Abstract
The analytical performances of the new Abbott RealTime hepatitis C virus (HCV) and human immunodeficiency virus type 1 viral load assays were compared at nine laboratories with different competitor assays. These included the Abbott LcX, Bayer Versant bDNA, Roche COBAS Amplicor, and Roche COBAS TaqMan assays. Two different protocols used during the testing period with and without a pre-m1000 RNA isolation spin were compared. The difference proved to be nonsignificant. A uracil-N-glycosylase (UNG) contamination control option in the HCV test for previous Roche COBAS Amplicor users was evaluated. It proved to decrease amplicon carryover by 100-fold independent of the amplicon input concentration. The protocol including UNG proved to overcome problems with false-positive negative controls. Comparison with other assays revealed only minor differences. The largest difference was observed between the Abbott HCV RealTime assay and the Roche COBAS Amplicor HCV Monitor version 2.0 assay.
Hepatitis C virus (HCV) and human immunodeficiency virus type 1 (HIV-1) are major causes of mortality in both developing and developed countries. For both viruses, relatively effective therapies have been available in developed countries for quite some time. HCV and HIV-1 viral loads are important parameters in patient management both before initiating therapy and during therapy. The decline of the HCV viral load during the first 3 months of therapy is, for instance, a strong indicator of the final outcome of therapy (9, 23). Moreover, in many countries, the HCV viral load in infected health care workers determines whether they are allowed to perform surgical procedures. HIV-1 viral load is monitored during therapy, and a viral load above a certain threshold, which may differ per treating physician, requires a switch in medication (6, 13).
The first tests that were described for determining the viral load were based on target amplification techniques like reverse transcriptase PCR (RT-PCR) and nucleic acid sequence-based amplification (NASBA). These tests had readout on agarose gel or readout with enzymatic detection of the amplicon after amplification with biotinylated primers (1, 3, 15, 17). Subsequently, signal amplification techniques were developed for the quantitative detection of HCV and HIV-1 (5, 16). Those techniques were improved and commercialized. The suppliers of the most widely used systems at the moment are Roche and Abbott, for RT-PCR-based systems (the COBAS Amplicor and LcX systems, respectively); bioMerieux (formerly Organon Technica), with a NASBA-based technique; and Bayer, with the Versant bDNA system being a signal amplification technique (4, 18, 19). Each technique has its own advantages and disadvantages. The relatively small dynamic range is a disadvantage that all current assay formats have in common. All assay formats are also rather sensitive to contamination, especially at the lower limit of detection (21). Handling of the sample after target amplification is a major cause of contamination for the NASBA- and RT-PCR-based techniques. Recently, amplification techniques that allow real-time detection of the amplicon have been developed for NASBA and RT-PCR. These new techniques make postamplification processing obsolete (14, 21). The major advantage of real-time RT-PCR and NASBA techniques is the lower sensitivity to contamination. These techniques are also less time-consuming and in general have a higher dynamic range. A serious drawback of most real-time techniques is the detection of amplicon with fluorescently labeled probes that are sensitive to point mutations within the target sequence. This is especially of concern with highly variable targets like HCV and HIV-1.
Abbott has recently CE (Conformité Européenne) marked its new HCV and HIV-1 viral load tests based on real-time RT-PCR (7, 8, 22). A major theoretical advantage of the real-time technique used is the probe system, which allows target detection by probe hybridization at low temperatures. This potentially reduces problems with genotype-dependent underquantification due to probe-target mismatches. Furthermore, the primers used for HIV-1 are located in a highly conserved region of the genome also used in the Abbott LcX HIV-1 assay. Before CE marking, Abbott initiated the Early Access Programme (EAP), involving 21 sites in Europe and 1 site in South Africa. This paper describes the validation of these HCV and HIV-1 viral load assays on the basis of data generated during the EAP.
MATERIALS AND METHODS
Collaborating sites.
Out of the 21 sites initially selected by Abbott Molecular to join the prerelease EAP, 9 sites participated in the joint evaluation by submitting data on patient samples. The assays and comparator assays run at the different sites are summarized in Table 1. Parts of the data from sites 4 and 5 have been published as part of separate studies (7, 8).
TABLE 1.
List of comparator assays run at the collaborating sitesa
| Site | Assay used
|
|
|---|---|---|
| HCV | HIV-1 | |
| 1 | bDNA | bDNA |
| 2 | CAM | CAM |
| 3 | In-house test | bDNA |
| 4 | LcX + in-house test | LcX |
| 5 | ND | LcX |
| 6 | CAM | CAM |
| 7 | ND | LcX |
| 8 | ND | LcX |
| 9 | CTM | CTM |
ND, not done; bDNA, Bayer Versant bDNA HIV-1 and HCV versions 3.0 (Bayer HealthCare) assays; CAM, Roche COBAS Amplicor HIV-1 Monitor version 1.5 and HCV Monitor version 2.0 (Roche Diagnostics, Basel, Switzerland) assays; LcX, the Abbott LcX HIV and HCV RNA quantitative assays (Abbott Molecular, Chicago, IL); CTM, Roche Cobas Ampliprep-Cobas TaqMan HIV-1 test (Roche Diagnostics, Basel, Switzerland).
Patient materials and quality control materials.
Patient plasma samples were run either fresh (site 7) or after storage at −80°C. In addition, all sites ran samples from the 2005 quality control program from the QCMD (www.QCMD.org). Four laboratories selected for the EAP who were not part of this joined evaluation ran QCMD samples and submitted data on these samples (Leeds, United Kingdom [A. Hale], Lille, France [L. Bocket], Lisbon, Portugal [R. Camacho], and Manchester, United Kingdom [M. Guiver]).
Abbott RealTime HIV-1 and HCV viral load assays.
The Abbott RealTime HIV-1 and HCV viral load assays were performed according to the instructions provided by Abbott Molecular. In short, HIV-1 RNA and HCV RNA were isolated from 1.0 and 0.5 ml of plasma, respectively, with the Abbott m1000 nucleic acid extraction system. The lower limits of detection reported by Abbott for the use of these isolation volumes were 40 HIV-1 RNA copies per ml and 12 HCV IU per ml. Fifty microliters of eluted RNA was subsequently mixed with 50 μl Mastermix. Real-time RT-PCR was run on the m2000rt RealTime PCR system from Abbott. Data from the comparator assays were generated during routine diagnostics for patient management.
Data analysis.
Samples below the limit of detection for both the Abbott RealTime assays and the comparator assays were not included in the study analysis. Samples below the limit of detection in one assay but positive in the other were discarded in the regression analysis. Data were analyzed with Sigmaplot 2001 for Windows.
RESULTS AND DISCUSSION
Performance of the Abbott RealTime system at the EAP sites.
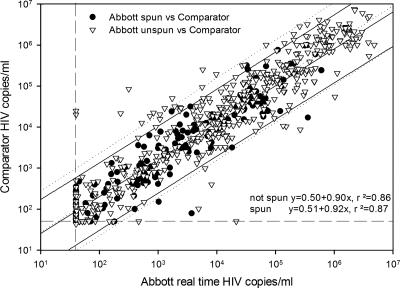
Several problems were encountered, which were solved during the EAP. First, it was found that the Abbott RealTime HCV and HIV-1 assays underquantified some samples that had been stored at −80°C (data not shown). Spinning the plasma sample before nucleic acid extraction could circumvent this “inhibitory” effect (data not shown). It was hypothesized that in certain samples, fibrin clotting after thawing might result in a loss of RNA-coated magnetic beads during the extraction procedure on the m1000 system. Part of the sample run during the EAP was therefore performed using an alternative isolation protocol. This protocol included a 5-min spin of the plasma at 2,000 × g after thawing but before m1000 extraction. Data obtained with and without spinning were compared (Fig. 1). The difference between the two protocols proved not to be significant (P value of 0.87 by t test). It was therefore decided that the difference in protocols should be ignored for further data analysis, except for individual samples.
FIG. 1.
Comparison of data obtained from samples that were spun and those obtained from samples that were not spun. Triangles and dotted lines are data points and regression curves/95% confidence intervals, respectively, from samples that had been frozen at −80°C and that had not been spun before m1000 RNA extraction. Circles and solid lines are data points and regression curves/95% confidence intervals, respectively, from samples that were either used fresh or had been frozen at −80°C and subsequently spun before m1000 RNA extraction. Horizontal and vertical dashed lines are the lower limits of quantification.
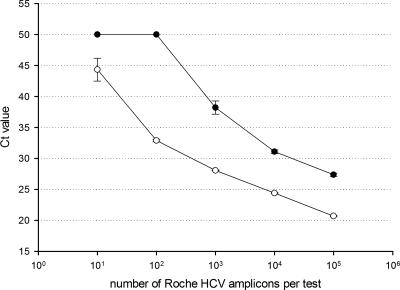
The second and probably more significant problem encountered was that virtually all laboratories that had previously used the Roche COBAS Amplicor HCV viral load test had significant problems with false-positive results on negative controls. The amplicon of the Roche COBAS Amplicor HCV test overlaps completely with the Abbott RealTime HCV viral load test. The Roche COBAS Amplicor assay is an open system after RT-PCR in the detection phase. Since significant amounts of amplicon are released in the laboratory, this system requires the dUTP/uracil-N-glycosylase (dUTP/UNG) contamination control system. The Abbott RealTime HCV viral load system is a closed system post-RT-PCR, and consequently, the generated amplicon will never be released in the laboratory under normal circumstances. Abbott chose not to implement the dUTP/UNG system in their assay since the dUTP/UNG contamination control system may lower the sensitivity of the assay. This is in part due to residual UNG activity after the 10-min 95°C inactivation step normally used. It was hypothesized that the Roche COBAS Amplicor HCV Monitor amplicons contaminated the Abbott RealTime HCV viral load assay in laboratories that had used the former test. A protocol that incorporated UNG without the use of dUTP to circumvent the problem of amplicon carryover from other systems like the COBAS Amplicor assay was developed. To test the efficiency of this protocol, an amplicon from the COBAS Amplicor HCV test with dUTP incorporated was quantified using the Abbott RealTime HCV viral load assay without UNG. This amplicon contained 4.2 × 1012 copies per ml. Tenfold dilutions from 106 to 10 copies per test were analyzed in quadruplicate by the Abbott RealTime HCV viral load assay with and without UNG (Fig. 2). UNG decreased the concentration by 100-fold irrespective of the amount of amplicon input. With the use of UNG, no further false-positive negative controls were encountered at sites where the Roche COBAS Amplicor HCV assay was used. This was anticipated, since UNG has been used successfully in the COBAS Amplicor assay for many years now.
FIG. 2.
Efficiency of UNG in preventing amplicon carryover. Black diamonds are the means of four tests with UNG added. White squares are the means of four tests without UNG added. Ct, cycle threshold.
With these changes relative to the original protocol incorporated, the HCV and HIV-1 RealTime viral load tests were generally perceived as being easy and convenient to use.
Comparison of the Abbott RealTime HCV and HIV-1 viral load tests with comparators.
The Abbott RealTime HIV-1 viral load assay was compared with the Roche COBAS Amplicor HIV Monitor version 1.5, the Roche COBAS TaqMan, the Bayer Versant bDNA version 3.0, and the Abbott LcX assays. Table 2 shows the following results: (i) the number of samples tested, (ii) regression coefficient (r2), (iii) the mean difference between the Abbott viral load and the viral load measured with the comparator (mean log10 Abbott − mean log10 comparator), (iv) the regression curve equation, and (v) the number of samples where the Abbott RealTime HIV viral load assay gave an underestimation (Abbott < comparator) or overestimation (comparator < Abbott) of more than 1 log10 plus the mean difference. Correlations between the Abbott RealTime HIV-1 viral load test and the comparators were generally good (r2 > 0.80). The Abbott RealTime HIV-1 viral load assay gave a lower viral load relative to those of all the comparators. No major differences were observed in the numbers of samples that differed more than 1 log10 relative to the comparator assays. Overall, the Abbott RealTime HIV-1 viral load assay scored well relative to the comparators, and only minor differences were observed. For subtypes B, C, and CRF02-AG, sufficient samples (65, 55, and 22 samples, respectively) could be tested for a comparison although not enough to compare the Abbott RealTime test separately with each comparator. Relative to the comparators, the Abbott RealTime HIV-1 viral load test quantified subtype B and C samples accurately. Mean differences (Abbott minus comparator) of the log10 results were −0.31 and −0.02, respectively. The Abbott RealTime HIV-1 assay generated higher viral load values; however, for subtype CRF02-AG, samples compared to the comparator assays (the mean difference [Abbott minus comparator] of the log10 results was 0.27). These data corroborate data from a recent report on the performance of the Abbott RealTime HIV-1 viral load test showing better performance on CRF02-AG recombinants than the comparator assays (12).
TABLE 2.
Comparison of the Abbott RealTime HCV and HIV-1 viral load assays with comparator assays
| Abbott RealTime assay | Assay used versus Abbott RealTime assay | No. of samples | r2a | Mean differenceb | Regression (y = ax + b)a
|
% of samples with difference
|
||
|---|---|---|---|---|---|---|---|---|
| a | b | Comparator < Abbottc | Abbott < comparatord | |||||
| HCV | LcX | 92 | 0.89 | −0.35 | 0.95 | 0.54 | 2.2 | 3.2 |
| COBAS Amplicor | 146 | 0.81 | −0.08 | 0.75 | 1.41 | 2.7 | 2.0 | |
| COBAS TaqMan | 161 | 0.83 | −0.25 | 0.84 | 1.02 | 4.3 | 4.3 | |
| bDNA | 36 | 0.98 | 0.06 | 0.93 | 0.12 | 0 | 0 | |
| HIV | LcX | 128 | 0.82 | −0.11 | 1.00 | 0.14 | 3.1 | 1.6 |
| COBAS Amplicor | 115 | 0.84 | −0.14 | 0.87 | 0.61 | 0.9 | 0.9 | |
| COBAS TaqMan | 55 | 0.76 | −0.24 | 0.93 | 0.62 | 1.8 | 1.8 | |
| bDNA | 168 | 0.90 | −0.14 | 0.93 | 0.38 | 0 | 1.8 | |
Spearman regression coefficient and equation of regression analysis.
The mean difference was calculated as follows: mean log10 Abbott RealTime assay − mean log10 comparator results.
Percentage of samples with a mean difference 1 log10 higher in the Abbott RealTime assay than in the comparator assay.
Percentage of samples with a mean difference 1 log10 higher in the comparator assay than in the Abbott RealTime assay.
The Abbott RealTime HCV viral load assay was compared with the Roche COBAS Amplicor HCV version 1.5, the Roche COBAS Taqman, the Bayer bDNA version 3.0, and the Abbott LcX assays. Table 2 shows the following results: (i) the number of samples tested, (ii) regression coefficient (r2), (iii) the mean difference between the Abbott viral load and the viral load measured with the comparator (mean log10 Abbott − mean log10 comparator), (iv) the regression curve equation, and (v) the number of samples where the Abbott RealTime HCV viral load assay gave an underestimation (Abbott < comparator) or overestimation (comparator < Abbott) of more than 1 log10 plus the mean difference. For HCV, the number of data points tested with the bDNA assay was too low for proper comparison. The regression equation of the comparison with the COBAS Amplicor HCV test differed significantly from similarity (a = 1 and b = 0). This proved to be due to higher values obtained with COBAS Amplicor assay at the low range and lower values in the high range. One sample that was below the limit of detection in the Abbott assay gave >105 IU/ml in the comparator assay (site 2, Roche COBAS Amplicor v1.5). The Abbott RealTime assay gave a lower viral load than all the comparators. No major differences were observed in the numbers of samples that differed more than 1 log10 relative to the comparator assays. Overall, the Abbott real-time HCV viral load assay scored well relative to the comparators, and only minor differences were observed. The number of data points generated on genotypes 2, 3, and 4 was rather low, and sufficient data were generated for comparison for genotype 1 only. The mean difference of the log10 results between the Abbott assay and the comparators for HCV genotype 1 samples was minor (−0.18 log10).
Performance of the Abbott RealTime HCV and HIV-1 viral load assays on the 2005 QCMD panels.
The QCMD quality control panels for the year 2005 were run at 11 EAP laboratories for further validation of the Abbott RealTime assays. Five sites ran the HCV panel, and eight sites ran the HIV-1 panel.
Three laboratories scored all samples correct in the qualitative analysis of the HCV panel. One laboratory scored the negative sample as positive but below the lower limit of detection. This was most probably due to contamination, since this laboratory ran the panel without using UNG, and the laboratory that made the QCMD HCV 2005 panel uses the COBAS Amplicor assay (M. Schutten, unpublished data). One laboratory reported the inhibition of one sample and one sample with a target concentration of 1.7 × 103 IU per ml as being negative.
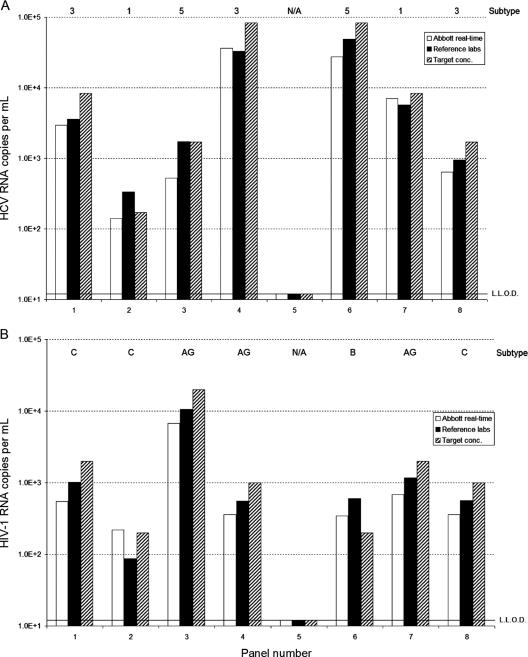
For HIV-1, all 10 laboratories had a 100% correct score on all samples for the qualitative analysis. The quantitative data generated by the laboratories with the Abbott RealTime HCV and HIV-1 kits were averaged and compared with the target concentration (what the QCMD intended to put into the sample) and with the mean of the predistribution test results performed by the QCMD reference laboratories (Fig. 3A and B). With respect to the predistribution testing results, it should be noted that for HCV and HIV-1, testing was done mainly with the COBAS Amplicor Monitor assays. For samples 2 and 8 of the HCV panel, only one quantitative result was available because the other predistribution tests were below the limit of detection. For both samples, one Versant HCV RNA version 3.0 test and one COBAS Amplicor HCV Monitor test proved to be false negative. For the HIV-1 panel, sample 2 had only two predistribution results because two Roche COBAS Monitor version 1.5 results were below the limit of detection. Apart from these predistribution test difficulties, the mean of the Abbott RealTime HCV and HIV-1 results and the target concentration and the predistribution mean results were relatively close.
FIG. 3.
Performance of the Abbott RealTime HCV (A) and HIV-1 (B) viral load assays on the 2005 QCMD HIV-1 and HCV panels. Abbott RealTime HCV and HIV data (white bars) are plotted against the anticipated input (hatched bars) and the data of the predistribution laboratory testing (solid bars). HIV-1 subtypes and HCV genotypes of the samples are indicated at the top. N/A, not applicable.
Recent problems with the introduction of a TaqMan-based system have shown that the development of a robust quantitative real-time-detected RT-PCR for highly variable targets like HCV and HIV-1 is not an easy task (2, 10-12, 20). From the data presented, it can be concluded that real-time RT-PCR systems based on hybridization probes rather than hydrolysis probes bear promise for the future. In conclusion, the RealTime HCV and HIV-1 viral load tests from Abbott are convenient, easy-to-use assays with good performance characteristics. It should be noted that laboratories that use the Roche COBAS Amplicor HCV Monitor assay are recommended to use the adapted protocol with UNG to prevent false-positive results due to amplicon carryover.
Acknowledgments
We thank A. Hale, L. Bocket, R. Camacho, and M. Guiver for the data generated on the QCMD panels. We also thank the QCMD Neutral Office for providing the data relating to the QCMD 2005 HCV and HIV Proficiency Programmes. The m1000 and m2000rt systems, including consumables and reagents for this study, were kindly provided by Abbott Molecular.
Analysis and presentation of the QCMD data were performed by the authors of this paper only and are their interpretation of the QCMD data provided. The paper does not express or constitute QCMD proficiency results and/or reports for these programs.
Footnotes
Published ahead of print on 4 April 2007.
REFERENCES
- 1.Abbott, M. A., B. J. Poiesz, B. C. Byrne, S. Kwok, J. J. Sninsky, and G. D. Ehrlich. 1988. Enzymatic gene amplification: qualitative and quantitative methods for detecting proviral DNA amplified in vitro. J. Infect. Dis. 158:1158-1169. [DOI] [PubMed] [Google Scholar]
- 2.Caliendo, A. M., A. Valsamakis, Y. Zhou, B. Yen-Lieberman, J. Andersen, S. Young, A. Ferreira-Gonzalez, G. J. Tsongalis, R. Pyles, J. W. Bremer, and N. S. Lurain. 2006. Multilaboratory comparison of hepatitis C virus viral load assays. J. Clin. Microbiol. 44:1726-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 350:91-92. [DOI] [PubMed] [Google Scholar]
- 4.de Baar, M. P., M. W. van Dooren, E. de Rooij, M. Bakker, B. van Gemen, J. Goudsmit, and A. de Ronde. 2001. Single rapid real-time monitored isothermal RNA amplification assay for quantification of human immunodeficiency virus type 1 isolates from groups M, N, and O. J. Clin. Microbiol. 39:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewar, R. L., H. C. Highbarger, M. D. Sarmiento, J. A. Todd, M. B. Vasudevachari, R. T. Davey, Jr., J. A. Kovacs, N. P. Salzman, H. C. Lane, and M. S. Urdea. 1994. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J. Infect. Dis. 170:1172-1179. [DOI] [PubMed] [Google Scholar]
- 6.EACS Euroguidelines Group. 2003. European guidelines for the clinical management and treatment of HIV-infected adults in Europe. AIDS 17(Suppl. 2):S3-S26. [PubMed] [Google Scholar]
- 7.Foulongne, V., B. Montes, M. N. ot-Rousseau, and M. Segondy. 2006. Comparison of the LCx human immunodeficiency virus (HIV) RNA quantitative, RealTime HIV, and COBAS AmpliPrep-COBAS TaqMan assays for quantitation of HIV type 1 RNA in plasma. J. Clin. Microbiol. 44:2963-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Diaz, A., G. S. Clewley, C. L. Booth, W. Labett, N. McAllister, and A. M. Geretti. 2006. Comparative evaluation of the performance of the Abbott real-time human immunodeficiency virus type 1 (HIV-1) assay for measurement of HIV-1 plasma viral load following automated specimen preparation. J. Clin. Microbiol. 44:1788-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gavier, B., M. A. Martinez-Gonzalez, J. I. Riezu-Boj, J. J. Lasarte, N. Garcia, M. P. Civeira, and J. Prieto. 1997. Viremia after one month of interferon therapy predicts treatment outcome in patients with chronic hepatitis C. Gastroenterology 113:1647-1653. [DOI] [PubMed] [Google Scholar]
- 10.Gelderblom, H. C., S. Menting, and M. G. Beld. 2006. Clinical performance of the new rRoche COBAS TaqMan HCV Test and High Pure System for extraction, detection and quantitation of HCV RNA in plasma and serum. Antivir. Ther. 11:95-103. [PubMed] [Google Scholar]
- 11.Giraldi, C., A. Noto, R. Tenuta, F. Greco, D. Perugini, M. Spadafora, A. M. Bianco, O. Savino, and A. Natale. 2006. A comparative evaluation between real time Roche COBas TAQMAN 48 HCV and bDNA Bayer Versant HCV 3.0. New Microbiol. 29:243-250. [PubMed] [Google Scholar]
- 12.Gueudin, M., J. C. Plantier, V. Lemee, M. P. Schmitt, L. Chartier, T. Bourlet, A. Ruffault, F. Damond, M. Vray, and F. Simon. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44:500-505. [DOI] [PubMed] [Google Scholar]
- 13.Hammer, S. M., M. S. Saag, M. Schechter, J. S. Montaner, R. T. Schooley, D. M. Jacobsen, M. A. Thompson, C. C. Carpenter, M. A. Fischl, B. G. Gazzard, J. M. Gatell, M. S. Hirsch, D. A. Katzenstein, D. D. Richman, S. Vella, P. G. Yeni, and P. A. Volberding. 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society—USA panel. Top. HIV Med. 14:827-843. [PubMed] [Google Scholar]
- 14.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 15.Kievits, T., B. van Gemen, D. van Strijp, R. Schukkink, M. Dircks, H. Adriaanse, L. Malek, R. Sooknanan, and P. Lens. 1991. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J. Virol. Methods 35:273-286. [DOI] [PubMed] [Google Scholar]
- 16.Lau, J. Y., G. L. Davis, J. Kniffen, K. P. Qian, M. S. Urdea, C. S. Chan, M. Mizokami, P. D. Neuwald, and J. C. Wilber. 1993. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet 341:1501-1504. [DOI] [PubMed] [Google Scholar]
- 17.Menzo, S., P. Bagnarelli, M. Giacca, A. Manzin, P. E. Varaldo, and M. Clementi. 1992. Absolute quantitation of viremia in human immunodeficiency virus infection by competitive reverse transcription and polymerase chain reaction. J. Clin. Microbiol. 30:1752-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy, D. G., L. Cote, M. Fauvel, P. Rene, and J. Vincelette. 2000. Multicenter comparison of Roche COBAS AMPLICOR MONITOR version 1.5, Organon Teknika NucliSens QT with Extractor, and Bayer Quantiplex version 3.0 for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 38:4034-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross, R. S., S. Viazov, S. Sarr, S. Hoffmann, A. Kramer, and M. Roggendorf. 2002. Quantitation of hepatitis C virus RNA by third generation branched DNA-based signal amplification assay. J. Virol. Methods 101:159-168. [DOI] [PubMed] [Google Scholar]
- 20.Sarrazin, C., B. C. Gärtner, D. Sizmann, R. Babiel, U. Mihm, W. P. Hofmann, M. von Wagner, and S. Zeuzem. 2006. Comparison of conventional PCR with real-time PCR and branched DNA-based assays for hepatitis C virus RNA quantification and clinical significance for genotypes 1 to 5. J. Clin. Microbiol. 44:729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schutten, M., and H. G. Niesters. 2001. Clinical utility of viral quantification as a tool for disease monitoring. Expert Rev. Mol. Diagn. 1:153-162. [DOI] [PubMed] [Google Scholar]
- 22.Swanson, P., S. Huang, V. Holzmayer, P. Bodelle, J. Yamaguchi, C. Brennan, R. Badaro, C. Brites, K. Abravaya, S. G. Devare, and J. Hackett, Jr. 2006. Performance of the automated Abbott RealTime HIV-1 assay on a genetically diverse panel of specimens from Brazil. J. Virol. Methods 134:237-243. [DOI] [PubMed] [Google Scholar]
- 23.Tong, M. J., L. M. Blatt, J. G. McHutchison, R. L. Co, and A. Conrad. 1997. Prediction of response during interferon alfa 2b therapy in chronic hepatitis C patients using viral and biochemical characteristics: a comparison. Hepatology 26:1640-1645. [DOI] [PubMed] [Google Scholar]