Abstract
The genetic diversity of 829 strains of Mycobacterium tuberculosis isolated during a 3-year period in Tuscany, Italy, a country with a low prevalence of tuberculosis, from 480 Italian-born and 349 foreign-born patients was determined by spoligotyping. The predominant spoligotype families were T (30.2% of isolates), Haarlem (19.9%), and the Latino-American and Mediterranean family (LAM) (11.2%); the remaining isolates were distributed among the Beijing (6.5%), S (4.2%), East Africa-India (EAI) (3.0%), Bovis (2.3%), Central Asia (CAS) (2.1%), Africanum (1.3%), and X (1.2%) families or were undefined (2.7%) or orphan (14.1%) isolates. Isolates of the families T, Haarlem, Bovis, and X were distributed among Italian- and foreign-born patients almost proportionally to the patients' numbers. Isolates of the LAM family were prevalent in foreign-born people (13.5%, versus 9.6% in Italian-born patients). Isolates of the S family were found almost exclusively in Italian-born patients, while strains of families EAI and CAS were isolated almost exclusively from foreign-born patients; Africanum isolates were all from African-born patients. The isolates of the Beijing family showed a trend to a steady increase during the survey. The prevalence of Beijing strains was 11.7% among foreign-born people and 2.7% among Italian-born patients. The Beijing strains were typed by the standardized IS6110 restriction fragment length polymorphism assay, which yielded a total of 38 distinct IS6110 patterns; 21 isolates (39.6%) occurred in six distinct clusters; of these, three contained two isolates and the other three contained four, five and six isolates, thus demonstrating that Beijing strains caused several tuberculosis outbreaks in the region. These findings indicate that transmission of Beijing strains between immigrants and the autochthonous population has occurred frequently and suggests an ongoing active transmission of the Beijing genotype in the region.
Tuberculosis (TB) is the world's second-most-common cause of death from an infectious disease, after human immunodeficiency virus/acquired immune deficiency syndrome, causing nearly 2 million deaths, 50 to 100 million infections, and 8 to 9 million new cases every year (13). As the global burden of TB falls principally on developing countries, where 95% of cases and 98% of deaths due to the disease occur (31), it is not surprising that much of TB in the Western world occurs in foreign-born immigrants from countries with a high prevalence of TB. Accurate estimates of the impact of immigration in low-prevalence countries and of other interdependent host-related factors, such as the distribution of human immunodeficiency virus/acquired immune deficiency syndrome in the population and the aging of the indigenous population (3, 25), are essential to design appropriate strategies for TB control.
The demographic characteristics of the population of Italy provide an excellent opportunity to investigate the impact of immigration on TB epidemiology in a low-prevalence Western European country. Italy, in fact, is a country with notification rates of bacteriologically confirmed TB cases in 1980 to 2004 ranging from 6 to 10 per 100,000 population (7 per 100,000 in 2001 to 2004) (30) but experiencing recent, steadily increasing immigration from high-prevalence countries.
Since the discovery of polymorphic DNA in Mycobacterium tuberculosis, molecular typing techniques have become valuable tools in the study of the epidemiology of TB (1, 19, 24). The aim of the present study was to perform a 3-year longitudinal molecular study of the genotypes of M. tuberculosis isolated in a region of central Italy, Tuscany, by PCR-based spacer oligonucleotide typing (spoligotyping) (18). Spoligotyping, in fact, has recently emerged as a rapid and reliable tool that is an alternative to the traditional IS6110 restriction fragment length polymorphism (RFLP) fingerprinting (28) method for the molecular epidemiology of TB (11, 12), as well as for the investigation of the evolutionary and population genetics of tubercle bacilli (9, 27). The last release of the international spoligotype database, SpolDB4, describes 1,939 shared types (a shared type, or ST, is defined as an identical spoligotype found in two or more individual patient isolates) and 3,370 orphan patterns consisting of entries occurring only once in the database. The STs and orphans of SpolDB4 are representative of a total of 39,295 M. tuberculosis strains from 122 countries. The STs are classified into 62 clades/lineages; some of these are ubiquitous and are spread all over the world, and others are potentially phylogeographically specific (4). Thus, analysis of the spoligotypes of the isolates may provide useful information to describe the population structure of M. tuberculosis in the region, to monitor moving and expanding genotypes, and potentially, to identify the origins and the risk factors of strains endowed with particular properties, such as multidrug resistance (MDR) or high virulence.
MATERIALS AND METHODS
Clinical isolates.
A total of 829 M. tuberculosis complex strains were collected during a 3-year study period from 1 January 2002 to 31 December 2004 from the same number of TB patients living in Tuscany, Italy, and admitted to 10 major community hospitals in the region. All isolates were identified by DNA molecular probes (AccuProbe [Gen-Probe, United States] and/or InnoLipa [Innogenetics, Belgium]). The drug susceptibilities of the isolates were determined by use of a radiometric BACTEC 460 TB or a fluorimetric MGIT 960 system (Becton Dickinson, Towson, MD) in accordance with the manufacturer's recommendations.
Spoligotyping.
The spoligotyping analysis of the strains was performed basically as described by Kamerbeek et al. (18). Briefly, genomic mycobacterial DNA was extracted from the bacteria grown on albumin-dextrose-catalase-supplemented Middlebrook 7H9 or Lowenstein-Jensen medium (Becton Dickinson) by the cetyltrimethylammonium bromide method. The oligonucleotides DRa and DRb were used as primers to amplify the whole direct-repeat (DR) region by PCR. The amplified biotinylated products were hybridized to a set of 43 immobilized oligonucleotides, each corresponding to one of the unique spacer DNA sequences within the DR locus. After hybridization, the membrane was washed, incubated in streptavidin-peroxidase conjugate (Boehringer, Germany), and finally developed by chemiluminescence. M. tuberculosis complex isolates were assigned to one of the 62 lineages/clades defined in the international spoligotype database SpolDB4 of the Pasteur Institute of Guadeloupe, France (4).
IS6110 RFLP analysis.
Extraction of DNA from mycobacterial strains and DNA fingerprinting with IS6110 as a probe were performed by the standardized method of van Embden et al. (28). In brief, after 3 to 4 weeks of growth on Lowenstein-Jensen medium, the bacteria were harvested and heat killed at 80°C for 20 min. DNA was extracted by the cetyltrimethylammonium bromide method and digested with PvuII. After electrophoresis of the digested DNA on an agarose gel, restriction fragments were blotted onto nylon sheets which were then probed with a peroxidase-labeled 245-bp IS6110-specific sequence and finally developed by chemiluminescence (Amersham). The films (Hyperfilm ECL; Amersham) were scanned, and the patterns were compared by the unweighted-pair group method using average linkages and the Dice coefficient by the use of the Gelcompar 4.1 software package (Applied Maths, Kortrijk, Belgium). Clinical isolates with identical IS6110 RFLP fingerprints or those that differed by one IS6110 band were considered to constitute one cluster.
RESULTS
Epidemiological data.
The epidemiological features of TB patients and isolates in Tuscany (Italy) from 2002 to 2004 are summarized in Table 1. The study population of 829 strains was constituted of 254 strains collected during 2002, 269 during 2003, and 306 during 2004. The male-to-female ratios were estimated as 1.3 in 2002 and 1.5 in 2003 and 2004 (1.4 for the whole 3-year period). The country of birth was known for 94% of patients; 480 (57.9%) were Italian-born, and 349 (42.1%) were foreign-born. Among these, approximately 34% were from Africa, 26% from Asia, 25% from eastern Europe, and 13% from the Americas, mostly from Peru.
TABLE 1.
Epidemiological features of study patients and isolates
| Epidemiological feature | No. (%) or ratio of patients or isolates
|
|||
|---|---|---|---|---|
| 2002 | 2003 | 2004 | Total | |
| All TB patientsa | ||||
| Total | 254 | 269 | 306 | 829 |
| No. of Italian-born patients | 157 | 153 | 170 | 480 |
| No. of foreign-born patients | 97 | 116 | 136 | 349 |
| Ratio of Italian-born/foreign-born patients | 1.6 | 1.3 | 1.2 | 1.4 |
| Ratio of patients, male/female | 1.3 | 1.5 | 1.5 | 1.4 |
| Italian-born patients in age group | ||||
| 0-14 yr | 0 | 0 | 2b | 2 (0.5) |
| 15-39 yr | 26 | 31 | 22 | 79 (21.1) |
| 40-64 yr | 28 | 37 | 41 | 106 (28.3) |
| >65 yr | 59 | 47 | 81 | 187 (50.0) |
| Foreign-born patients in age group | ||||
| 0-14 yr | 1 | 0 | 3 | 4 (1.4) |
| 15-39 yr | 69 | 78 | 91 | 238 (80.7) |
| 40-64 yr | 8 | 18 | 23 | 49 (16.6) |
| >65 yr | 0 | 2 | 2 | 4 (1.4) |
| Foreign-born patients from: | ||||
| Africa | 36 | 35 | 30 | 101 (33.7) |
| Asia | 19 | 25 | 34 | 78 (26.0) |
| Eastern Europe | 13 | 24 | 39 | 76 (25.3) |
| Americas (Central and South) | 11 | 14 | 14 | 39 (13.0) |
| Other countries | 1 | 2 | 3 | 6 (2.0) |
| Drug susceptibilities of isolates | ||||
| No. tested | 239 | 262 | 302 | 803 |
| Any drug resistance | 32 (13.4) | 41 (15.6) | 40 (13.2) | 113 (14.1) |
| MDR | 5 (2.1) | 5 (1.9) | 4 (1.3) | 14 (1.7) |
Country of birth and age were known for 94.1% and 80.7% of TB patients, respectively.
Parents of both patients (2-year-olds) are foreign-born.
During the study years, the population of Tuscany was approximately 3.5 × 106 to 3.6 × 106 persons, of whom approximately 1.7 × 106 to 2.2 × 105 (4.8 to 6.1%) were foreign-born. Assuming that no bacteriologically confirmed TB case occurring in the region was missed in our survey, the rate of culture-positive TB cases can be estimated to range from 7.2 to 8.5 per 100,000 population. These rates are obviously lower than the real incidence of the disease, which, based on notifications made to regional health authorities, was calculated to be approximately 11 per 100,000 population during the 3-year survey. Nonetheless, while the incidence of culture-positive TB was generally low (4.5 to 5.0 per 100,000) among the Italian-born population, the incidence of TB among foreign-born people was more than 10-fold higher, with values as high as 47, 49, and 55 per 100,000 population during 2002, 2003, and 2004, respectively.
The distribution of TB in the different age groups showed striking differences between Italian-born and foreign-born patients. The age group of 15 to 39 years was largely prevalent among foreign-born patients and represented 88.4% of isolates in 2002, 79.6% in 2003, and 76.5% in 2004 (80.7% of isolates, on the whole). The age group of 65 and older was significantly more represented (50%) in Italian-born patients (52% in 2002, 41% in 2003, and 55% in during 2004). When tested by spoligotyping (see below), 67.4% of M. tuberculosis isolates from these patients yielded spoligotypes which were unique (44.9%) or occurred in small clusters of two to four isolates (22.5%), which indicates that TB in the Italian-born patients 65 and older is largely due to the reactivation of latent remote infection.
The drug susceptibility results showed nearly the same percentages of isolates resistant to at least one antibiotic in 2002, 2003, and 2004, with 13.4%, 15.6%, and 13.2% resistant isolates, respectively. In general, no differences in the percentages of isolates resistant to first-line anti-TB drugs were observed between Italian-born and foreign-born patients, with the exception of resistance to isoniazid, which was significantly higher in foreign-born patients (12.7% of resistant isolates) than in Italian-born patients (7.1% of resistant isolates). Isoniazid resistance was randomly distributed among M. tuberculosis spoligotypes, thus ruling out that a particular genotype(s) might account for the higher incidence of resistance among foreign-born patients (data not shown). MDR was only observed in five isolates in 2002 and 2003 (2.1% and 1.9%, respectively) and in four isolates (1.3%) in 2004. All but two MDR isolates yielded distinct spoligotype patterns. The two clustered isolates were from patients born in Ecuador and shared the drug resistance pattern, thus suggesting a possible epidemiological link (data not shown).
Spoligotyping.
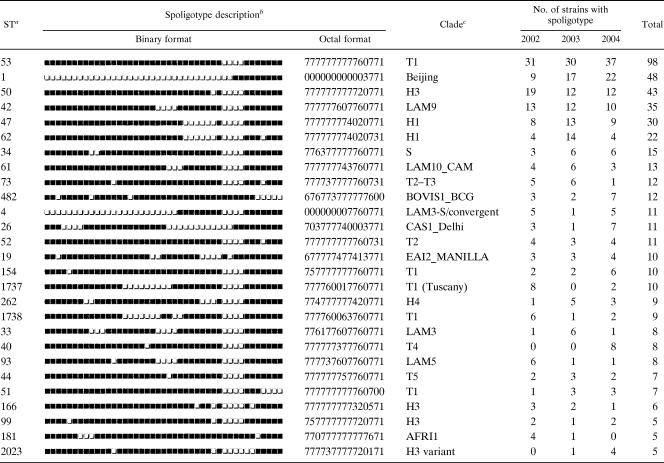
The 829 M. tuberculosis complex isolates, genotyped by the standard spoligotyping technique, generated 299 distinct patterns. The spoligotype ST designations were attributed by comparing the patterns obtained with those included in the SpolDB4 database (4). A total of 616 isolates (74.3%) were grouped in 86 distinct clusters (2 to 98 isolates per cluster). Of these, 59 included two to four isolates and were considered as minor spoligotypes, while 27 clusters containing five or more isolates each were considered as major spoligotypes and comprised 468 (75.9%) of the clustered isolates. Among these, the predominant spoligotypes, shown in Table 2, include (i) ST53, with a total of 98 isolates, a ubiquitous spoligotype belonging to clade T1; (ii) ST1, with 48 isolates, which represents the predominant spoligotype of the Beijing family; (iii) ST50, ST47, and ST62, belonging to the Haarlem family, with 43, 30, and 22 isolates, respectively; and (iv) ST42 of the LAM9 clade, with 35 isolates. Among the other clustered major spoligotypes, it is worthwhile to mention the occurrence of spoligotype ST1737, clade T1 Tuscany, with 10 isolates which likely represent autochthonous spoligotypes, as previously reported (20), or of spoligotypes which are endemic in distant geographical areas, such as ST26 of the CAS1_Delhi clade, with 11 isolates; ST19 of the EAI2_Manilla clade, with 10 isolates; and ST181 of the Africanum type 1 clade, with 5 isolates. Among the remaining 213 (25.7%) unclustered isolates, 99 (11.9%) were not yet reported to the SpolDB4 database and represented the true orphan isolates, indicated with ST0; the remaining 114 (13.8%) unclustered isolates, though unique, were already reported in SpolDB4, which allowed the attribution of an ST number.
TABLE 2.
Description of the major clustered isolates (5 or more cases) determined by spoligotyping
aSpoligotype designation were assigned according to the definition in the SpolDB4 database.
bThe black and white boxes indicate the presence and absence, respectively, of the specific spacer at position 1 to 43 in the DR locus.
cClades are defined according to the definition in the SpolDB4 database.
Table 3 reports the distribution in the 3-year survey of the M. tuberculosis families, defined by the spoligotype analysis, with respect to patients' country of birth. Three predominant families, i.e., T, Haarlem, and LAM, included over 60% of the isolates. The remaining isolates were distributed among the Beijing, S, EAI, Bovis, CAS, Africanum, and X families or were undefined or orphan isolates. Isolates of the families T, Haarlem, Bovis, and X were distributed among Italian- and foreign-born patients almost proportionally to the patients' numbers. Isolates of the LAM family, representing 11.2% of total isolates, were equally distributed among Italian- and foreign-born patients but, considering the higher percentage of Italian-born patients, they were prevalent in foreign-born people (13.5%, versus 9.6% in Italian-born patients). Isolates of the S family were almost exclusively found in Italian-born patients, while strains of families EAI and CAS were almost exclusively isolated from foreign-born (mostly Asian) patients; Africanum isolates were all from African-born patients.
TABLE 3.
Distribution of major M. tuberculosis families among STs isolated during 2002 to 2004 in Tuscany, Italy
| Spoligotype family | No. (%) of isolates
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2002
|
2003
|
2004
|
Total
|
|||||||||
| Italian-born | Foreign-born | Total | Italian-born | Foreign-born | Total | Italian-born | Foreign-born | Total | Italian-born | Foreign-born | Total | |
| T | 58 | 20 | 78 (30.7) | 51b | 21 | 72 (26.8) | 62 | 38 | 100 (32.7) | 171 | 79 | 250 (30.2) |
| Haarlem | 38a | 17 | 55 (21.7) | 38 | 26 | 64 (23.8) | 26 | 20 | 46 (15.0) | 102 | 63 | 165 (19.9) |
| LAM | 13 | 19 | 32 (12.6) | 14 | 13 | 27 (10.0) | 19 | 15 | 34 (11.1) | 46 | 47 | 93 (11.2) |
| Beijing | 3 | 8 | 11 (4.3) | 5 | 12 | 17 (6.3) | 5 | 21 | 26 (8.5) | 13 | 41 | 54 (6.5) |
| S | 15 | 0 | 15 (5.9) | 9 | 0 | 9 (3.3) | 10 | 1 | 11 (3.6) | 34 | 1 | 35 (4.2) |
| EAI | 1 | 7 | 8 (3.1) | 1 | 8 | 9 (3.3) | 0 | 8 | 8 (2.6) | 2 | 23 | 25 (3.0) |
| Bovis | 3 | 0 | 3 (1.2) | 4 | 4 | 8 (3.0) | 7 | 1 | 8 (2.6) | 14 | 5 | 19 (2.3) |
| CAS | 0 | 7 | 7 (2.8) | 0 | 3 | 3 (1.1) | 0 | 7 | 7 (2.3) | 0 | 17 | 17 (2.1) |
| Africanum | 0 | 7 | 7 (2.8) | 0 | 2 | 2 (0.7) | 0 | 2 | 2 (0.7) | 0 | 11 | 11 (1.3) |
| LAM or S | 3 | 2 | 5 (2.0) | 1 | 0 | 1 (0.4) | 2 | 3 | 5 (1.6) | 6 | 5 | 11 (1.3) |
| X | 4 | 0 | 4 (1.6) | 2 | 2 | 4 (1.5) | 2 | 0 | 2 (0.7) | 8 | 2 | 10 (1.2) |
| Undefinedc | 6 | 1 | 7 (2.8) | 4 | 2 | 6 (2.2) | 4 | 5 | 9 (2.9) | 14 | 8 | 22 (2.7) |
| True orphans and othersd | 13 | 9 | 22 (8.7) | 24 | 23 | 47 (17.5) | 33 | 15 | 48 (15.7) | 70 | 47 | 117 (14.1) |
| Total | 254 | 269 | 306 | 829 | ||||||||
This group contains one isolate defined as “H1-S” in SpolDB4.
This group contains one isolate defined as “T1 (H?)” in SpolDB4.
Strains with a signature that is as yet undefined in SpolDB4.
Strains with ST0 (orphans) and strains with a spoligotype >1939 (not included in SpolDB4).
For all but one of the families mentioned above, the numbers of isolates remained substantially constant during the years of the survey. In contrast, the isolates of the Beijing family, which includes 48 isolates of ST1 and 6 Beijing-like isolates (defined on the basis of deletion of spacers 1 to 34), showed a trend to a steady increase during the survey (11 isolates in 2002, 17 in 2003, and 26 in 2004). The overall prevalence of Beijing strains was 6.5%, but it reached 11.7% among foreign-born people (versus 2.7% among Italian-born patients). The Beijing strains were isolated from 41 foreign-born patients (23 from China, 4 from ex-Soviet Union states, 4 from Peru, 1 each from Japan, India, Pakistan, Chile, Morocco, and Senegal, and 4 from unknown countries) and from 13 Italian-born patients.
IS6110 RFLP typing of Beijing isolates.
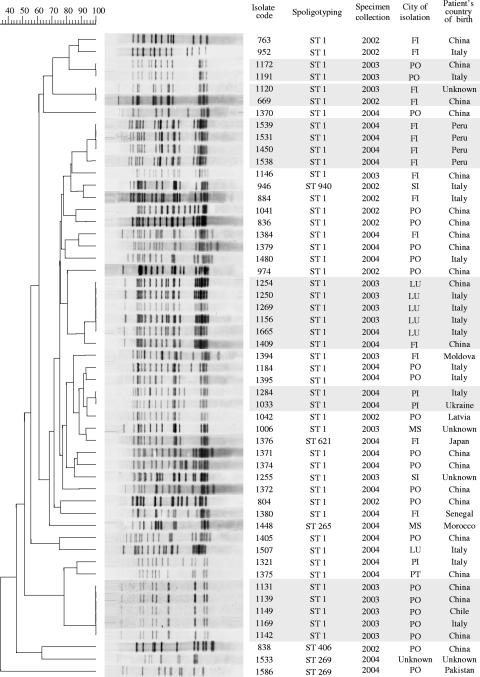
Due to their potential epidemiological and pathogenetic importance (10, 15), the Beijing strains isolated in the 3-year survey were typed by the IS6110 RFLP assay. Figure 1 shows the molecular traits of the isolates with respect to the year of specimen collection, the city of isolation, and the patient's country of birth. Fifty-three isolates were available for fingerprinting and yielded a total of 38 distinct IS6110 patterns; 21 isolates (39.6%) occurred in six distinct clusters. Of these, three contained two isolates, and the other three contained four, five, and six isolates each. In general, each cluster, which is considered to contains strains involved in epidemiologically linked TB outbreaks, included strains isolated in the same city (with only one exception) in a 1- or 2-year period. Assuming that a cluster includes one reactivation case (n-1 method) (26), the “active transmission rate” of Beijing strains in the study area can be calculated as (21 − 6)/53; that is, 28.3%.
FIG. 1.
IS6110 fingerprints of 53 clinical isolates of M. tuberculosis of the Beijing genotype isolated in Tuscany, Italy, during 2002 to 2004. The IS6110 RFLP patterns were compared, and the dendrogram was constructed by using the unweighted-pair group method using average linkages and the Dice coefficient with the Gelcompar 4.1 software package (Applied Maths, Kortrijk, Belgium). The scale on the upper left depicts similarity coefficients. Clustered identical isolates are highlighted in light-gray boxes. The isolate code, spoligotype designation, year and city of culture isolation, and patient's country of birth are shown on the right. City abbreviations: FI, Firenze (Florence); PO, Prato; SI, Siena; LU, Lucca; PI, Pisa; MS, Massa; PT, Pistoia.
DISCUSSION
The results reported in this paper demonstrate a distinct epidemiological pattern of TB in Tuscany, Italy, between Italian-born and immigrant patients. In foreign-born people, the incidence of disease was in fact 10-fold higher and the disease was highly prevalent in young people (15 to 39 years old). The spoligotype analysis of the isolates allowed us to obtain an outline of the population structure of M. tuberculosis, to detect groups of related strains, and to assess links with strains that are predominant in specific geographic areas. In our study, the majority of isolates (more than 60%) belonged to three wide-spread ST families, i.e., T, Haarlem, and LAM. The ill-defined T family, according to the SpolDB4 spoligotype database, includes ubiquitous spoligotypes which likely represent relatively old genotypes prevalent in Europe (11); the Haarlem family, of European origin, is highly prevalent in northern Europe. Isolates of these families, together with isolates of families Bovis and X, probably of Anglo-Saxon origin and known to contain strains with low IS6110 copy numbers (12), were distributed almost proportionally among Italian- and foreign-born patients. The LAM family, prevalent in Latin America and the Mediterranean region (12), was prevalent in foreign-born patients, mostly from central Africa (15 patients), North and Mediterranean Africa (10 patients), and South America (5 patients). In contrast, isolates of the S family were found almost exclusively in Italian-born patients, while strains of families EAI, prevalent in southeast Asia, east Africa, and some parts of Europe, and CAS, essentially present in central and middle eastern Asia, were almost exclusively isolated from foreign-born patients (mostly from Asia); isolates of the Africanum family were all from African-born patients. These findings indicate that at the time of the study, at least for the ST families that are phylogeographically specific to distant high-prevalence areas (i.e., EAI, CAS, and Africanum), there had been little spread of TB from the immigrant communities to the native population. This could reflect the likelihood of infection having occurred in the country of origin prior to entry to Italy or within particular ethnic communities in the host country. This is in agreement with the widely accepted hypothesis that in low-prevalence countries, foreign-born people are infected in their country of origin and then develop disease shortly after immigration (5, 7, 8, 21). Similar observations of limited transmission from foreign-born patients to United States-born individuals have been reported from molecular cluster analyses in San Francisco (6, 17) and, more recently, by Hirsh et al. (16) using deletion analysis. In this respect the hypothesis formulated by Hirsh et al. (16) that adaptation of strains of M. tuberculosis to different host populations plays a role in restricting the spread of the organism between different groups is tempting, although the factors responsible for the maintenance of stable associations between host and pathogen populations are yet to be defined.
It is noteworthy that a total of 54 of 829 (6.5%) TB cases were due to the Beijing genotype. This prevalence, although markedly lower than that reported in the former Soviet Union, where more than 40% of isolates belong to this genotype, is higher than the prevalence of Beijing isolates reported in some western European countries (10). Moreover, the Beijing family was the only one that showed a trend to a steady expansion during the years of our survey. These findings raise concerns because the Beijing genotype, which is known to be spreading worldwide (10, 15), is considered hypervirulent and an “escape mutant” from Mycobacterium bovis BCG vaccination (29). In our setting, Beijing strains were prevalent in foreign-born patients (41 out of 54 patients, i.e., 76%), mostly from China. This was expected, as Asia has been the main source of importation of Beijing strains to European countries, such as The Netherlands and Denmark (2, 22). However, our data also showed that five cases of Beijing TB occurred in patients from South America (four from Peru and one from Chile); this finding seems not to be casual, as the South American route of importation of Beijing strains to Europe has been already reported from Spain (14). In contrast to the other phylogeographically specific M. tuberculosis families, EAI, CAS, and Africanum, which are found almost exclusively in foreign-born patients as discussed above, 24% of Beijing TB cases in our survey occurred in Italian-born patients. This indicates that transmission of Beijing strains between immigrants and the autochthonous population has occurred frequently, thus suggesting an ongoing active transmission of the Beijing genotype in the region. IS6110 fingerprint analysis of the isolates supports this possibility, as it demonstrates that Beijing strains caused several TB outbreaks in the region in a relatively short time period; although it is indeed possible that some of these clustered cases were acquired in the country of birth, it is perhaps more likely that TB is being transmitted in the host country. Moreover, the rate of active transmission of Beijing strains in our 3-year survey (28.3%) appears to be significantly higher than that of non-Beijing strains (14.9%) reported in a previous paper of ours (20), thus further supporting the concept that Beijing strains are associated with a high active transmission.
There is also concern that the Beijing family may have a predilection for drug resistance, especially MDR (10, 15). In fact, the Beijing genotype of M. tuberculosis, with a higher prevalence of drug resistance mutations than non-Beijing strains, has been identified in 40 to 50% of clinical isolates recently studied in Russia (23). In our survey, only one Beijing isolate, from a patient from the former Soviet Union, was MDR (i.e., simultaneous resistance to isoniazid and rifampin) and no substantial resistance to any other anti-TB drug was observed (data not shown). This observation is not unusual, as the association of the Beijing genotype with drug resistance varies according to the country studied (10).
In conclusion, this study gives an outline of the M. tuberculosis strains circulating in Tuscany, Italy, and describes the distribution of the major phylogenetic families. It contributes to a better understanding of the current trend of TB epidemiology in a low-incidence Western country.
Acknowledgments
This work was financially supported by MIUR (PRIN-2004) and, partly, by the Italian Istituto Superiore di Sanità (National Research Program on AIDS—2006, ISS grant 50G.18).
We acknowledge Thierry Zozio's contribution for reconfirmation of the data comparison in the SpolDB4 database.
Footnotes
Published ahead of print on 25 April 2007.
REFERENCES
- 1.Barnes, P. F., and M. D. Cave. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 349:1149-1156. [DOI] [PubMed] [Google Scholar]
- 2.Borgdorff, M. W., P. de Haas, K. Kremer, and D. van Soolingen. 2003. Mycobacterium tuberculosis Beijing genotype, The Netherlands. Emerg. Infect. Dis. 9:1310-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broekmans, J. 2000. Tuberculosis control in low prevalence countries, p. 75-92. In L. Reichman and E. Hershfield (ed.), Tuberculosis: a comprehensive international approach. Dekker, New York, NY.
- 4.Brudey, K., J. R. Driscoll, L. Rigouts, W. Prodinger, A. Gori, S. A. M. Al-Hajoj, C. Allix, L. Aristimuño, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, H. M. Ly, C. Martin, C. Martin, I. Mokrousov, O. Narvskaïa, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, M. Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rüsch-Gerdes, A. Sajduda, S. Samper, P. Seth, I. Shemyakin, U. B. Singh, A. Somoskovi, R. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudey, K., M. Gordon, P. Mostrom, L. Svensson, B. Jonsson, C. Sola, M. Ridell, and N. Rastogi. 2004. Molecular epidemiology of Mycobacterium tuberculosis in western Sweden. J. Clin. Microbiol. 42:3046-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin, D. P., K. DeRiemer, P. M. Small, A. P. de Leon, R. Steinhart, G. F. Schecter, C. L. Daley, A. R. Moss, E. A. Paz, R. M. Jasmer, C. B. Agasino, and P. C. Hopewell. 1998. Differences in contributing factors to tuberculosis incidence in U.S.-born and foreign-born persons. Am. J. Respir. Crit. Care Med. 158:1797-1803. [DOI] [PubMed] [Google Scholar]
- 7.Dale, J. W., G. H. Bothamley, F. Drobniewski, S. H. Gillespie, T. D. McHugh, and R. Pitman. 2005. Origins and properties of Mycobacterium tuberculosis isolates in London. J. Med. Microbiol. 54:575-582. [DOI] [PubMed] [Google Scholar]
- 8.Diel, R., S. Rusch-Gerdes, and S. Niemann. 2004. Molecular epidemiology of tuberculosis among immigrants in Hamburg, Germany. J. Clin. Microbiol. 42:2952-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duchene, V., S. Ferdinand, I. Filliol, J. F. Guegan, N. Rastogi, and C. Sola. 2004. Phylogenetic reconstruction of Mycobacterium tuberculosis within four settings of the Caribbean region: tree comparative analyse and first appraisal on their phylogeography. Infect. Genet. Evol. 4:5-14. [DOI] [PubMed] [Google Scholar]
- 10.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. 2006. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg. Infect. Dis. 12:736-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filliol, I., J. R. Driscoll, D. Van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valétudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. Haas, H. Heersma, G. Källenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Moström, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. Ngo Niobe Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. de Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. A. Dang, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, M. L. Ho, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frieden, T. R., T. R. Sterling, S. S. Munsiff, C. J. Watt, and C. Dye. 2003. Tuberculosis. Lancet 362:887-899. [DOI] [PubMed] [Google Scholar]
- 14.García de Viedma, D., F. Chaves, and J. Iñigo. 2006. New route of importation of Mycobacterium tuberculosis Beijing genotype. Emerg. Infect. Dis. 12:169-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jasmer, R. M., A. Ponce de Leon, P. C. Hopewell, R. G. Alarcon, A. R. Moss, E. A. Paz, G. F. Schecter, and P. M. Small. 1997. Tuberculosis in Mexican-born persons in San Francisco: reactivation, acquired infection and transmission. Int. J. Tuberc. Lung Dis. 1:536-541. [PubMed] [Google Scholar]
- 18.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agderveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. M. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. A. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lari, N., L. Rindi, C. Sola, D. Bonanni, N. Rastogi, E. Tortoli, and C. Garzelli. 2005. Genetic diversity, determined on the basis of katG463 and gyrA95 polymorphisms, spoligotyping, and IS6110 typing, of Mycobacterium tuberculosis complex isolates from Italy. J. Clin. Microbiol. 43:1617-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillebaek, T., A. B. Andersen, J. Bauer, A. Dirksen, S. Glismann, P. de Haas, and A. Kok-Jensen. 2001. Risk of Mycobacterium tuberculosis transmission in a low-incidence country due to immigration from high-incidence areas. J. Clin. Microbiol. 39:855-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lillebaek, T., A. B. Andersen, A. Dirksen, J. R. Glynn, and K. Kremer. 2003. Mycobacterium tuberculosis Beijing genotype. Emerg. Infect. Dis. 9:1553-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mokrousov, I., T. Otten, A. Vyazovaya, E. Limeschenko, M. L. Filipenko, C. Sola, N. Rastogi, L. Steklova, B. Vyshnevskiy, and O. Narvskaya. 2003. PCR-based methodology for detecting multidrug-resistant strains of Mycobacterium tuberculosis Beijing family circulating in Russia. Eur. J. Clin. Microbiol. Infect. Dis. 22:342-348. [DOI] [PubMed] [Google Scholar]
- 24.Moström, P., M. Gordon, C. Sola, M. Ridell, and N. Rastogi. 2002. A survey of methods used in molecular epidemiology of tuberculosis. Clin. Microbiol. Infect. 8:694-704. [DOI] [PubMed] [Google Scholar]
- 25.Rajagopalan, S. 2001. Tuberculosis and aging: a global health problem. Clin. Infect. Dis. 33:1034-1039. [DOI] [PubMed] [Google Scholar]
- 26.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 27.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 28.van Embden, J. D. A., M. D. Cave, J. T. Crowford, J. W. Dale, K. D. Eisenhach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. S. Shinnick, and P. M. Small. 1993. Strain indentification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization. 2005. Summary by WHO region. http://www.who.int/tb/publications/global_report/2006/pdf/eur.pdf.
- 31.Zumla, A., P. Mwaba, S. B. Squire, and J. M. Grange. 1999. The tuberculosis pandemic—which way now? J. Infect. 38:74-79. [DOI] [PubMed] [Google Scholar]