Abstract
We developed a simple, specific, and sensitive two-multiplex-PCR assay that enabled the detection of all known group B streptococcal (GBS) capsular polysaccharides. This test is well adapted for GBS capsular polysaccharide typing in large-scale epidemiological studies.
Group B streptococci (GBS; Streptococcus agalactiae) are a leading cause of invasive infections in neonates and a serious cause of mortality or morbidity in adults with underlying diseases (13). Nine distinct capsular polysaccharide (CPS) serotypes have been described (6, 7). The CPS is commonly used for strain typing. The commercial kits most widely used are based on latex agglutination (LA), but these tests are only moderately reliable, resulting in nontypeability (NT) or erroneous serotyping of the isolates. Therefore, molecular capsular typing techniques are attractive because they are reproducible, specific, and easy to perform. Different genotypic methods have been described for the molecular capsular typing of GBS (1, 2, 8, 9, 14, 17). However, while these techniques are relatively easy to perform in the routine laboratory, they all involve the conjunction of two different techniques, e.g., PCR plus sequencing, PCR plus hybridization, or PCR plus enzymatic restriction. We report in this work on a simple multiplex PCR assay which enables the detection of all known GBS CPSs.
The DNA sequences of the cps operons of all GBS CPSs that have been described have recently been made available (3). The nine cps DNA sequences were analyzed by using Beacon Designer 5.1 software to generate CPS-specific primer pairs, which enabled the amplification of fragments of different sizes that could be easily discriminated by agarose gel electrophoresis. Primer specificity was tested against the sequences in the GenBank database by using BLAST searches to verify the absence of serendipitous similarities. PCR simulations were carried out by using AmplifX 1.37 software. Primers that met these criteria and that were specific for sequences corresponding to CPS types Ia, Ib, II, IV,V, VI, VII, and VIII were identified; and the most appropriate pairs, which were selected on the basis of similar melting temperatures and the ability to generate distinguishable amplicon sizes, were retained (Table 1). Due to the high degree of sequence similarity of these loci, we failed to define primers specific for CPS type III. We therefore selected the pairs with the lowest potential for cross-hybridization with other cps operons. As shown in Table 1, all but one of the primer pairs were predicted to be CPS type specific, whereas the primer pair used to detect type III strains was expected to cross-react with type Ia and II strains. Moreover, the size differences between the amplicons allowed us to readily identify each CPS type, based on the electrophoretic mobility of the corresponding PCR product (Table 1).
TABLE 1.
CPS type-specific primers and prediction of PCR products by computer simulation
| Primer name | Sequence (5′ to 3′) | Gene target(s) | Amplicon size(s) (bp) | GenBank accession no. of targeted operon |
|---|---|---|---|---|
| Ia-F | GGTCAGACTGGATTAATGGTATGC | cps1aH | AB028896 | |
| Ia-R | GTAGAAATAGCCTATATACGTTGAATGC | cps1aH | 521 and 1,826 | |
| Ib-F | TAAACGAGAATGGAATATCACAAACC | cps1bJ | AB050723 | |
| Ib-R | GAATTAACTTCAATCCCTAAACAATATCG | cpsIbK | 770 | |
| II-F | GCTTCAGTAAGTATTGTAAGACGATAG | cps2K | AY375362 | |
| II-R | TTCTCTAGGAAATCAAATAATTCTATAGGG | cps2K | 397 | |
| III-Fa | TCCGTACTACAACAGACTCATCC | cps1a/2/3I | AF163833 | |
| III-Ra | AGTAACCGTCCATACATTCTATAAGC | cps1a/2/3J | 1,826 | |
| IV-F | GGTGGTAATCCTAAGAGTGAACTGT | cps4N | AF355776 | |
| IV-R | CCTCCCCAATTTCGTCCATAATGGT | cps4N | 578 | |
| V-F | GAGGCCAATCAGTTGCACGTAA | cps5O | AF349539 | |
| V-R | AACCTTCTCCTTCACACTAATCCT | cps5O | 701 | |
| VI-F | GGACTTGAGATGGCAGAAGGTGAA | cps6I | AF337958 | |
| VI-R | CTGTCGGACTATCCTGATGAATCTC | cps6I | 487 | |
| VII-F | CCTGGAGAGAACAATGTCCAGAT | cps7M | AY376403 | |
| VII-R | GCTGGTCGTGATTTCTACACA | cps7M | 371 | |
| VIII-F | AGGTCAACCACTATATAGCGA | cps8J | AY375363 | |
| VIII-R | TCTTCAAATTCCGCTGACTT | cps8J | 282 | |
| dltS-F | AGGAATACCAGGCGATGAACCGAT | dltS | AL766853 | |
| dltS-R | TGCTCTAATTCTCCCCTTATGGC | dltS | 952 |
Primers III-F and III-R never yielded the expected amplicon with DNA templates extracted from all GBS serotype II strains tested in this study.
The specificity and efficiency of each primer pair used separately were determined by PCR with DNA extracted from 33 GBS strains representative of all nine serotypes (n = 5 strains each for types Ia, Ib, II, III, IV, and V and n = 3 strains each for types VI, VII, and VIII). This analysis included the sequenced strains A909 (type Ia), NEM316 (type III), and 2603 V/R (type V) (5, 15, 16). The expected PCR patterns were obtained with all primer pairs and strains except primers III-F and III-R, which did not yield the expected 1,826-bp fragment with the five serotype II strains tested (data not shown). The specificities of the PCRs were assessed by sequencing the PCR products derived from the 33 strains. As expected, all sequenced amplicons displayed >98% identity with the corresponding CPS reference sequence.
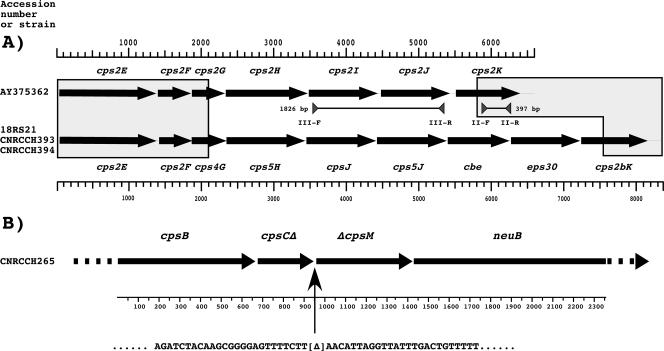
Our finding that primers III-F and III-R did not hybridize with any serotype II clinical isolates suggested that the sequence with GenBank accession number AY375362 (3) might not be representative of cps operons encoding the serotype II CPS. Sequencing of the central region of the cps operons containing the genes presumably targeted by primers III-F and III-R of two unrelated CPS type II GBS isolates (isolates CNRCCH393 and CNRCCH394) revealed that these loci were identical to that of type II strain 18RS21 (Fig. 1) (4, 15). However, comparison of the strain 18RS21 sequence and the sequence with GenBank accession number AY375362 revealed that only their 5′ and 3′ extremities were identical, whereas the internal segments were different in size and gene content (Fig. 1A). Based on our results and those presented in previous reports (10, 17), we propose that the strain 18RS21 sequence should be considered the cps2 sequence prototype. The 18RS21 and AY375362 sequences could be designated cps2a and cps2b, respectively. Thus, at least two different cps2 loci, cps2a and cps2b, could encode serotype II CPS.
FIG. 1.
Schematic representation of GBS cps loci. (A) Sequence analysis of two unrelated cps loci encoding the GBS serotype II capsular polysaccharide in strains CNRCCH393 and CNRCCH394. The gene nomenclatures are those used in the published sequence with GenBank accession number AY375362, thought to constitute the prototype sequence for type II cps loci (3), and the published sequence with GenBank accession number AAJO01000077 (strain 18RS21). The gray boxes indicate regions of sequence identity. Note that primers II-F and II-R match the 3′ moiety of cps2K present in all sequences, whereas primers III-F (specific for the 5′ extremity of cps2I) and III-R (specific for the 3′ extremity of cps2J) appear to be specific for the sequence with GenBank accession number AY375362. Sequencing of the strain CNRCCH393 and CNRCCH394 cps2 loci was done by chromosome walking between the 5′ and 3′ ends of cps2E and cps2K, respectively. Scales are indicated in base pairs. (B) Sequence analysis of the cps locus of NT strain CNRCCH265. Sequencing of this locus was done by chromosome walking between the 5′ and 3′ ends of cpsB and neuB, respectively. This strain should synthesize a COOH-truncated form of CpsC (CpsCΔ) but should not express any CpsM derivatives (ΔCpsM) because the appropriate translational signals have been deleted. The scale is indicated in base pairs.
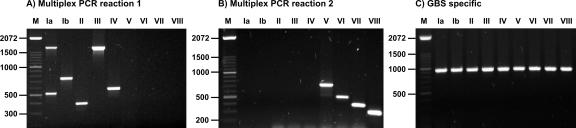
Our aim was to develop a simple multiplex PCR assay that enables accurate GBS CPS typing. Two primer mixes (with mix I containing primer pairs specific for CPS types Ia, Ib, II, III, and IV and mix II containing primer pairs specific for CPS types V, VI, VII, and VIII) were used in separate PCRs. In every reaction, the CPS type could be unambiguously determined, with each type possessing a characteristic electrophoretic pattern (Fig. 2A and B). In this assay, the systematic use of two PCR mixes per strain provided an internal negative control, as only one mix should produce a PCR fragment. A third PCR (with primer pair dltS-F and dltS-R) targeting the GBS-specific dltS gene (11) was also included as an internal positive control (Fig. 2C).
FIG. 2.
Representative PCR multiplex reactions. The CPS types of the strains analyzed are indicated above their respective lanes. (A) Multiplex PCR for detection of CPS types Ia, Ib, II, III, and IV; (B) multiplex PCR for detection of CPS types V, VI, VII, and VIII; (C) GBS-specific PCR. Lanes M, molecular size standard (100-bp ladder; Invitrogen). The numbers on the right of each panel are in base pairs.
Four hundred twenty-six human nonredundant GBS isolates collected in different French geographical areas between 2004 and 2006 were studied. Fifty-three (12.4%) strains were isolated from patients with GBS invasive diseases: 47 (11%; 16 from neonates, 2 from children, and 29 from adults) were from blood cultures and 6 (1.4%; all from neonates) were from cerebrospinal fluid. Two hundred thirty-two strains (54%) were isolated from vaginal samples from pregnant women, and 70 strains (16%) were from colonized but noninfected neonates. Seventy-one additional strains (16.6%) were obtained from urine samples (n = 56) or pus from various sites (n = 15). All GBS isolates were serotyped by agglutination with a commercial standard kit from Essum AB (Umea, Sweden) containing immunoglobulin G binding particles coated with rabbit antibodies specific to capsular serotypes Ia, Ib, II, III, IV, and V. LA serotyping (LAS) allowed us to identify the capsular serotypes of 93% (397/426) of the strains studied (Table 2). All GBS strains were also tested by using the multiplex PCR system illustrated in Fig. 2. Following PCR with the dltS primer pair, all strains yielded the expected PCR product, which confirmed that the DNA preparations were devoid of PCR inhibitors and that the corresponding strains were GBS. A capsular genotype was assigned to 99.7% (425/426) of the isolates (Table 2). Among the 397 strains typeable by both methods, the results of PCR CPS typing and LAS were in agreement for 394 isolates (99%). The three discordant strains (LAS and PCR CPS typing results for the three strains, III and IV, respectively; V and Ib, respectively; and V and III, respectively; Table 2) were each retested in three independent LAS experiments, and the PCR products were checked by sequencing. The same discordances between the phenotypic and the genotypic assays were obtained. The CPS types of these three strains determined by PCR were considered to be correct, based on the assumption that genotypic methods are more reliable than phenotypic methods.
TABLE 2.
Concordance matrix comparing PCR typing and LAS for 426 GBS clinical isolates from various sources
| Serotype obtained by LAS | No. of strains typed as follows by PCRa:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | VI | VII | VIII | NT | Total | |
| Ia | 32 | 32 | |||||||||
| Ib | 13 | 13 | |||||||||
| II | 28 | 28 | |||||||||
| III | 189 | 1 | 190 | ||||||||
| IV | 22 | 22 | |||||||||
| V | 1 | 1 | 110 | 112 | |||||||
| VI | 0 | 0 | |||||||||
| VII | 0 | 0 | |||||||||
| VIII | 0 | 0 | |||||||||
| NT | 16 | 1 | 2 | 1 | 2 | (1) | (5) | 1 | 29 | ||
| Total | 48 | 15 | 30 | 190 | 24 | 112 | 1 | 5 | 0 | 1 | 426 |
The six strains in parentheses were typed as CpsVI and CpsVII by PCR but were considered NT by LAS because the kit used did not contain the corresponding specific antisera.
Among the 29 (7%) NT strains that did not react or that gave weak polyagglutination by LAS, 22 (5.1%) were assigned by PCR to type Ia (n = 16), Ib (n = 1), II (n = 2), IV (n = 1), and V (n = 2). Six strains (1.4%) were assigned by PCR to type VI (n = 1) and type VII (n = 5) and were classified as NT by LAS due to the absence of the corresponding antisera in our kit. A single strain, CNRCCH265, could not be typed by either LAS or the PCR assay and was further studied. We searched by PCR for the presence of genes cpsA to -E; cpsL; and neuB, -A, -C, and -D, which are conserved in all nine GBS cps operons (3). This analysis indicated that the genes cpsC to -E and cpsL were apparently missing from this strain (data not shown). Sequence analysis revealed that a single large deletion that resulted in an out-of-frame fusion between the first half of cpsC and the second half of cpsM occurred in strain CNRCCH265 (Fig. 1B). The loss of the genes cpsC to -M is consistent with its NT phenotype. GBS isolates that are NT due to the presence of mutations or insertion sequences in the cps biosynthetic genes have been described (2, 3, 8, 9, 12, 14, 17). However, we describe here the first molecular characterization of a GBS clinical isolate bearing a large deletion within the cps operon.
In conclusion, the multiplex PCR assay described in this work provides a simple tool for GBS CPS typing. This sensitive and specific method enables the characterization of all known GBS CPSs, thereby reducing the rate of detection of NT isolates. This assay is therefore particularly well adapted for GBS CPS typing in large-scale epidemiological studies.
Nucleotide sequence accession numbers.
The GenBank/EMBL accession numbers of the sequences derived from strains 18RS21, CNRCCH393, and CNRCCH265 are AAJO01000077, AM498296, and AM498295, respectively.
Acknowledgments
We thank Edouard Bingen, René Courcol, Nicolas Fortineau, Thierry Lambert, Patrice Nordmann, Marie-Cécile Ploy, and Isabelle Podglajen for providing GBS strains from their collections and Gregory Tirell (Provincial Laboratory for Public Health and National Centre for Streptococcus, Canada) and Luc Parisé (BD Diagnostics—GeneOhm) for the gifts of GBS strains of serotype VI, VII, and VIII. We thank Franck Letourneur and the Sequencing Platform of the Institut Cochin. We are grateful to Alexandra Gruss for critical reading of the manuscript.
This work was supported by research funds from INSERM, CNRS, University Paris Descartes, l'Institut de Veille Sanitaire, la Fondation pour la Recherche Médicale, and the Institut Pasteur (GPH no. 9).
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Amundson, N. R., A. E. Flores, S. L. Hillier, C. J. Baker, and P. Ferrieri. 2005. DNA macrorestriction analysis of nontypeable group B streptococcal isolates: clonal evolution of nontypeable and type V isolates. J. Clin. Microbiol. 43:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borchardt, S. M., B. Foxman, D. O. Chaffin, C. E. Rubens, P. A. Tallman, S. D. Manning, C. J. Baker, and C. F. Marrs. 2004. Comparison of DNA dot blot hybridization and Lancefield capillary precipitin methods for group B streptococcal capsular typing. J. Clin. Microbiol. 42:146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cieslewicz, M. J., D. Chaffin, G. Glusman, D. Kasper, A. Madan, S. Rodrigues, J. Fahey, M. R. Wessels, and C. E. Rubens. 2005. Structural and genetic diversity of group B streptococcus capsular polysaccharides. Infect. Immun. 73:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Cueninck, B. J., T. F. Greber, T. K. Eisenstein, R. M. Swenson, and G. D. Shockman. 1983. Isolation, chemical composition, and molecular size of extracellular type II and type Ia polysaccharides of group B streptococci. Infect. Immun. 41:527-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 6.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, A. Schuchat, et al. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 7.Hickman, M. E., M. A. Rench, P. Ferrieri, and C. J. Baker. 1999. Changing epidemiology of group B streptococcal colonization. Pediatrics 104:203-209. [DOI] [PubMed] [Google Scholar]
- 8.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong, F., L. Ma, and G. L. Gilbert. 2005. Simultaneous detection and serotype identification of Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization. J. Med. Microbiol. 54:1133-1138. [DOI] [PubMed] [Google Scholar]
- 10.Manning, S. D., D. W. Lacher, H. D. Davies, B. Foxman, and T. S. Whittam. 2005. DNA polymorphism and molecular subtyping of the capsular gene cluster of group B streptococcus. J. Clin. Microbiol. 43:6113-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poyart, C., M. C. Lamy, C. Boumaila, F. Fiedler, and P. Trieu-Cuot. 2001. Regulation of d-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J. Bacteriol. 183:6324-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramaswamy, S. V., P. Ferrieri, A. E. Flores, and L. C. Paoletti. 2006. Molecular characterization of nontypeable group B streptococcus. J. Clin. Microbiol. 44:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sellin, M., C. Olofsson, S. Hakansson, and M. Norgren. 2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarit y Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen, L., Q. Wang, Y. Li, F. Kong, G. L. Gilbert, B. Cao, L. Wang, and L. Feng. 2006. Use of a serotype-specific DNA microarray for identification of group B streptococcus (Streptococcus agalactiae). J. Clin. Microbiol. 44:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]