Abstract
The emergence of virulent Pseudomonas aeruginosa clones is a threat to cystic fibrosis (CF) patients globally. Characterization of clonal P. aeruginosa strains is critical for an understanding of its clinical impact and developing strategies to meet this problem. Two clonal strains (AES-1 and AES-2) are circulating within CF centers in eastern Australia. In this study, phenotypic characteristics of 43 (14 AES-1, 5 AES-2, and 24 nonclonal) P. aeruginosa isolates were compared to gain insight into the properties of clonal strains. All 43 isolates produced bands of the predicted size in PCRs for vfr, rhlI, rhlR, lasA, lasB, aprA, rhlAB, and exoS genes; 42 were positive for lasI and lasR, and none had exoU. Thirty-seven (86%) isolates were positive in total protease assays; on zymography, 24 (56%) produced elastase/staphylolysin and 22 (51%) produced alkaline protease. Clonal isolates were more likely than nonclonal isolates to be positive for total proteases (P = 0.02), to show elastase and alkaline protease activity by zymography (P = 0.04 and P = 0.01, respectively), and to show elastase activity by the elastin-Congo red assay (P = 0.04). There were no other associations with genotype. Overall, increasing patient age was associated with decreasing elastase activity (P = 0.03). Thirty-two (74%) isolates had at least one N-acylhomoserine lactone (AHL) by thin-layer chromatography. rhl-associated AHL detection was associated with the production and level of total protease and elastase activity (all P < 0.01). Thirty-three (77%) isolates were positive for ExoS by Western blot analysis, 35 (81%) produced rhamnolipids, and 34 (79%) showed chitinase activity. Findings suggest that protease activity during chronic infection may contribute to the transmissibility or virulence of these clonal strains.
Pseudomonas aeruginosa lung infection is a major determinant of morbidity and mortality in cystic fibrosis (CF) patients (16). The pathogenesis of P. aeruginosa infection depends on multiple cell-associated and extracellular virulence factors including proteases (elastase [LasB], alkaline protease [AprA], and staphylolysin [LasA]), hemolysins (rhamnolipids), and toxins such as exoenzyme S (ExoS) and exotoxin A (13). Proteases contribute to pathogenesis in the lung through the induction of tissue necrosis and inflammation, destruction of surface receptors on neutrophils resulting in the inhibition of chemotaxis, phagocytosis, and the oxidative burst and degradation of surfactant proteins (27, 32).
Many P. aeruginosa virulence factors, including the proteases, are regulated by cell-to-cell communication systems that rely on diffusible N-acylhomoserine lactones (AHLs) to monitor population size in a process known as “quorum sensing” (QS) (7, 31). P. aeruginosa has two AHL-regulated circuits: las, consisting of the transcriptional activator LasR and the AHL synthase LasI, which directs the synthesis of N-(3-oxododecanoyl)-l-homoserine lactone (OdDHL), and rhl, consisting of RhlR and RhlI, which directs the synthesis of N-butanoyl-l-homoserine lactone (BHL) and N-hexanoyl-l-homoserine lactone (HHL) (44). The las circuit also regulates the expression of the rhl circuit. A third, more recently described cell-signaling system, MvfR (multiple virulence factor R), regulates Pseudomonas quinolone signal (PQS) biosynthesis and is intertwined with the las and rhl systems (31).
Early P. aeruginosa infections in lungs of CF patients are typically intermittent. However, by the teenage years, up to 80% of CF patients have become chronically infected (16). Longitudinal studies have shown that the organism adapts for long-term survival in the CF lung by the down-regulation of virulence factors required for acute infection, including the proteases and the type III secretion system (20), and the up-regulation of genes required for biofilm development (15).
It is widely accepted that most CF patients harbor unique (nonclonal) P. aeruginosa strains acquired from the environmental pool. However, over recent years, clonal strains have been identified among CF patients in several centers in the United Kingdom (21, 36), Australia (2, 30), and Canada (S. Aaron, personal communication). Two clonal strains, Australian epidemic strain 1 (AES-1) (otherwise known as m16, the Melbourne epidemic strain [MES], or PI) and AES-2 (PII), currently infect from 6% to 40% of CF patients in five clinics in eastern Australia (2, 30). CF clonal strains have not been isolated from the environment to date, suggesting person-to-person transmission, but acquisition from a common source has not been excluded.
There is now consensus among the international CF community that the emergence of clonal strains poses a potential major threat to patients’ well-being. With few exceptions, clonal P. aeruginosa strains identified in CF clinics in Australia and overseas have been associated with increased virulence compared with unique strains. This increased virulence is not fully explained by greater antibiotic resistance. AES-1 has been associated with the unexpected deaths of five children with CF who were under 5 years of age, raising particular concern over its virulence in young children (3). Subsequently, patients with the AES-1 strain have been shown to have lower lung function and increased hospitalizations compared with those with unique strains. Patients with AES-2 tend to be younger and to have significantly lower pulmonary function (30). Transmission of AES-1 to a patient with non-CF bronchiectasis has been associated with rapidly declining clinical course, causing further concern (33).
Insight into the properties of clonal strains is critical for the development of effective strategies to address this problem. The CF Liverpool epidemic strain (LES) has shown high levels of expression of virulence factors, including elastase and staphylolysin, associated with the premature activation of QS systems (34), and a virulent epidemic clone isolated from patients with microbial keratitis has been characterized by high activities of an elastase variant (26). Our previous study of AES-1, AES-2, and nonclonal P. aeruginosa isolates from subjects with CF showed that clonal isolates were more likely to have protease IV activity than nonclonal isolates (40). Enhanced protease IV activity may promote tissue destruction and inflammation and facilitate host-to-host spread. The major aim of this study was to gain further insight into properties that mediate increased infectivity or virulence in our clonal P. aeruginosa strains.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Investigations were carried out using 43 P. aeruginosa isolates from the sputa of 43 patients attending the adult Royal Prince Alfred Hospital (RPAH) CF Clinic, Sydney, Australia, between 2001 and 2004. The male-to-female ratio of the 43 patients was 21:22. The mean age was 27 years, and the age range was 18 to 58 years (Table 1). These values are consistent with those for the RPAH CF Clinic population as a whole. The study was approved by the Institutional Ethics Committee (approval X02-0320), and subjects provided informed consent for the work. All patients had been chronically infected with P. aeruginosa for a minimum of 4 years. Eighteen of the 43 (42%) isolates were derived from sputa collected at the time of exacerbation, which is defined according to criteria established by Fuchs and colleagues (11), and the remaining samples were obtained during routine clinic visits.
TABLE 1.
Characteristics of the study population
| Isolate group | Age (yr) | Exacerbation status | Mucoidy | AHL productiona (BHL/HHL/OHHL/ODHL/OdDHL) | Mean protease activity (mU/ml/OD600)b ± SD
|
ExoS | ||
|---|---|---|---|---|---|---|---|---|
| Total protease | Elastase | Staphylolysin | ||||||
| AES-1 | ||||||||
| 1 | 20 | No | ++ | +/+/+/−/− | 60 ± 14 | 0 ± 0 | 88 ± 0 | + |
| 2 | 24 | Yes | ++ | −/−/−/−/− | 35 ± 4 | 0 ± 0 | 1 ± 1 | + |
| 3 | 22 | No | ++ | +/+/+/−/− | 1,244 ± 188 | 367 ± 168 | 18 ± 6 | + |
| 4 | 31 | No | + | +/+/−/−/− | 28 ± 17 | 5 ± 6 | 2 ± 3 | + |
| 5 | 24 | Yes | ++ | −/−/−/−/− | 51 ± 24 | 5 ± 2 | 2 ± 1 | + |
| 6 | 20 | No | ++ | −/+/+/−/− | 66 ± 18 | 12 ± 10 | 2 ± 0 | + |
| 7 | 19 | Yes | + | −/+/−/−/− | 1,024 ± 103 | 75 ± 53 | 0 ± 0 | − |
| 8 | 25 | Yes | ++ | −/+/+/−/− | 830 ± 115 | 42 ± 12 | 0 ± 0 | + |
| 9 | 22 | No | − | +/+/−/−/− | 1,232 ± 50 | 81 ± 61 | 16 ± 0 | − |
| 10 | 20 | Yes | ++ | +/+/−/−/− | 1,205 ± 70 | 171 ± 121 | 51 ± 13 | + |
| 11 | 22 | No | ++ | −/−/−/−/− | 17 ± 5 | 22 ± 17 | 1 ± 0 | + |
| 12 | 29 | Yes | ++ | −/+/−/−/− | 129 ± 19 | 107 ± 98 | 1 ± 1 | + |
| 13 | 32 | No | ++ | −/+/−/−/− | 1,157 ± 46 | 135 ± 17 | 3 ± 0 | + |
| 14 | 22 | No | + | −/+/−/−/− | 160 ± 16 | 24 ± 14 | 5 ± 3 | + |
| AES-2 | ||||||||
| 15 | 18 | No | ++ | −/+/+/+/+ | 997 ± 92 | 170 ± 51 | 3 ± 4 | + |
| 16 | 28 | No | ++ | +/+/−/−/− | 92 ± 12 | 3 ± 0 | 2 ± 0 | − |
| 17 | 29 | No | + | +/+/+/+/+ | 878 ± 35 | 184 ± 35 | 6 ± 0 | + |
| 18 | 26 | No | ++ | +/+/+/+/+ | 81 ± 33 | 6 ± 7 | 0 ± 0 | + |
| 19 | 37 | Yes | ++ | +/+/−/−/− | 366 ± 76 | 68 ± 0 | 3 ± 1 | + |
| Nonclonal | ||||||||
| 20 | 20 | No | +++ | +/+/−/+/+ | 1,198 ± 84 | 282 ± 57 | 81 ± 0 | + |
| 21 | 21 | No | ++ | −/+/−/+/+ | 699 ± 259 | 73 ± 23 | 88 ± 2 | + |
| 22 | 28 | No | ++ | +/+/−/−/− | 857 ± 47 | 53 ± 21 | 9 ± 3 | − |
| 23 | 29 | No | ++ | −/−/−/−/− | 0 ± 0 | 0 ± 0 | 2 ± 0 | − |
| 24 | 21 | Yes | ++ | +/+/−/+/+ | 1,346 ± 74 | 91 ± 5 | 31 ± 0 | + |
| 25 | 19 | No | +++ | −/+/−/−/− | 784 ± 78 | 149 ± 137 | 91 ± 100 | − |
| 26 | 48 | No | +++ | −/+/−/−/− | 901 ± 351 | 53 ± 59 | 2 ± 3 | + |
| 27 | 35 | Yes | − | −/−/−/−/− | 74 ± 77 | 0 ± 0 | 2 ± 1 | + |
| 28 | 34 | No | +++ | +/+/−/+/− | 1,082 ± 26 | 98 ± 29 | 69 ± 1 | + |
| 29 | 36 | Yes | ++ | −/−/−/−/− | 66 ± 9 | 0 ± 0 | 1 ± 0 | + |
| 30 | 33 | Yes | +++ | −/−/−/−/− | 0 ± 0 | 0 ± 0 | 2 ± 1 | + |
| 31 | 26 | No | ++ | −/+/−/−/− | 78 ± 16 | 7 ± 9 | 3 ± 0 | − |
| 32 | 19 | No | + | −/−/−/−/− | 0 ± 0 | 0 ± 0 | 5 ± 3 | + |
| 33 | 20 | Yes | +++ | +/+/−/+/− | 935 ± 30 | 155 ± 31 | 4 ± 1 | + |
| 34 | 58 | Yes | + | +/+/−/−/− | 49 ± 18 | 8 ± 22 | 2 ± 1 | + |
| 35 | 22 | Yes | ++ | +/+/+/+/+ | 90 ± 70 | 10 ± 14 | 2 ± 0 | + |
| 36 | 26 | No | + | +/+/−/−/− | 818 ± 34 | 52 ± 74 | 29 ± 0 | + |
| 37 | 38 | Yes | +++ | −/+/−/+/+ | 47 ± 52 | 7 ± 10 | 7 ± 5 | − |
| 38 | 31 | No | ++ | −/−/−/−/− | 13 ± 1 | 0 ± 0 | 3 ± 0 | − |
| 39 | 19 | No | ++ | +/+/−/−/− | 0 ± 0 | 16 ± 13 | 2 ± 1 | − |
| 40 | 30 | No | + | +/+/+/−/− | 873 ± 100 | 51 ± 25 | 61 ± 1 | + |
| 41 | 34 | Yes | ++ | −/−/−/−/− | 0 ± 0 | 0 ± 0 | 5 ± 1 | + |
| 42 | 32 | Yes | + | −/−/−/+/− | 0 ± 0 | 0 ± 0 | 9 ± 12 | + |
| 43 | 25 | Yes | ++ | −/−/−/−/− | 77 ± 9 | 0 ± 0 | 3 ± 3 | + |
| PAO1 | − | +/+/+/+/+ | 957 ± 91 | 277 ± 20 | 87 ± 5 | + | ||
Composite AHL results for C. violaceum CV026 and A. tumefaciens A136 when bacteria were grown in TSB and in supplemented AB medium. Spots were presumptively identified as representative of individual AHL molecules, following published results (47).
One unit of enzyme activity was determined as the amount of activity required to change the final absorbance by 1.0 per ml of culture. Individual enzyme activities were normalized to the OD600 of the culture at harvest. Mean values and standard deviations were determined across three experiments.
Isolates were routinely identified as being P. aeruginosa by growth on cetrimide fucidin cephaloridine (Oxoid, Adelaide, Australia), acetamide, and Pseudosel agars (both BBL, Liverpool, Australia); a positive oxidase reaction (MedVet Science, Adelaide, Australia); growth on Columbia horse blood agar (Oxoid, Adelaide, Australia) at 42°C; and resistance to 9-chloro-9-(4-diethylaminophenyl)-10-phenylacridan (Dutec Diagnostics, Croydon, Australia). The identification of isolates showing inconsistent results on these tests was confirmed using the API 20E system. Genotyping using a restriction enzyme, SpeI, followed by pulsed-field gel electrophoresis (PFGE) previously showed that two clones (designated AES-1 and AES-2) infected 40% and 6% of patients, respectively (P. Tingpej, B. Rose, and C. Harbor, unpublished data). In this study, a clonal strain was defined as one showing the same PFGE banding pattern (or a difference of three bands or less) in three or more patients (43). Figure 1 shows DNA banding patterns from AES-1 and AES-2 and nonclonal strains. DNA macrorestriction patterns of AES-1 and AES-2 were found to share 67% similarity. Isolates used in this study were randomly selected from the clonal and nonclonal pools in proportion to their prevalence in the RPAH Clinic (14 AES-1, 5 AES-2, and 24 nonclonal isolates). The well-characterized wound-derived P. aeruginosa PAO1 (17) was included as a control. The AHL reporter strains Chromobacterium violaceum CV026 and Agrobacterium tumefaciens A136 were used for AHL assays, and Staphylococcus aureus strain ATCC 12600 was used for the staphylolysin assay. Pseudomonas putida was used as a negative control for dot hybridization. For protease production, bacteria were grown in tryptone soya broth (TSB; Oxoid, Adelaide, Australia) to late exponential phase (optical density at 600 nm [OD600] ≈ 1.6) at 37°C with rapid agitation. For ExoS Western blot assays, strains were grown in TSB with 1 mM EGTA to an OD600 of 1.0 (20). For AHL analyses, isolates were grown in TSB to late exponential phase or in AB medium supplemented with 0.2% glucose to an OD600 of 1.0 (4, 12). A. tumefaciens was grown in supplemented minimal A medium (A+) (29), and C. violaceum was grown in TSB, both at 30°C. Supernatants were collected and filtered through a 0.22-μm filter for protease, exoenzyme, and AHL detection. Cell pellets were collected for gene amplification and dot hybridization studies.
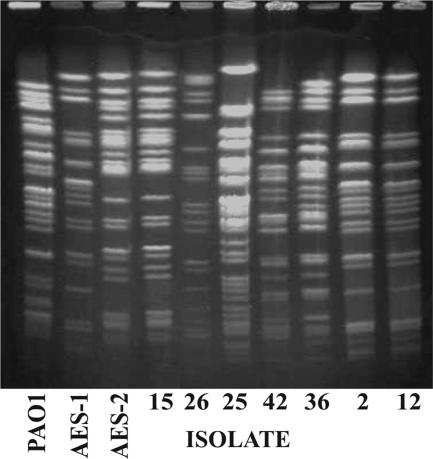
FIG. 1.
DNA banding pattern following macrorestriction using a rare-cutting restriction enzyme, SpeI, and PFGE analysis using our previously described methods (1). Lanes 1, 2, and 3 are PAO1, AES-1, and AES-2 controls, respectively. Lanes 4 to 10 represent P. aeruginosa strains isolated from individual patients (Table 1). Lanes 5 to 8 have unique banding patterns and hence represent nonclonal strains. Isolates in lanes 9 and 10 (isolates 2 and 12) have a macrorestriction pattern identical to that of AES-1, and lane 4 (isolate 15) is identical to AES-2.
Gene detection by PCR.
DNA was extracted from cell pellets using the microLYSIS kit (Microzone Ltd., Sussex, United Kingdom). PCRs were carried out for the presence of five QS genes, vfr (5′-TGTTCTTCCAGGAGCGTGG-3′/3′-TCGCAAAATCACATCGAC-5′), lasI, lasR, rhlI, and rhlR (45); three protease genes, lasA (26), lasB, and aprA (45); the rhamnolipid gene rhlAB (45); and two exotoxin genes, exoU and exoS (46), as previously described.
Dot blot hybridization.
When PCR screening identified a strain that apparently did not possess the lasI or lasR gene, dot blot hybridization was performed to confirm the negative findings. Bacterial cell pellets were washed and resuspended in phosphate-buffered saline to an OD660 of 0.2. Digoxigenin-labeled DNA probes for the lasI and lasR genes were generated from the PAO1 strain using the primers for the PCRs (described above) according to the manufacturer's instructions (Roche Diagnostics GmbH, Mannheim, Germany). Bacterial cell suspensions were dot blotted onto a Hybond N+ membrane (General Electric Healthcare, Fairfield, CT). Blots were then hybridized using DIG EasyHyb buffer (Roche Diagnostics GmbH, Mannheim, Germany). Hybrids were visualized by standard alkaline phosphatase colorimetric detection. P. putida cells were used as a negative control.
Exoprotease assays. (i) Total protease activity.
Total protease activity was determined as described previously (47). The protease activity was normalized to the densities (OD600) of the cultures grown. Results were mean values ± standard deviations (SD) from three replicate experiments carried out on three separate occasions.
(ii) Zymography.
Gelatin zymography for protease activity was carried out using 10% polyacrylamide gels containing 0.1% (wt/vol) gelatin (Bio-Rad, Regents Park, Australia). Unconcentrated supernatants were denatured in sodium dodecyl sulfate (SDS) sample buffer. Following electrophoresis, gels were washed in 2.5% Triton X-100 (Sigma-Aldrich, MO) to renature proteins, incubated overnight at 37°C to induce protease activity, and then stained with 0.25% Coomassie blue for 2 h with gentle shaking (10). Zymograms were imaged using an Olympus (Tokyo, Japan) C-3040Zoom digital image acquisition system. Protease activities were represented by translucent bands against a Coomassie blue background. All isolates were tested in triplicate on at least three occasions.
(iii) Elastase-specific activity.
lasB-mediated elastolytic activity was determined by the elastin-Congo red (ECR) assay as described previously (35). Uninoculated TSB was included as a negative control. All isolates were tested in triplicate on at least two occasions. Results are represented as mean values ± SD.
(iv) Staphylolysin activity.
lasA-mediated staphylolytic activity was determined as described previously (23). All isolates were tested in triplicate at least twice, and results shown are mean values ± SD.
Chitinase activity.
Chitinase activity was determined using the carboxymethyl-chitin-remazol brilliant violet assay adapted from methods described by the manufacturer (Loewe Biochemica, Munich, Germany) and Folders et al. (9). Remazol brilliant violet liberated by chitinase activity was measured at 550 nm and normalized to the cell density. Uninoculated TSB was used as a negative control. All isolates were tested in triplicate on at least two occasions, and results are represented as mean values ± SD.
Rhamnolipid production.
The presence or absence of rhamnolipids was assessed by the rhamnolipid-mediated alkane degradation method (49). PAO1 cultures grown under the same conditions as the test isolates and uninoculated TSB were used as positive and negative controls, respectively.
Western blots for exoenzyme S.
Supernatants were boiled for 5 min prior to 12% (wt/vol) nonreducing SDS-polyacrylamide gel electrophoresis (PAGE) at 100 mA for 2 h. Separated protein was electrophoretically blotted at 30 V onto a Tris-glycine buffer-equilibrated polyvinylidene difluoride membrane (Immuno-blot; Bio-Rad, Regents Park, Australia) using the Bio-Rad Mini-Transblot system. Blots were blocked in 5% (wt/vol) bovine serum albumin in phosphate-buffered saline, subsequently incubated overnight with chicken polyclonal exoenzyme S antibody (Abcam, Cambridge, United Kingdom) at a 1:10,000 dilution, and developed in anti-chicken antibody alkaline phosphatase conjugate (1:1,000) (Sigma-Aldrich, MO) using BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate dipotassium-nitrotetrazolium blue chloride) solution (Sigma-Aldrich, MO). Tests were carried out in duplicate on two different occasions.
Thin-layer chromatography.
AHLs were identified by analytical thin-layer chromatography (TLC) initially from bacteria grown in TSB as described previously for our studies of microbial keratitis (48). Chromatograms were developed using C18 reversed-phase silica gel TLC plates (Whatmann, Clifton, NJ) with methanol-water (60:40, vol/vol). Plates were overlaid with a thin film of TSB or A+ agar seeded with either C. violaceum CV026 or A. tumefaciens A136 and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml), respectively. Short-chain AHLs (BHL and HHL) were detected by the production of violacein by C. violaceum CV026, resulting in purple spots at the site of AHL migration. Long-chain AHLs (OdDHL, N-3-oxodecanoyl-l-homoserine lactone [ODHL], and N-3-oxohexanoyl-l-homoserine lactone [OHHL]) were detected by the traG:lacZ/traR reporter in A. tumefaciens A136, inducing β-galactosidase expression and the development of blue/green dots at the site of AHL migration. HHL was also detected using the A. tumefaciens reporter. The purified BHL, HHL, and OdDHL used as positive controls were kindly supplied by Naresh Kumar (Department of Chemistry, The University of New South Wales, Sydney, Australia). It was reported previously that P. aeruginosa strains from lungs of CF patients produce the highest amounts of long-chain AHL in AB medium containing 0.2% glucose (12). Thus, bacteria that tested negative for long-chain AHLs when grown in TSB medium were retested following culture in AB medium supplemented with 0.2% glucose. All samples were tested at least three times.
Statistical analyses.
Pearson's chi-square test was used to analyze the secretion of the virulence factors and AHL molecules between clonal and nonclonal groups. P values of <0.05 were considered to be significant. Relative risk (RR) and 95% confidence intervals (CIs) were used to determine the strengths of the associations. The levels of virulence factor production were analyzed using a standard t test for normally distributed data and using the Mann-Whitney U test for nonparametric data. Relationships between protease, elastase activity, and patient age were calculated using Pearson's correlation coefficient (r).
RESULTS
Mucoidy.
The degree of mucoidy of the isolates was assessed at isolation on horse blood agar by visual inspection and designated as negative, +(low), ++(medium), or +++(high). Forty-one of the 43 (95%) isolates were mucoid. Of these, nine (21%) showed + mucoidy, 25 (58%) showed ++ mucoidy, and seven (16%) showed +++ mucoidy (Table 1). All colonies of each mucoid isolate were mucoid.
Presence of virulence factor and QS genes.
All 43 isolates produced bands of the predicted sizes in the vfr, rhlI, rhlR, lasA, lasB, aprA, and rhlAB PCRs. One isolate (nonclonal) was negative for both lasI and lasR. The absence/mutation of both genes was confirmed by dot hybridization. There was no evidence of large-scale insertions or deletions within the amplified regions of any of the genes. All 43 samples were positive for the exoS gene and negative for the exoU gene.
Exoprotease production.
Thirty-seven of the 43 (86%) isolates produced positive results in the total protease assay. Levels were variable and in some cases very low (Table 1). SDS-10% PAGE zymography produced a protease profile of up to three bands for each sample (Fig. 2). Bands were detected at molecular masses of approximately 120 kDa, 98 kDa, and 51 kDa. Elastase and staphylolysin were represented by bands at 120 kDa and 98 kDa, respectively, and alkaline protease was represented by the band at 51 kDa (48). In some samples, there were two bands at 51 kDa. Although the protease IV enzyme has a molecular mass of 26 kDa, it aggregates under mild SDS denaturing conditions and is resolved at a molecular mass of 350 kDa (4). It was therefore barely detectable on the 10% zymograms used in this study. Twenty-four of the 43 (56%) isolates produced elastase/staphylolysin, and 22 of 43 (51%) isolates produced alkaline protease (Table 2). The amount of protease secreted by individual isolates as indicated by band intensity was variable. Thirty-two of the 43 (74%) isolates exhibited elastase activity using the ECR assay. Levels ranged from 3 mU/ml to 367 mU/ml (Table 1). All 24 isolates that produced elastase on zymography also showed elastase activity in the ECR assay. Eight isolates that were negative by zymography showed low levels of activity using the ECR assay. As expected, there was a strong correlation between total protease and elastase activities (r = 0.847; P < 0.001). Forty of the 43 (93%) isolates produced staphylolysin, but activities were generally low, ranging from 1 mU/ml to 91 mU/ml (Table 1).
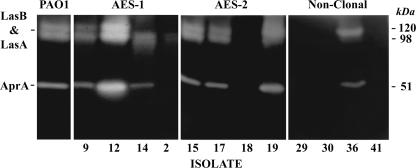
FIG. 2.
Protease activity on 1% gelatin zymograms using 10% SDS-PAGE for AES-1 (isolates 9, 12, 14, and 2), AES-2 (isolates 15, 17, 18, and 19), and nonclonal strains (isolates 29, 30, 36, and 41) (Table 1). Up to three bands were detected, corresponding to molecular masses of 51, 98, and 120 kDa. Bands at 98 and 120 kDa represented aggregate activity of elastase and staphylolysin, while the band at 51 kDa represented alkaline protease. Isolates included in the figure are representative of isolates in each group. PAO1 was included as a control.
TABLE 2.
Frequency of detection of virulence factors and AHLs in clonal and nonclonal isolates
| Virulence factor | No. of isolates (%)
|
Statistics (clonal vs nonclonal)
|
|||
|---|---|---|---|---|---|
| Total (n = 43) | Clonal (n = 19) | Nonclonal (n = 24) | P value | RR (95% CI) | |
| Total protease | 37 (86) | 19 (100) | 18 (75) | 0.019 | 1.3 (1.1-1.7) |
| ECR elastase | 32 (74) | 17 (89) | 15 (63) | 0.044 | 1.4 (1.0-2.0) |
| Zymography | |||||
| Elastase/staphylolysin | 24 (56) | 14 (74) | 10 (42) | 0.036 | 1.8 (1.0-3.0) |
| Alkaline protease | 22 (51) | 14 (74) | 8 (33) | 0.009 | 2.2 (1.2-4.1) |
| Staphylolysin | 40 (93) | 16 (84) | 24 (100) | 0.079 | |
| Chitinase | 34 (79) | 16 (84) | 18 (75) | 0.461 | |
| Rhamnolipids | 35 (81) | 16 (84) | 19 (79) | 0.673 | |
| ExoS | 33 (77) | 16 (84) | 17 (71) | 0.302 | |
| AHL detection | |||||
| rhl-directed AHLs | 31 (72) | 16 (84) | 15 (63) | 0.115 | |
| las-directed AHLs | 16 (37) | 7 (37) | 9 (38) | 0.965 | |
| At least one AHL | 32 (74) | 16 (84) | 16 (67) | 0.190 | |
Chitinase activity and rhamnolipid production.
Thirty-four of the 43 (79%) isolates showed chitinase activity; levels ranged from 6 mU/ml to 168 mU/ml (Table 2). Thirty-five of the 43 (81%) isolates produced rhamnolipids (Table 2).
Exoenzyme S production.
Thirty-three of the 43 (77%) isolates grown in type III secretion system-inducing low-calcium medium showed a single band of 36 kDa corresponding to the ExoS protein on the Western blots. The intensities of the bands were variable. Results for individual isolates are shown in Table 1.
AHL production.
When results from the two AHL biosensors were combined, the number of spots varied from none to five. Thirty-two of the 43 (74%) samples produced spots corresponding to one or more AHLs (Table 2). Using the reporter strain C. violaceum CV026, two spots, most likely representing BHL and HHL (12, 47), were noted. Thirty-one of the 43 (72%) isolates produced rhl-associated AHLs. Of these isolates, 19 (44%) produced a spot corresponding to both HHL and BHL (Fig. 3), and 12 (28%) had a spot corresponding to HHL but not BHL. Spot densities and diameters varied among isolates. Using the A. tumefaciens A136 reporter strain that detects las-directed AHLs with a high degree of sensitivity (37), up to four spots were detected, most likely corresponding to OdDHL, ODHL, HHL, and OHHL (47). Following growth in the TSB medium used previously in our microbial keratitis study (47), at least one las-directed AHL was detected in 16 (37%) isolates. Of these isolates, OdDHL was detected in 8 isolates, ODHL was detected in 11 isolates, and OHHL was detected in 6 isolates. Three additional isolates yielded OHHL spots in assays using supplemented AB medium. The HHL results using the A. tumefaciens A136 reporter were consistent with those using the C. violaceum CV026 reporter. As expected, the isolate that tested negative for the lasI and lasR genes had undetectable levels of AHL.
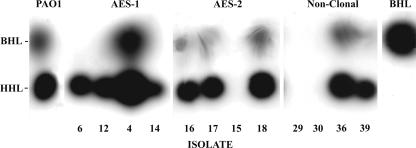
FIG. 3.
Representative TLC analyses of AHLs produced by PAO1, AES-1 (isolates 6, 12, 4, and 14), AES-2 (isolates 16, 17, 15, and 18), and nonclonal (isolates 29, 30, 36, and 39) (Table 1) P. aeruginosa isolates from the lungs of CF patients and purified BHL. AHLs were resolved from unconcentrated supernatants using methanol-water, and representative spots were visualized with the biosensor C. violaceum (CV026). Up to two dots were noted and identified as being BHL and HHL as shown. Isolates included in the figure are representative of all isolates in each group. PAO1 and purified BHL were included as controls.
Overall relationships between phenotypic and patient characteristics.
Isolates with positive results for one or both rhl-associated AHLs were significantly more likely to show total protease (RR = 1.7 [CI = 1.0 to 2.7]; P = 0.004) and elastase (RR = 5.8 [CI = 1.6 to 20.6]; P = 0.006) activities than those testing negative for these AHLs. The levels in each case were also significantly higher (both P < 0.01). However, when the las-directed AHL data were included, the associations between AHL and protease production were no longer significant. There were no relationships between the production of proteases or AHLs and exoenzyme S, rhamnolipids, and chitinase and no association between mucoidy and other phenotypic characteristics. There was a trend for decreasing protease and elastase activity with increasing patient age, but the relationship reached statistical significance for elastase only (r = −0.34; P = 0.03). Patient age may provide some indication of the duration of infection. There were no relationships between exacerbation and any of the phenotypic characteristics.
Clonal versus nonclonal groups.
Data for AES-1 and AES-2 were combined for these analyses because of small numbers. The male-to-female ratios in the combined clonal versus nonclonal groups were similar, 9:10 and 12:12, respectively, as were the mean ages (and age ranges), 25 (18 to 37) and 29 (19 to 58) years, respectively. A greater proportion of nonclonal isolates produced +++ mucoidy than clonal isolates (7 of 24 versus 0 of 19), but proportions of isolates with mucoidy at the + and ++ levels were similar between clonal and nonclonal groups, and there was no overall relationship between mucoidy and genotype. There was no significant difference in the probability of having a clonal strain among patients with exacerbation compared with those without (7 of 18 [39%] versus 12 of 25 [48%]).
As shown in Table 2, clonal isolates were significantly more likely than nonclonal isolates to produce proteases including elastase, alkaline protease, and total protease. Clonal isolates also tended to have greater total protease and elastase activities than nonclonal isolates, although the differences were not statistically significant in this relatively small sample.
There was no difference in relationships between patient age and total protease production in the clonal versus nonclonal groups. Elastase levels were more likely to decrease with increasing patient age in the nonclonal group (r = −0.38; P = 0.07) than in the clonal group (r = −0.08, P = 0.75), but this relationship did not reach statistical significance. Clonal isolates were also more likely to have at least one detectable AHL and to exhibit BHLs/HHLs, but these trends did not reach statistical significance. There were no relationships between genotype and the production of ExoS, rhamnolipids, or chitinase. Among the nonclonal group, there was a significant correlation between elastase production and mucoidy (r = 0.46; P = 0.03). This relationship was not evident in the clonal group, but these findings should be interpreted with caution because of the limited sample size and because mucoidy was evaluated visually rather than by quantitation.
DISCUSSION
Our finding that the clonal strains were more likely to have protease activity than nonclonal strains was of particular interest. These results are consistent with those of our previous study of protease IV activity using the same isolates (40). The focus on proteases reflects previous reports that both the United Kingdom CF LES (34) and an epidemic ocular P. aeruginosa strain had increased protease activity (26). It therefore seems possible that genetically distinct clones of P. aeruginosa from different environmental sources have evolved common mechanisms to facilitate spread and/or confer increased virulence.
Nonetheless, the role of protease activity in transmissibility or increased virulence in CF lung infection has not been established. It was reported previously that specific antibodies are produced against virulence factors such as the proteases in the lung (18). Overall findings from the nonclonal group support current concepts that the progressive down-regulation of proteases promotes long-term survival in lungs of CF patients. Nonetheless, it is also clear from this and other studies that a proportion of nonclonal isolates from well-established chronic infections secrete proteases (some at a high level) (14, 24). Conversely, some AES-1 and AES-2 isolates showed only low protease activity. It is not known whether the lack of protease activity affects the transmissibility or virulence of these isolates. High protease activity has been linked with lung exacerbation in some surveys (14, 19), but in the present study, there was no relationship between protease activity and clinical state in either the clonal or nonclonal group. One reason for the lack of an association here may be that only one isolate from each patient was tested. The protease activity of this isolate may not have been representative of that of other phenotypes possibly present (42). The link between high levels of mucoidy and protease production noted in our nonclonal group was reported previously (22).
It is not known whether AES-1 and AES-2 are generally less susceptible to the processes that down-regulate proteases during chronic infection than nonclonal strains or whether these clones have undergone changes that allow the ongoing secretion of these virulence factors over extended periods. There was no evidence of major genetic changes within the amplified regions of protease genes to correspond with the elastase size variant identified in the ocular strain (26).
It is now widely accepted that proteases, like many other P. aeruginosa virulence factors, are QS controlled. However, relationships between protease activity and QS production during chronic CF lung infection have not been clearly defined. Several studies demonstrated the presence of P. aeruginosa AHLs in CF lung secretions (5, 38), but there have been few studies of AHL production in CF lung isolates. The results of one recent small longitudinal series suggested that phenotypic adaptations associated with chronic infection do not affect either the nature or the amount of AHL produced (12). However, a comparative genomic analysis of two isolates of the same strain from the same CF patient collected 8 years apart revealed the development of mutations in lasR but not in the rhlI and rhlR genes (39). In the present study, the majority of isolates in both the clonal and nonclonal groups were positive for the rhl-mediated BHLs/HHLs, but less than half had detectable levels of las-directed AHL. Overall, there was a good correlation between protease and AHL production; isolates with undetectable AHL invariably produced very low or undetectable levels of proteases, and conversely, very high total protease activity (in excess of 900 mU/ml) was associated with the presence of at least three AHLs.
Intriguingly, OdDHL and ODHL were not detected in any AES-1 isolates even upon repeated testing under the growth conditions that were previously reported to promote long-chain AHL production (12). There was no evidence of major genetic changes with the amplified regions of the lasI/lasR gene to account for these findings. However, minor genetic changes in the las signaling pathway have been linked with increased protease expression in the United Kingdom LES (34) and with changes in virulence factor expression in other studies (44). We speculate that in samples with undetectable levels of OdDHL, protease activity is mediated by BHL/HHL in association with the PQS system. PQS regulates the expression of lasB and, in combination with BHL, has been shown to have a synergistic effect on the expression of lasB compared with BHL or PQS alone (28).
There was no evidence of any association between genotype, ExoS, or rhamnolipid production or chitinase activity. As expected from previous studies, all 43 CF isolates (clonal and nonclonal) had an invasive exoS phenotype (8, 25). However, our finding that the majority of isolates had easily detectable levels of ExoS in Western blots was surprising, since a loss of type III effectors over the chronic phase of infection in the CF lung has been widely reported (20, 25).
There have been few studies of rhamnolipid production or chitinase activity in CF lung isolates. Rhamnolipids are a heterogeneous group of glycolipidic surface-active molecules thought to have multiple virulence activities related mainly to their surfactant properties (41). Rhamnolipids promote early infiltration of airway epithelia (49) and may have a central role in biofilm architecture (6). The role of chitinase in lung infection is unknown. The lack of association between rhamnolipids or chitinase production and protease activity was not surprising. However, since rhamnolipids and chitinase have been reported to be QS controlled (9, 41), the production of these virulence factors in a small proportion of samples without detectable AHL could reflect the complexities of virulence factor-regulatory mechanisms or the presence of a mutation(s) leading to the decoupling of normal regulation.
Our comparisons of the phenotypic characteristics of clonal and nonclonal strains extend the general understanding of the pathobiology of P. aeruginosa infection in the CF lung. However, the major aim of this study was to gain insight into the properties that distinguish clonal from nonclonal P. aeruginosa strains in CF. Our findings support evidence from studies of other clonal P. aeruginosa strains showing that protease activity may mediate increased infectivity and/or virulence. Advances in the understanding of the properties underpinning the increased infectivity of epidemic P. aeruginosa strains are critical to the development of new therapeutics to treat infected patients and strategies to prevent transmission. Key traits may also serve as targets for vaccines. Findings may also be relevant for non-CF bronchiectasis, for the immunocompromised, and for intensive care and burn units, where clonal P. aeruginosa is also a problem.
Acknowledgments
We are grateful to the Australian Cystic Fibrosis Research Trust for its support.
Footnotes
Published ahead of print on 28 March 2007.
REFERENCES
- 1.Anthony, M., B. Rose, M. B. Pegler, M. Elkins, H. Service, K. Thamotharampillai, J. Watson, M. Robinson, P. Bye, J. Merlino, and C. Harbour. 2002. Genetic analysis of Pseudomonas aeruginosa isolates from the sputa of Australian adult cystic fibrosis patients. J. Clin. Microbiol. 40:2772-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, D., S. Bell, M. Robinson, P. Bye, B. Rose, C. Harbour, C. Lee, H. Service, M. Nissen, M. Syrmis, and C. Wainwright. 2003. Evidence for patient-to-patient spread of a clonal strain of Pseudomonas aeruginosa in different cystic fibrosis clinics in Australia. J. Clin. Microbiol. 41:2266-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, D., G. M. Nixon, R. Carzino, A. Bigham, J. B. Carlin, R. M. Robins-Browne, and K. Grimwood. 2002. Detection of a widespread clone of Pseudomonas aeruginosa in a pediatric cystic fibrosis clinic. Am. J. Respir. Crit. Care Med. 166:983-987. [DOI] [PubMed] [Google Scholar]
- 4.Caballero, A. R., J. M. Moreau, L. S. Engel, M. E. Marquart, J. M. Hill, and R. J. O'Callaghan. 2001. Pseudomonas aeruginosa protease IV enzyme assays and comparison to other Pseudomonas proteases. Anal. Biochem. 290:330-337. [DOI] [PubMed] [Google Scholar]
- 5.Chambers, C. E., M. B. Visser, U. Schwab, and P. A. Sokol. 2005. Identification of N-acylhomoserine lactones in mucopurulent respiratory secretions from cystic fibrosis patients. FEMS Microbiol. Lett. 244:297-304. [DOI] [PubMed] [Google Scholar]
- 6.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberl, L. 1999. N-Acylhomoserine lactone-mediated gene regulation in gram-negative bacteria. Syst. Appl. Microbiol. 22:493-506. [DOI] [PubMed] [Google Scholar]
- 8.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 9.Folders, J., J. Tommassen, L. C. van Loon, and W. Bitter. 2000. Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J. Bacteriol. 182:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frederiks, W. M., and O. Mook. 2004. Metabolic mapping of proteinase activity with emphasis on in situ zymography of gelatinases: review and protocols. J. Histochem. Cytochem. 52:711-722. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs, H. J., D. S. Borowitz, D. H. Christiansen, E. M. Morris, M. L. Nash, B. W. Ramsey, B. J. Rosenstein, A. L. Smith, M. E. Wohl, et al. 1994. Effect of aerosolized recombinant human DNase on exacerbations of respiratory symptoms and on pulmonary function in patients with cystic fibrosis. N. Engl. J. Med. 331:637-642. [DOI] [PubMed] [Google Scholar]
- 12.Geisenberger, O., M. Givskov, K. Riedel, N. Hoiby, B. Tummler, and L. Eberl. 2000. Production of N-acyl-L-homoserine lactones by Pseudomonas aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Microbiol. Lett. 184:273-278. [DOI] [PubMed] [Google Scholar]
- 13.Goodman, A. L., and S. Lory. 2004. Analysis of regulatory networks in Pseudomonas aeruginosa by genomewide transcriptional profiling. Curr. Opin. Microbiol. 7:39-44. [DOI] [PubMed] [Google Scholar]
- 14.Grimwood, K., R. A. Semple, H. R. Rabin, P. A. Sokol, and D. E. Woods. 1993. Elevated exoenzyme expression by Pseudomonas aeruginosa is correlated with exacerbations of lung disease in cystic fibrosis. Pediatr. Pulmonol. 15:135-139. [DOI] [PubMed] [Google Scholar]
- 15.Head, N. E., and H. Yu. 2004. Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect. Immun. 72:133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoiby, N., and C. Koch. 1990. Pseudomonas aeruginosa infection in cystic fibrosis and its management. Thorax 61:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollsing, A. E., M. Granstrom, M. L. Vasil, B. Wretlind, and B. Strandvik. 1987. Prospective study of serum antibodies to Pseudomonas aeruginosa exoproteins in cystic fibrosis. J. Clin. Microbiol. 25:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffar-Bandjee, M. C., A. Lazdunski, M. Bally, J. Carrere, J. P. Chazalette, and C. Galabert. 1995. Production of elastase, exotoxin A, and alkaline protease in sputa during pulmonary exacerbation of cystic fibrosis in patients chronically infected by Pseudomonas aeruginosa. J. Clin. Microbiol. 33:924-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain, M., D. Ramirez, R. Seshadri, J. F. Cullina, C. A. Powers, G. S. Schulert, M. Bar-Meir, C. L. Sullivan, S. A. McColley, and A. R. Hauser. 2004. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 42:5229-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, A., J. Govan, C. Doherty, M. Dodd, B. Isalka, T. Stanbridge, and A. Webb. 2001. Spread of a multiresistant strain of Pseudomonas aeruginosa in an adult cystic fibrosis clinic. Lancet 358:557-558. [DOI] [PubMed] [Google Scholar]
- 22.Kamath, S., V. Kapatral, and A. M. Chakrabarty. 1998. Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol. Microbiol. 30:933-941. [DOI] [PubMed] [Google Scholar]
- 23.Kessler, E., M. Safrin, J. C. Olson, and D. E. Ohman. 1993. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protase. J. Biol. Chem. 268:7503-7508. [PubMed] [Google Scholar]
- 24.Lanotte, P., L. Mereghetti, B. Lejeune, P. Massicot, and R. Quentin. 2003. Pseudomonas aeruginosa and cystic fibrosis: correlation between exoenzyme production and patient's clinical state. Pediatr. Pulmonol. 36:405-412. [DOI] [PubMed] [Google Scholar]
- 25.Lee, V. T., R. S. Smith, B. Tummler, and S. Lory. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect. Immun. 73:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomholt, J. A., K. Poulsen, and M. Kilian. 2001. Epidemic population structure of Pseudomonas aeruginosa: evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infect. Immun. 69:6284-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mariencheck, W. I., J. F. Alcorn, S. M. Palmer, and J. R. Wright. 2003. Pseudomonas aeruginosa elastase degrades surfactant proteins A and D. Am. J. Respir. Cell Mol. Biol. 28:528-537. [DOI] [PubMed] [Google Scholar]
- 28.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. 1976. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.O'Carroll, M. R., M. W. Syrmis, C. E. Wainwright, R. M. Greer, P. Mitchell, C. Coulter, T. P. Sloots, M. D. Nissen, and S. C. Bell. 2004. Clonal strains of Pseudomonas aeruginosa in paediatric and adult cystic fibrosis units. Eur. Respir. J. 24:101-106. [DOI] [PubMed] [Google Scholar]
- 31.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. L. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signalling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 33.Robinson, P., R. Carzino, D. Armstrong, and A. Olinsky. 2003. Pseudomonas cross-infection from cystic fibrosis patients to non-cystic fibrosis patients: implications for inpatient care of respiratory patients. J. Clin. Microbiol. 41:5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salunkhe, P., C. H. M. Smart, A. W. Morgan, S. Panagea, M. J. Walshaw, C. A. Hart, R. Geffers, B. Tummler, and C. Winstanley. 2005. A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J. Bacteriol. 187:4908-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schad, P. A., R. A. Bever, T. I. Nicas, F. Leduc, L. F. Hanne, and B. H. Iglewski. 1987. Cloning and characterization of elastase genes from Pseudomonas aeruginosa. J. Bacteriol. 169:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott, F. W., and T. L. Pitt. 2004. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J. Med. Microbiol. 53:609-615. [DOI] [PubMed] [Google Scholar]
- 37.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 39.Smith, E. E., D. G. Buckley, S. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 103:8487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, L., B. Rose, P. Tingpej, H. Zhu, T. C. Conibear, J. Manos, P. Bye, M. Elkins, M. D. Willcox, S. Bell, C. Wainwright, and C. Harbour. 2006. Protease IV activity of Pseudomonas aeruginosa from the lungs of adults with cystic fibrosis. J. Med. Microbiol. 55:1641-1644. [DOI] [PubMed] [Google Scholar]
- 41.Soberon-Chavez, G., F. Lepine, and E. Deziel. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 68:718-725. [DOI] [PubMed] [Google Scholar]
- 42.Struelens, M. J., V. Schwam, A. Deplano, and D. Baran. 1993. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J. Clin. Microbiol. 31:2320-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. Salmond, B. W. Bycroft, A. Lazdunski, G. S. Stewart, and P. Williams. 1995. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, H., R. Bandara, T. C. Conibear, S. J. Thuruthyil, S. A. Rice, S. Kjelleberg, M. Givskov, and M. D. Willcox. 2004. Pseudomonas aeruginosa with LasI quorum-sensing deficiency during corneal infection. Investig. Ophthalmol. Vis. Sci. 45:1897-1903. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, H., T. C. Conibear, R. Bandara, Y. Aliwarga, F. Stapleton, and M. D. Willcox. 2006. Type III secretion system-associated toxins, proteases, serotypes and antibiotic resistance of Pseudomonas aeruginosa isolates associated with keratitis. Curr. Eye Res. 31:297-306. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, H., S. J. Thuruthyil, and M. D. Willcox. 2002. Determination of quorum-sensing signal molecules and virulence factors of Pseudomonas aeruginosa isolates from contact lens-induced microbial keratitis. J. Med. Microbiol. 51:1063-1070. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, H., S. J. Thuruthyil, and M. D. Willcox. 2001. Production of N-acyl homoserine lactones by gram-negative bacteria isolated from contact lens wearers. Clin. Exp. Ophthalmol. 29:150-152. [DOI] [PubMed] [Google Scholar]
- 49.Zulianello, L., C. Canard, T. Kohler, D. Caille, J. S. Lacroix, and P. Meda. 2006. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect. Immun. 74:3134-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]