Abstract
The recent discovery of a mobile genetic element encoding a pilus-like structure in Streptococcus pneumoniae and the demonstration of a role for the pilus in virulence in mice have led to the proposal of the use of the pilus as a candidate pneumococcal vaccine. We examined the frequency of occurrence of the pneumococcal pilus, as determined by the presence of the rrgC gene, and analyzed its association with virulence, capsular serotypes, and multilocus sequence types in the American Indian pneumococcal collection and isolates of S. pneumoniae from blood cultures collected at Children's Hospital Boston. Overall, 21.4% of strains in the American Indian collection had the rrgC gene, but there was no difference between isolates obtained from the nasopharynx and those obtained from sterile sites (blood or cerebrospinal fluid). Vaccine-type strains were significantly more likely than non-vaccine-type strains to have this pilus gene (P < 0.001). Among isolates with identical multilocus sequence types, there was a high concordance (95%) between the multilocus sequence type and the presence or the absence of rrgC. Finally, in the era of the pneumococcal conjugate vaccine, the frequency of rrgC in isolates from Children's Hospital Boston has decreased significantly (42.8% before 2000 versus 21.3% after 2000; P = 0.019). Therefore, our data show that the pilus is present in a minority of strains and is associated with certain serotypes and that its frequency has been reduced by the conjugate pneumococcal vaccine.
Pilus-like surface structures in gram-positive bacteria were first identified in Corynebacterium diphtheriae by electron microscopy and have also been characterized genetically and biochemically (10, 25-27). Most recently, pilus-like structures have been identified in common human pathogens such as group A and B streptococci as well as Streptococcus pneumoniae (1, 7, 15-17, 19). Gram-positive pili (recently reviewed in reference 23) are extended polymers formed by a transpeptidase reaction involving sortase-mediated covalent cross-linking of LPXTG motifs in subunit proteins.
Mouse models of pneumococcal pneumonia and bacteremia have suggested a role for the pilus in virulence (1, 12). Competition experiments with models of pneumonia and bacteremia showed that a piliated strain could outcompete an otherwise isogenic pilus-deficient mutant. Immunization of mice with pilus antigens was recently shown to induce protection against lethal challenge by piliated strains, raising the possibility that the inclusion of pilus proteins in a multivalent pneumococcal vaccine may be advantageous (11).
The prevalence of the pilus in the pneumococcal population, however, is not known. Genes of the pneumococcal rlrA islet, which encodes the pilus, are present in one of the sequenced genomes (that of strain TIGR4) but not the other (that of strain R6) (1). A previous study evaluated the distribution of sortase genes in 82 strains; srtD, a component of the pneumococcal pilus islet, was present in 17% of strains (22).
A better understanding of the distribution of the pilus in the general population of pneumococci would help with the evaluation of its potential as a candidate vaccine. In the present study, we determined the prevalence of the rrgC gene from two different collections of pneumococcal clinical isolates: nasopharyngeal (NP), blood, and cerebrospinal fluid pneumococcal strains isolated from individuals of the Navajo and White Mountain Apache communities in the southwestern United States between April 1997 and October 2000 and pneumococci isolated from blood cultures at the Infectious Diseases Diagnostic Division at Children's Hospital Boston from 1995 to 2006. In addition, among the isolates in the American Indian collection, we evaluated the frequency of the rrgC gene by serotype, multilocus sequence type, and site of isolation (NP or blood culture) to evaluate whether the pilus is associated with increased virulence in humans.
MATERIALS AND METHODS
Pneumococcal strain collections.
Two different collections were analyzed in this study. The first collection (the American Indian pneumococcal collection) consisted of a selection of strains from the American Indian trial of the seven-valent pneumococcal vaccine (PCV7), conducted with individuals in the Navajo and White Mountain Apache communities in the southwestern United States between April 1997 and October 2000. The details of this trial have been described previously (20, 21). Briefly, this was a group-randomized, controlled, efficacy trial of a seven-valent pneumococcal conjugate vaccine (PnCRM7; Wyeth Vaccines), with a study of NP colonization nested within the trial. NP swabs were obtained from the study participants at approximately 7, 12 to 15, and 18 to 24 months of age (between February 1998 and May 2000). Pneumococcal identification was confirmed by optochin susceptibility and bile solubility assays. Pneumococci were serotyped by a novel immunoblot method to detect multiple serotypes in NP specimens (3). Included in this study were 361 NP isolates.
Invasive disease isolates (n = 123) from individuals of all ages in the Navajo and White Mountain Apache communities were collected throughout the study period. These pneumococcal isolates were collected and serotyped at the Arctic Investigations Program of the CDC, Anchorage, AK.
Isolates from Children's Hospital Boston.
Invasive pneumococcal isolates that were cultured from blood samples at Children's Hospital between 1998 and 2006 (n = 159) and that were still viable were included. Pneumococci were stored in tubes containing glass beads prior to 2002; after that date, the freezing procedure included 20% glycerol. The strains were streaked onto tryptic soy agar with 5% sheep's blood and incubated overnight at 37°C in 5% CO2. Initial pneumococcal identification was made from blood cultures by colony morphology, α-hemolysis, and optochin susceptibility or bile solubility or by detection of the pneumococcal capsule by the Pneumoslide assay (Becton Dickinson and Company).
Colony PCR for the presence of pilus islet or pneumolysin genes.
The S. pneumoniae isolates were streaked overnight on blood agar plates. The bacteria were washed in phosphate-buffered saline, boiled for 10 min at 94°C in ThermoPol buffer (1×; New England Biolabs, Beverly MA), and then centrifuged and cooled on ice. In every PCR, 1 μl of the supernatant of the boiled bacteria was used. PCRs were performed with 0.5 μl of Taq polymerase (New England Biolabs), each deoxynucleoside triphosphate at a concentration of 250 μM, and each primer at a concentration of 0.3 μM in a final volume of 25 μl. The reaction conditions consisted of 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min/kb, followed by 5 min at 72°C. The primers were designed to amplify the three genes that encode proteins that constitute the core of the pilus: rrgA (primers pQE30A5 [5′-ATTAGGATCCAAGATATTTCAGAAGGCAGTTGCA-3′] and pQE30A3 [5′-TTAAGCATGCTTCTCTCTTTGGAGGAATAGGTTC-3′]), rrgB (primers pQE30B5 [5′-ATTAGGATCCCTTGCTGCCTTATTACTGA-3′] and pQE30B3 [5′-TATTGCATGCGATAGTGATTTTTTTGTTGAC-3′]), and rrgC (primers pQE30C5 [5′-ATCAGGATCCGCTCTGTGTTTTTCTCTTGTATGG-3′] and pQE30C3 [5′-ATCCGCATGCATCAATCCGTGGTCGCTTGTTATTTTTA-3′]). In addition, we also designed primers TTM301 and TTM302 (5′-ACTTGTTGATGTTGGTAGCCTGTT-3′ and 5′-TTCCGACGGTAAAATAATCTCTGT-3′, respectively) for detection of the sortase D gene (srtD), which is part of the pilus islet. As a positive control, PCR with primers Plyup and Plydw, described by Whatmore et al. (28), for detection of the ply gene was also performed with each strain.
MLST.
All isolates from the American Indian data set were subjected to multilocus sequence typing (MLST; http://spneumoniae.mlst.net/) by using two forward sequences and two reverse sequences for every gene (16a). The term sequence type is used to indicate a distinct MLST type pattern. Sequence type complexes were assigned by using the eBURST algorithm (9) with the software available from the MLST home page. The MLST type pattern information for all strains typed in this study is in the MLST database at http://spneumoniae.mlst.net.
Statistical analysis.
Differences in the frequencies of the presence of the pilus genes between groups were analyzed by Fisher's exact test. Analyses were performed by using SPSS for Macintosh 11.0 software. For all comparisons, a P value of <0.05 was considered to represent statistical significance.
RESULTS
Development of PCR screening for presence of pilus genes.
Data on the genetic similarity of the pneumococcal pilus genes are limited. In pilot studies, we evaluated the performance of PCRs for the detection of the rrgA, rrgB, rrgC, and srtD genes in over 80 clinical isolates, using strains TIGR4 and Rx1 as positive and negative controls, respectively. The PCR for the rrgC gene gave the most sensitive results; in no case did we find a strain for which the PCR for rrgA, rrgB, or srtD was positive and the rrgC PCR was negative (data not shown). These results are consistent with those obtained in a recent study in which the amino acid sequence identities of the three pilus proteins RrgA, RrgB, and RrgC between two clinical isolates (isolate TIGR4 and a 6B clinical isolate) were compared; that study showed that RrgC had the highest degree of similarity between the two strains (amino acid sequence identities of RrgA, RrgB, and RrgC between the two strains, 83%, 47%, and 99%, respectively) (11). Therefore, we defined a strain as having the pilus if the PCR for rrgC was positive; PCRs for rrgC and ply were thus performed with all strains. A strain was considered pilus negative if the rrgC PCR of the colony showed a negative result and the ply PCR of the same colony was positive.
Frequency of the rrgC gene in the American Indian pneumococcal collection.
Overall, 104 (21.5%) of 484 strains tested positive for the pilus rrgC gene. The frequency of this gene was similar between the NP isolates (74 of 361; 20.5%) and the invasive isolates, which included 2 isolates from cerebrospinal fluid (30 of 123; 24.4%). This difference was not statistically significant (relative risk of having the rrgC gene for invasive versus that for NP strains, 1.05; 95% confidence interval, 0.94 to 1.18; P = 0.375, Fisher's exact test). Hence, the presence of rrgC per se does not appear to be associated with increased virulence, since rrgC is present at similar frequencies in NP and bacteremic isolates.
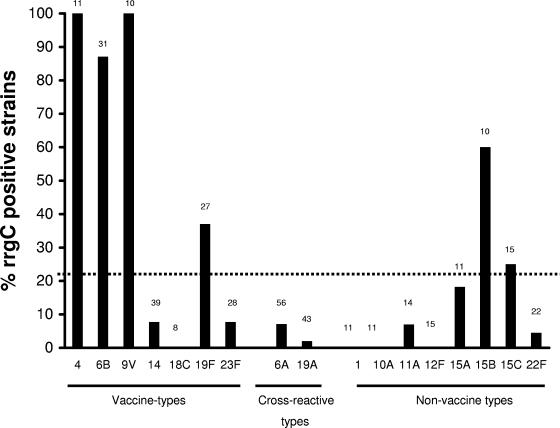
Next we evaluated the frequency of rrgC by capsular serotype (including all the vaccine serotypes and, for cross-reactive or nonvaccine serotypes, only those serotypes for which the collection contained ≥10 strains). As shown in Fig. 1, the prevalence of rrgC differed by serotype. In particular, strains of several vaccine serotypes (serotypes 4, 6B, and 9V) were far more likely to have the rrgC gene than strains of the non-vaccine type (frequencies of rrgC-positive strains in vaccine-type strains and non-vaccine-type strains in the entire data set, 41.7% and 15.7%, respectively; P < 0.001). Strains of potentially cross-reactive vaccine serotypes in the data set (serotypes 6A, 9A, 9L, 18A, 19A, 23A, and 23B) were also significantly less likely to carry this gene than vaccine-type strains (8 of 123 strains; 6.5% of strains were rrgC positive; P < 0.001 compared to the results for the vaccine-type strains by Fisher's exact test).
FIG. 1.
Frequency of rrgC gene among the isolates from the American Indian pneumococcal collection, by serotype. For vaccine-type serotypes, data from all isolates are shown; for non-vaccine-type serotypes, only data from serotypes that included 10 or more isolates are represented. The total number of isolates for each serotype is shown at the top of each column.
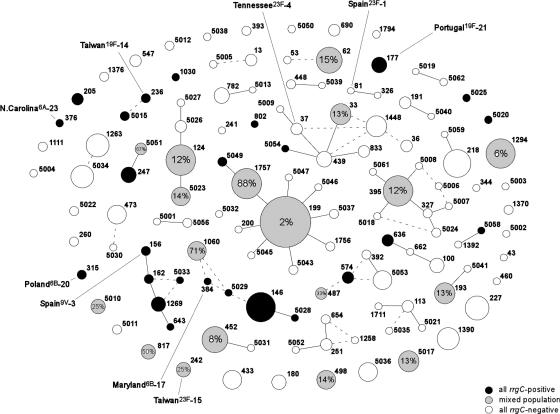
Among the 430 isolates belonging to an MLST type for which there was more than one isolate in our collection (and, thus, for which we could evaluate concordance with respect to the presence of rrgC), the genotypes of only 5.4% of the strains were discordant from the predominant genotype (rrgC positive or negative). Figure 2 shows the frequency of the rrgC gene in each sequence type in the American Indian collection. Of the 26 defined pneumococcal molecular epidemiology network epidemic sequence types (18), 9 were represented in the American Indian collection; of these, the frequency of the rrgC gene was 100% in 6 types (Spain 9V-3 sequence type 81 [ST81], Maryland 6B-17 ST384, Taiwan 19F-14 ST236, Poland 6B-20 ST315, Portugal 19F-21 ST315, and North Carolina 6A-23 ST376).
FIG. 2.
Frequency of the pilus rrgC gene among the 484 isolates from the American Indian pneumococcal collection obtained by use of the output of eBURST, version 3. Each circle represents a single MLST, with the area proportional to the number of isolates of that type. White, black, and gray circles, MLST types with 0% rrgC positive isolates, 100% rrgC positive isolates, and a mixed population, respectively. The number within the gray circles represents the percentage of rrgC-positive isolates within the MLST types with mixed populations. Black lines represent single-locus variants, while dashed lines represent two-locus variants.
Frequency of rrgC in strains from the Children's Hospital collection: reduction in the era of immunization with the pneumococcal conjugate vaccine.
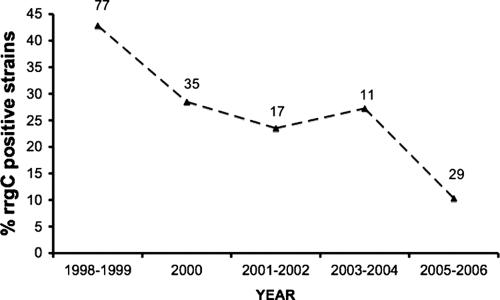
The strong association between the presence of the rrgC gene and certain PCV7 serotypes raised the possibility that, following the advent of universal immunization with pneumococcal conjugate vaccination in U.S. children, there may have been a significant reduction in the overall frequency of the rrgC gene. To examine this, we compared the frequencies of rrgC-positive strains from a collection of isolates from blood cultures collected at times spanning the years before and after the introduction of the conjugate vaccine. A total of 159 blood culture isolates were analyzed for the presence of the rrgC gene by PCR. Overall, 53 (33%) strains were positive for the rrgC gene. The frequency of the pilus rrgC gene did indeed decline from a frequency of 43% (33/77) before universal vaccination (1998 and 1999) to a frequency of 21% (10/47) after universal vaccination (P = 0.019 by Fisher's exact test). The year-by-year frequencies are shown in Fig. 3.
FIG. 3.
Frequency of the rrgC gene among isolates from the Children's Hospital pneumococcal collection, by year of isolation (dashed line). The total number of strains isolated from blood cultures per year is indicated above the line. Immunization with the universal pneumococcal conjugate vaccine in the United States began in 2000.
DISCUSSION
The advent of the heptavalent conjugate vaccine has dramatically changed the epidemiology of pneumococcal disease in the United States. Vaccine-associated reductions in pneumococcal disease can be seen not only in immunized children but also in adults. It has been estimated that the conjugate vaccine in the United States has prevented more than twice as many cases of invasive pneumococcal disease through its indirect effects on pneumococcal transmission (i.e., herd immunity) as it has through its direct effect of protecting vaccinated children (4). While this vaccine is highly effective against the vaccine serotypes, this vaccine strategy may eventually be limited by the phenomenon of serotype replacement (6, 8, 13, 14), limited serotype coverage, high cost, and difficulties in production. Thus, alternative vaccines that can offer protection against pneumococcal colonization and/or disease are being sought. Several pneumococcal proteins have been proposed as potential vaccine candidates, based primarily on their demonstrated role in virulence in animal models (2). Most recently, the three proteins that constitute the structure of the pilus have been shown to protect mice against invasive disease and have thus been proposed as potential vaccine candidates (11).
Our data suggest several limitations for the usefulness of a pilus-based vaccine. Our study documents the frequency of strains with the rrgC gene in two large collections of pneumococci. In the American Indian collection, the rrgC gene was not prominently represented in the pneumococcal population (it was present in only 21.4% of strains). Nevertheless, if the pilus was associated with increased virulence for invasive disease in humans, a vaccine based on pilus proteins may be advantageous, as it would theoretically have an impact on the most invasive strains. However our data do not support this view. There was no difference in the frequency of rrgC-positive strains in NP and bacteremic isolates in the American Indian pneumococcal collection. Our data suggest that the pilus does not in fact represent an important virulence factor for invasive disease in humans, despite the data obtained from mouse studies (1, 12). Finally, regardless of these considerations, the conjugate pneumococcal vaccine has had a surprisingly selective impact on pneumococci carrying the rrgC gene, further reducing the potential of a strategy based on immunization with pilus proteins for countries that have implemented universal vaccination with the conjugate.
Our study raises interesting questions regarding the pilus and its role in pathogenesis. First, the pilus appears to be more prevalent in some serotypes than in others. From our data, it does not appear that the pilus is associated with increased virulence, which could have otherwise explained its overrepresentation in some vaccine-type strains. Conversely, one could imagine that strains that are overrepresented in the NP would be more likely to acquire the pilus from other strains. Again, this does not appear to be the case: serotype 6A was the most commonly isolated serotype among the NP isolates (n = 50), with only 8% being pilus positive; in contrast, serotype 4 was rare among the NP isolates (n = 4), but all had the pilus rrgC gene. Why the pilus would be more common in certain capsular serotypes (and, in particular, in vaccine-type strains) is not clear at this time.
The similar frequency of rrgC-positive strains among NP and bacteremic isolates implies that the pilus does not play an important role in virulence. This result could be due, in part, to the increased susceptibility of Native Americans to invasive pneumococcal disease (5), making it less likely that an effect of the pilus on virulence could be appreciated. However, the frequency of the rrgC gene in vaccine-type strains in the American Indian pneumococcal collection (41.7%) is remarkably similar to the frequency of that gene in the Children's Hospital Boston collection in the pre-PCV7 (Prevnar) era (42.8%), during which time >80% of invasive strains were of the vaccine type. These data, obtained from pneumococci isolated from very different populations, thus provide further support for our findings.
Our study has some limitations. First, the criterion that we used to define a strain as having the pilus (positive by PCR for the rrgC gene) may be too stringent. It is conceivable that rrgC-negative strains could nevertheless express the pilus. While the exact function of the protein encoded by rrgC is not known at this time, it is likely that this protein may be added first during pilus formation (1); and studies have shown that while deletion of rrgB and rrgC prevents pilus formation, deletion of rrgC alone does not (16). Nevertheless, as indicated above, we did not find rrgC-negative strains that carry other genes of the pilus islet. Second, because the heterogeneity of this gene is unknown, it is possible that the PCR primers that we used did not amplify the rrgC gene in some strains, which would underestimate the prevalence of the pilus in our two collections. As is the case with another pneumococcal protein, pneumococcal surface protein A, there may be sufficient heterogeneity in the rrgC gene such that a PCR amplification strategy may be insensitive (24). Barring the availability of genomic sequences from a large number of strains, this possibility cannot be formally excluded. However, the fact that we never identified strains, in a subset of isolates tested, that were positive for any of the three other pneumococcal pilus genes tested (rrgA, rrgB, and srtD) would suggest that the primers used to amplify rrgC were sufficiently permissive.
Conversely, it is also conceivable that our criterion may be too broad; perhaps some strains that carry the rrgC gene do not have the other genes essential for pilus expression. A thorough evaluation of this possibility would require either assays for each gene needed for pilus expression in all these strains, which is impractical, or the availability of highly sensitive and specific antibodies that can detect the intact pilus, but such antibodies are not currently available. Although we cannot completely exclude this possibility, it seems unlikely. First, we found a high concordance between the presence of rrgC and srtD, a gene essential for pilus expression. Furthermore, the genetic structure of the rlrA islet suggests that it constitutes a mobile element which comprises all the genes needed for pilus expression (12), making it likely that a strain having the rrgC gene would also have the other pilus genes.
Our study provides data regarding the frequency of the rrgC gene in two collections of pneumococcal isolates and information regarding the possible role of the pilus in invasive disease in humans. Studies with mice have suggested that the pilus plays an important role in virulence (1, 12); however, the pneumococcal pilus does not appear to be associated with increased virulence in humans. Furthermore, because of the association of the rrgC gene with certain vaccine serotypes and the success of the conjugate vaccine in reducing the frequency of disease caused by these strains, it appears that the frequency of the pilus in the pneumococcal population has been significantly reduced where PCV7 has been used. Therefore, our data do not support the inclusion of pilus antigens in a candidate protein-based vaccine against pneumococcus.
Acknowledgments
We gratefully acknowledge the support from the Pamela and Jack Egan Fund. R.M.'s work is supported by grants from the National Institutes of Health (grants AI067737-01 and AI066013-01A2) and from PATH. M.L.'s and K.T.'s work was supported by grant 5 R01 AI048935 from the National Institutes of Health. The American Indian PnCRM7 Efficacy Trial and the nested NP study were supported by the National Institutes of Health, Wyeth Vaccines, USAID, and the Centers for Disease Control and Prevention (funding for M.S. and K.L.O.).
We thank the field workers, nurses, and staff of the Center for American Indian Health for their dedicated work on the PnCRM7 Efficacy Trial, from which the isolates used in this study were collected. We also acknowledge the willing participation and commitment of the families who participated in that trial. We thank Janet Matsubara and all the members of the Children's Hospital Boston Division of Infectious Diseases Diagnostics who provided isolates from Children's Hospital Boston. We are also very grateful to Porter Anderson and Michael Wessels for helpful discussions and suggestions during the course of this work.
The opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the Indian Health Service.
We declare that they have no competing interests.
Footnotes
Published ahead of print on 28 March 2007.
REFERENCES
- 1.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 103:2857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briles, D. E., S. Hollingshead, A. Brooks-Walter, G. S. Nabors, L. Ferguson, M. Schilling, S. Gravenstein, P. Braun, J. King, and A. Swift. 2000. The potential to use PspA and other pneumococcal proteins to elicit protection against pneumococcal infection. Vaccine 18:1707-1711. [DOI] [PubMed] [Google Scholar]
- 3.Bronsdon, M. A., K. L. O'Brien, R. R. Facklam, C. G. Whitney, B. Schwartz, and G. M. Carlone. 2004. Immunoblot method to detect Streptococcus pneumoniae and identify multiple serotypes from nasopharyngeal secretions. J. Clin. Microbiol. 42:1596-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2005. Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease—United States, 1998-2003. Morb. Mortal. Wkly. Rep. 54:893-897. [PubMed] [Google Scholar]
- 5.Cortese, M. M., M. Wolff, J. Almeido-Hill, R. Reid, J. Ketcham, and M. Santosham. 1992. High incidence rates of invasive pneumococcal disease in the White Mountain Apache population. Arch. Intern. Med. 152:2277-2282. [PubMed] [Google Scholar]
- 6.Dagan, R., N. Givon-Lavi, O. Zamir, M. Sikuler-Cohen, L. Guy, J. Janco, P. Yagupsky, and D. Fraser. 2002. Reduction of nasopharyngeal carriage of Streptococcus pneumoniae after administration of a 9-valent pneumococcal conjugate vaccine to toddlers attending day care centers. J. Infect. Dis. 185:927-936. [DOI] [PubMed] [Google Scholar]
- 7.Dramsi, S., E. Caliot, I. Bonne, S. Guadagnini, M. C. Prevost, M. Kojadinovic, L. Lalioui, C. Poyart, and P. Trieu-Cuot. 2006. Assembly and role of pili in group B streptococci. Mol. Microbiol. 60:1401-1413. [DOI] [PubMed] [Google Scholar]
- 8.Eskola, J., T. Kilpi, A. Palmu, J. Jokinen, J. Haapakoski, E. Herva, A. Takala, H. Kayhty, P. Karma, R. Kohberger, G. Siber, and P. H. Makela. 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N. Engl. J. Med. 344:403-409. [DOI] [PubMed] [Google Scholar]
- 9.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaspar, A. H., and H. Ton-That. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 188:1526-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gianfaldoni, C., S. Censini, M. Hilleringmann, M. Moschioni, C. Facciotti, W. Pansegrau, V. Masignani, A. Covacci, R. Rappuoli, M. A. Barocchi, and P. Ruggiero. 2007. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 75:1059-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, S. S., R. Platt, S. L. Rifas-Shiman, S. I. Pelton, D. Goldmann, and J. A. Finkelstein. 2005. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics 116:e408-e413. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan, S. L., E. O. Mason, Jr., E. R. Wald, G. E. Schutze, J. S. Bradley, T. Q. Tan, J. A. Hoffman, L. B. Givner, R. Yogev, and W. J. Barson. 2004. Decrease of invasive pneumococcal infections in children among 8 children's hospitals in the United States after the introduction of the 7-valent pneumococcal conjugate vaccine. Pediatrics 113:443-449. [DOI] [PubMed] [Google Scholar]
- 15.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B Streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 16.LeMieux, J., D. L. Hava, A. Basset, and A. Camilli. 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 74:2453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Lipsitch, M., K. O'Neill, D. Cordy, B. Bugalter, K. Trzcinski, C. M. Thompson, R. Goldstein, S. Pelton, H. Huot, V. Bouchet, R. Reid, M. Santosham, and K. L. O'Brien. Strain characteristics of Streptococcus pneumoniae isolated from carriage and invasive disease during a cluster-randomized clinical trial of the 7-valent pneumococcal conjugate vaccine. J. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 17.Maisey, H. C., M. Hensler, V. Nizet, and K. S. Doran. 2007. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 189:1464-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGee, L., H. Wang, A. Wasas, R. Huebner, M. Chen, and K. P. Klugman. 2001. Prevalence of serotypes and molecular epidemiology of Streptococcus pneumoniae strains isolated from children in Beijing, China: identification of two novel multiply-resistant clones. Microb. Drug Resist. 7:55-63. [DOI] [PubMed] [Google Scholar]
- 19.Mora, M., G. Bensi, S. Capo, F. Falugi, C. Zingaretti, A. G. Manetti, T. Maggi, A. R. Taddei, G. Grandi, and J. L. Telford. 2005. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. USA 102:15641-15646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moulton, L. H., K. L. O'Brien, R. Kohberger, I. Chang, R. Reid, R. Weatherholtz, J. G. Hackell, G. R. Siber, and M. Santosham. 2001. Design of a group-randomized Streptococcus pneumoniae vaccine trial. Control. Clin. Trials 22:438-452. [DOI] [PubMed] [Google Scholar]
- 21.O'Brien, K. L., L. H. Moulton, R. Reid, R. Weatherholtz, J. Oski, L. Brown, G. Kumar, A. Parkinson, D. Hu, J. Hackell, I. Chang, R. Kohberger, G. Siber, and M. Santosham. 2003. Efficacy and safety of seven-valent conjugate pneumococcal vaccine in American Indian children: group randomised trial. Lancet 362:355-361. [DOI] [PubMed] [Google Scholar]
- 22.Paterson, G. K., and T. J. Mitchell. 2006. The role of Streptococcus pneumoniae sortase A in colonisation and pathogenesis. Microbes Infect. 8:145-153. [DOI] [PubMed] [Google Scholar]
- 23.Scott, J. R., and D. Zahner. 2006. Pili with strong attachments: gram-positive bacteria do it differently. Mol. Microbiol. 62:320-330. [DOI] [PubMed] [Google Scholar]
- 24.Swiatlo, E., A. Brooks-Walter, D. E. Briles, and L. S. McDaniel. 1997. Oligonucleotides identify conserved and variable regions of pspA and pspA-like sequences of Streptococcus pneumoniae. Gene 188:279-284. [DOI] [PubMed] [Google Scholar]
- 25.Swierczynski, A., and H. Ton-That. 2006. Type III pilus of corynebacteria: pilus length is determined by the level of its major pilin subunit. J. Bacteriol. 188:6318-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53:251-261. [DOI] [PubMed] [Google Scholar]
- 27.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 28.Whatmore, A. M., S. J. King, N. C. Doherty, D. Sturgeon, N. Chanter, and C. G. Dowson. 1999. Molecular characterization of equine isolates of Streptococcus pneumoniae: natural disruption of genes encoding the virulence factors pneumolysin and autolysin. Infect. Immun. 67:2776-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]