMV viral load testing is well established and is considered the standard of care in monitoring the response to antiviral therapy. However, the lack of standardization among CMV viral load assays continues to be problematic for clinical laboratories. In this study we evaluated the performance characteristics of the Abbott test compared to those of the more widely used Amplicor CMV Monitor and Hybrid Capture assays. In addition, we assessed the impact of different extraction methods on viral loads obtained by both the Abbott and QIAGEN tests. The goal was to determine if these tests could serve as a basis for standardizing CMV viral load testing.
The performance characteristics of the Abbott test using plasma specimens make it well suited for clinical use, with a limit of detection of 2.3 log10 copies/ml and a linear range of at least 5.0 log10 copies/ml (2.0 log10 copies/ml to at least 6.0 log10 copies/ml). The reproducibility is similar to that observed for other real-time PCR tests (1, 5), with the greatest variability observed near the limit of detection. In addition, there is close agreement between viral loads obtained by the Abbott and Monitor tests when plasma samples are tested. Based on these data, it appears that the studies that have been published establishing the clinical utility of the Amplicor CMV Monitor assay can be extrapolated to the Abbott test (11, 12, 14).
When a cell-based standard was used, the limit of detection was 1.7 log10 copies/ml; the linear range appeared to be more limited (1.7 to 5.2 log10 copies/ml) than that seen when plasma specimens were used. This may reflect the large amount of intracellular viral DNA in the form of concatemers present in the standard testing material at higher levels of human CMV infectivity. This may interfere with primer and probe binding, resulting in a reduction in the detectable copy numbers (3, 13). Therefore, the performance characteristics of the Abbott test require further evaluation using whole-blood specimens.
Both the Abbott and the QIAGEN reagents were manufactured by artus and are essentially the same product, providing clinical laboratories with equivalent options for developing CMV viral load tests. When tests based on these reagents are coupled with either the QIAGEN, the MagNA Pure, or the easyMAG extraction method, the viral loads are remarkably similar. As expected, when the Abbott and QIAGEN tests were compared using the same extraction method, the viral loads were in close agreement (mean difference, 0.144 log10 copies/ml). A larger mean difference in viral loads (0.144 to 0.370 log10 copies/ml) was observed when the QIAGEN test was evaluated using three different extraction methods, although the greatest difference between extraction methods was only 2.3-fold. Further studies are needed to determine if this close agreement is seen when a wider range of extraction methods is used. These data support the concept that the Abbott and QIAGEN reagents provide laboratories with additional reagents that can be used to for CMV viral load testing and that may improve assay standardization.
Acknowledgments
We thank Abbott Molecular for supplying the reagents for the study. This project was supported in part by the National Institutes of Health contract NO1-AI35172 to J.W.B. and the Emory Center for AIDS Research (P30 AI050409).
N.S.L. serves as a technical consultant to Abbott Molecular, and A.M.C. has served on scientific advisory boards for Abbott Molecular, QIAGEN, and Roche Diagnostics.
Footnotes
Published ahead of print on 4 April 2007.
REFERENCES
- 1.Beuselinck, K., M. van Ranst, and J. van Eldere. 2005. Automated extraction of viral-pathogen RNA and DNA for high-throughput quantitative real-time PCR. J. Clin. Microbiol. 43:5541-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed] [Google Scholar]
- 3.Burd, E. M., J. S. Pulido, D. G. Puro, and W. J. O'Brien. 1996. Replication of human cytomegalovirus in human retinal glial cells. Ophthalmol. Vis. Sci. 10:1957-1966. [PubMed] [Google Scholar]
- 4.Caliendo, A. M., K. St. George, S.-Y. Kao, J. Allega, B.-H. Tan, R. LaFontaine, L. Bui, and C. R. Rinaldo. 2000. Comparison of quantitative cytomegalovirus (CMV) PCR in plasma and CMV antigenemia assay: clinical utility of the prototype AMPLICOR CMV MONITOR test in transplant recipients. J. Clin. Microbiol. 38:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caliendo, A. M., A. Valsamakis, Y. Zhou, B. Yen-Lieberman, J. Andersen, S. Young, A. Ferreira-Gonzalez, G. T. Tsongalis, R. Pyles, J. Bremer, and N. S. Lurain. 2006. Multilaboratory comparison of hepatitis C virus viral load assays. J. Clin. Microbiol. 44:1726-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caliendo, A. M., B. Yen-Lieberman, J. Baptista, J. Andersen, C. Crumpacker, R. Schuurman, S. A. Spector, J. Bremer, and N. S. Lurain. 2003. Comparison of molecular tests for detection and quantification of cell-associated cytomegalovirus DNA. J. Clin. Microbiol. 41:3509-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emery, V. C., C. A. Sabin, A. V. Cope, D. Gor, A. F. Hassan-Walker, and P. D. Griffiths. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 355:2032-2036. [DOI] [PubMed] [Google Scholar]
- 8.Fiebelkorn, K. R., B. G. Lee, C. E. Hill, A. M. Caliendo, and F. S. Nolte. 2002. Clinical evaluation of an automated nucleic acid isolation system. Clin. Chem. 48:1613-1615. [PubMed] [Google Scholar]
- 9.Gerna, G., D. Zipeto, M. Parea, M. G. Revello, E. Silini, E. Percivalle, M. Zavattoni, P. Grossi, and G. Milanesi. 1991. Monitoring of human cytomegalovirus infections and ganciclovir treatment in heart transplant recipients by determination of viremia, antigenemia and DNAemia. J. Infect. Dis. 164:488-498. [DOI] [PubMed] [Google Scholar]
- 10.Humar, A., D. Gregson, A. M. Caliendo, A. McGeer, G. Malkan, M. Krajden, P. Corey, P. Greig, S. Walmsley, G. Levy, and T. Mazzulli. 1999. Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation 68:1305-1311. [DOI] [PubMed] [Google Scholar]
- 11.Humar, A., D. Kumar, G. Boivin, and A. M. Caliendo. 2002. Cytomegalovirus (CMV) viral load kinetics to predict recurrent disease in solid organ transplant patients with CMV disease. J. Infect. Dis. 186:829-833. [DOI] [PubMed] [Google Scholar]
- 12.Humar, A., G. Malkan, G. Moussa, P. Greig, G. Levy, and T. Mazzulli. 2000. Human herpesvirus-6 is associated with cytomegalovirus reactivation in liver transplant recipients. J. Infect. Dis. 181:1450-1453. [DOI] [PubMed] [Google Scholar]
- 13.LaFamina, R. L., and G. S. Hayward. 1983. Replicative forms of human cytomegalovirus DNA with joined termini are found in permissively infected human cells but not in non-permissive Balb/c-3T3 mouse cells. J. Gen. Virol. 64:373-389. [DOI] [PubMed] [Google Scholar]
- 14.Sia, I. G., J. A. Wilson, C. M. Groettum, M. J. Espy, T. F. Smith, and C. V. Paya. 2000. Cytomegalovirus (CMV) DNA load predicts relapsing CMV infection after solid organ transplantation. J. Infect. Dis. 181:717-720. [DOI] [PubMed] [Google Scholar]
- 15.Tanabe, K., T. Tokumoto, N. Ishikawa, I. Koyama, K. Takahashi, S. Fuchinoue, T. Kawai, S. Koga, T. Yagisawa, H. Toma, K. Ota, and H. Nakajima. 1997. Comparative study of cytomegalovirus (CMV) antigenemia assay, polymerase chain reaction, serology, and shell vial assay in the early diagnosis and monitoring of CMV infection after renal transplantation. Transplantation 64:1721-1725. [DOI] [PubMed] [Google Scholar]