Abstract
A simple method for the enumeration of viable Mycobacterium paratuberculosis cells was developed and evaluated using the MGIT 960 culture system. For each of 12 M. paratuberculosis strains isolated from either cattle or humans, single-cell suspensions of M. paratuberculosis cells were adjusted to an optical density at 600 nm of 1.00 (107.6 to 108.2 cells/ml), and serial dilutions were prepared. Standard curves were established by relating the MGIT time-to-detection data to the log10 CFU for these suspensions using standard plate counting and BACTEC 460 results as reference methods. Universal and strain-specific standard quantification curves were generated. A one-phase exponential decay equation best fit the universal standard curve and strain-specific curves (R2 of 0.96 and >0.99, respectively). Two subgroups within the universal curves were distinguished: one for laboratory-adapted strains and the other for recently isolated low-passage bovine strains. The predictive errors for log10 estimations using the universal standard curve, each subgroup's standard curve, and strain-specific curves were ±0.87, ±0.45, and ±0.31 log10 units, respectively. CFU estimations by all three standard curves were highly reproducible, regardless of the M. paratuberculosis strain or inoculum volume. In comparison with the previously described BACTEC 460 M. paratuberculosis counting method, quantification with MGIT 960 was less expensive, more rapid, more accurate, and more sensitive (<10 CFU). This MGIT counting method has broad applications for studies requiring the quantification of viable M. paratuberculosis cells, such as drug susceptibility testing or environmental survival studies.
Quantification of viable bacteria is a crucial foundation for many types of research. This seemingly simple task can be challenging, expensive, and imprecise for Mycobacterium paratuberculosis, a slowly growing organism (>24-h generation time) with a strong tendency to form large clumps (15). Studies of environmental survival, resistance to pasteurization or disinfectants, and quantification of the pathogen in milk and feces from infected animals are just a few examples that require precise and sensitive quantification of viable M. paratuberculosis cells.
The first liquid culture system used for the detection of M. paratuberculosis in clinical samples was the BACTEC 460 system (Becton Dickinson, Sparks, MD), based on a modified 12B (radiometric) culture medium (8). An algorithm for converting cumulative growth index units in the BACTEC 460 system into the number of inoculated M. paratuberculosis cells was previously reported (21). This algorithm has been used both in its original form as well as in a more simplified form for multiple studies of M. paratuberculosis and has proven to be invaluable for studies of M. paratuberculosis survival in the face of multiple chemical, physical, and environmental challenges (16, 21, 26, 28-31, 34).
The BACTEC 460 TB system has been replaced with the new nonradiometric BACTEC MGIT (mycobacterial growth indicator tube) 960 system (9, 15, 20), designed for the detection of mycobacterial species commonly found in human clinical samples (5, 15, 17). The system uses an oxygen-quenching fluorescent sensor in conjunction with software algorithms to determine when tubes are “positive,” i.e., when significant bacterial growth has occurred. This system has been adapted for the detection of M. paratuberculosis in veterinary clinical samples by using a new culture medium specific for M. paratuberculosis, called MGIT ParaTB medium (Becton Dickinson, Sparks, MD), and a modified algorithm built into the MGIT 960 instrument for the interpretation of fluorescence measurements made hourly for each culture tube (4).
The advantages of the MGIT 960 system for culture, the recovery of slowly growing mycobacteria from clinical samples, have already been reported (18, 22, 25). The instrument reports results as time to detection (TTD) in days and hours. Just as with the BACTEC system, additional testing is required to confirm the identity of mycobacterial isolates, usually acid-fast staining and PCR (24, 32). The quantitative capabilities of the MGIT 960 system have not previously been explored.
The goal of this study was to create a method for counting M. paratuberculosis cells analogous to what was previously reported for the BACTEC 460 system (21) using MGIT TTD data.
MATERIALS AND METHODS
Bacterial strains, cultures, and preparation of M. paratuberculosis single-cell suspensions.
A total of 12 M. paratuberculosis strains isolated from cattle or humans were used in this study: ATCC 19698, K-10, JTC100, JTC102, JTC114, JTC303, USF-5, UCF-7, B213, B236, B238, and B244. Strains ATCC 19698 and K-10 are type strains; those with the JTC prefix are clinical isolates recovered from bovine fecal or tissue samples by the Johne's Testing Center (School of Veterinary Medicine, University of Wisconsin—Madison). Strains UCF-5 and UCF-7 are M. paratuberculosis strains of human origin that were recently isolated from Crohn's disease patients. These strains and four bovine strains, B213, B236, B238, and B244, were kindly provided by Saleh A. Naser (University of Central Florida, Orlando, FL). All strains were cultivated in 7H9 broth supplemented with 10% oleic acid-albumin-dextrose-catalase (Difco Laboratories, MD) and 2 μg/ml of mycobactin J (Allied Monitor, Fayette, MO) for 1 month at 37°C. The identity of the organisms was verified by multiplex PCR (2, 3) for the insertion elements IS900, IS901, IS1311, and IS1245 as well as by high-performance liquid chromatography of cell wall mycolic acids by a reference laboratory (State Laboratory of Hygiene, Madison, WI).
Preparation of mycobacterial cells.
Single-cell suspensions of each strain were prepared as previously described, with slight modifications (21). Briefly, mycobacterial cells grown in mycobactin-supplemented 7H9 broth were harvested by centrifugation at 10,000 × g for 20 min and washed three times in 10 mM phosphate-buffered saline (PBS) (pH 7.2). Cell pellets were homogenized using an overhead stirrer (Wheaton Instrument, Milville, NJ) for 1 min on ice to minimize clumping of cells. The homogenized mycobacterial cells were passed through an 8-μm-pore-size filter (Millipore Corp., Bedford, MA). The predominance of single cells in the final preparation was confirmed by an examination of acid-fast-stained slides. Seedlots of each strain were then kept in 1.5-ml aliquots at −80°C until use.
Colony counts.
The number of viable mycobacterial cells in each single-cell suspension was determined by standard plate counting as a reference method. Briefly, the undiluted stock cell suspension (1.0 ml) was added to 9.0 ml of 10 mM PBS (pH 7.2). Tenfold serial dilutions were made in 10 mM PBS (pH 7.2), with vortexing between each dilution step. One hundred microliters from each dilution was plated onto each of three 7H10 agar plates supplemented with 10% oleic acid-albumin-dextrose-catalase and 2 μg/ml of mycobactin J. Colony counts (CFU) were determined after the incubation of plates at 37°C for 10 weeks. MGIT ParaTB tubes were inoculated in triplicate with 100 μl of the same serial dilutions. To evaluate the effect of the MGIT tube inoculum volume, one set of tubes was inoculated with 100 μl and another was inoculated with 1,000 μl from each of the serial dilutions of multiple M. paratuberculosis strains.
Experiment I: generation of standard curves.
Serial dilutions of single-cell suspensions of each of the 12 M. paratuberculosis strains were prepared, and 100 μl was inoculated into ParaTB MGIT medium (Becton Dickinson, Sparks, MD) as described above. Each tube contained 7 ml of modified Middlebrook 7H9 broth base with mycobactin J and a fluorescent indicator embedded in silicon on the bottom of the tube. Eight hundred microliters of MGIT ParaTB supplement (Becton Dickinson, Sparks, MD), 500 μl of egg yolk suspension (Becton Dickinson, Sparks, MD), and 100 μl of VAN cocktail (Becton Dickinson, Sparks, MD) were added to each tube, resulting in final concentrations of 10 μg/ml vancomycin, 40 μg/ml amphotericin B, and 60 μg/ml nalidixic acid, as per the manufacturer's instructions. The tubes were inoculated in triplicate, incubated at 37°C in an MGIT 960 instrument, and removed when the instrument signaled positive. Acid-fast staining (Ziehl-Neelsen) was performed on smears made from each signal-positive tube to confirm the presence of mycobacteria. This experiment was completed twice. The TTD (days) for each tube was recorded and plotted against log10 CFU inoculum to generate a standard curve for each strain tested (i.e., the strain-specific standard curve). In addition, the data for all 12 M. paratuberculosis strains were pooled to create a universal standard curve. The model that best fit the data was determined using statistical software (GraphPad Prism version 4.03 for Windows; GraphPad Software, San Diego, CA). Two methods were used to compare the universal standard curve to the strain-specific curves. First, cross-validation for each strain was performed based on pooled data for the other 11 strains. Second, the standard error for the strain-specific curve was compared with that of the universal standard curve at 95% prediction intervals (PI) for log10 CFU estimates.
Experiment II: utilization of the MGIT 960 TTD counting method.
The ability of the MGIT ParaTB quantification standard curves to accurately predict M. paratuberculosis CFU was determined with randomly selected dilutions prepared from seven strains (ATCC 19698, JTC303, JTC114, UCF-5, UCF-7, B238, and B244). Plate-counted CFU were compared to predicted CFU based on both the strain-specific and universal standard curves. These counts were obtained independently from the results from experiment I. The relationship between the observed and predicted CFU counts was evaluated by correlation coefficients.
Comparison of M. paratuberculosis quantification by the BACTEC 460 and MGIT 960 methods.
Using plate counts as the reference method, CFU predictions for the same M. paratuberculosis cell suspensions were obtained by both the BACTEC 460 and MGIT 960 methods. The BACTEC 460 method was performed as previously described (21), with the following slight modifications. Briefly, 100 μl of 10-fold dilutions from each strain was inoculated into three BACTEC 12B vials containing 4 ml Middlebrook 7H12B medium supplemented with 2 μg/ml of mycobactin J, 1.0 ml of egg yolk (Becton Dickinson, Sparks, MD), and the same final concentrations of antibiotics as those used for MGIT tubes. All vials were incubated at 37°C without shaking. The growth index for each vial was measured every 24 h for 8 weeks by the BACTEC 460 instrument using a normal atmosphere for gas exchange. The upper limit for the designation of no growth for inoculated vials as well as negative controls was 30 growth units after 8 weeks. The instrument was calibrated weekly according to the manufacturer's instructions. When cumulative growth index readings exceeded 2,000 units, a previously described algorithm was used to estimate CFU of M. paratuberculosis inoculated.
Comparison of MGIT and spectrophotometric enumeration methods.
Five M. paratuberculosis strains were selected (ATCC 19698, UCF-5, UCF-7, B236, and B238). Stock suspensions of each strain were suspended in 10 mM PBS (pH 7.2) to an optical density at 600 nm (OD600) of 1.00 using a spectrophotometer (Biomate 3; Thermo Electron Corp., Madison, WI). Tenfold dilutions (100 μl of 10° to 106 CFU) were inoculated into triplicate supplemented MGIT ParaTB tubes. The tubes were incubated in the MGIT 960 instrument, and the TTD was recorded for each tube. CFU inoculated per tube were predicted by both the universal standard curve and the matching strain-specific standard curve. The predicted log10 CFU were compared with observed colony counts on plates. Initial M. paratuberculosis cell concentrations in stock suspensions were calculated by multiplying CFU by the relevant dilution factors.
RESULTS
Generation of standard curves.
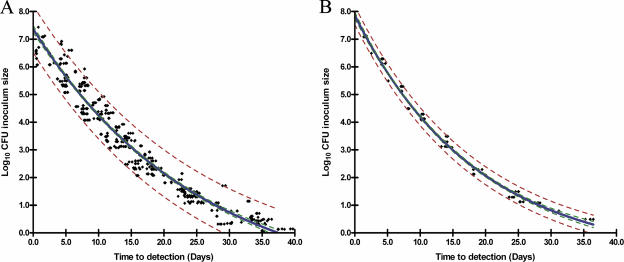
Standard curves (CFU versus TTD) for each of the 12 M. paratuberculosis strains (strain-specific standard curves) (Fig. 1B) as well as the pooled data for all 12 strains (universal standard curve) (Fig. 1A) were fitted to a one-phase exponential decay model with three parameters: log10 inoculum size = span × e(−K × TTD) + plateau. Span is the difference between TTD at time zero and the plateau, K is degree of decay for the log10 CFU, and plateau is the value for log10 CFU curve flattening. Estimations for span, K, and plateau were obtained by a nonlinear least-squares approach using GraphPad Prism 4.03 software. The R2 of goodness-of-fit values between TTD and log10 CFU in the universal standard curve and strain-specific curves were 0.96 and >0.99, respectively.
FIG. 1.
Generation of standard curves for M. paratuberculosis using the MGIT 960 culture system. (A) Universal standard curve for the pooled data from 12 strains of M. paratuberculosis [log10 CFU inoculum size = 9.515 × e(−0.03977 × TTD) − 2.143]. (B) Representative strain-specific standard curve of M. paratuberculosis ATCC 19698 [log10 CFU inoculum size = 8.802 × e(−0.0538 × TTD) − 0.93851]. The blue line shows the best-fitting curve for all strains. The red dotted lines and green dotted lines show 95% PIs and 95% CIs, respectively.
Comparison between universal standard curve and strain-specific standard curves.
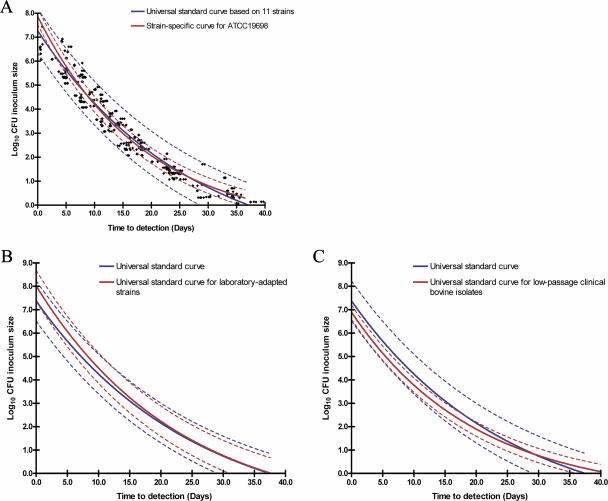
The validity of applying the universal standard curve for each of 12 different strains was determined by cross-validation and prediction errors. In the cross-validation assessment, the CFU counts obtained from the strain-specific curve for each of the 12 strains were within the 95% PI compared with the universal standard curve based on the other 11 strains (Fig. 2A). This was true for every M. paratuberculosis strain tested in this study. As expected, a strain-specific curve was more accurate for itself (very narrow 95% PI) than was the universal standard curve. Mean CFU prediction errors for the universal standard curve and strain-specific curves were ±0.87 log10 CFU and ±0.31 log10 CFU, respectively. CFU predictions were higher (>1.0 log10 CFU) for the universal standard curve when >106.5 CFU M. paratuberculosis were inoculated into an MGIT ParaTB tube.
FIG. 2.
Validity of the universal standard curve and generation of subgroup universal standard curves for laboratory-adapted strains and low-passage clinical bovine isolates. (A) Representative curve for the cross-validation for M. paratuberculosis ATCC 19698 in a universal standard curve based on the other 11 M. paratuberculosis strains [log10 CFU inoculum size = 9.699 × e(−0.03728 × TTD) − 2.430]. (B) Subgroup universal standard curves for group I strains (type strains and laboratory-adapted strains) [log10 CFU inoculum size = 9.927 × e(−0.0437 × TTD) − 1.910]. (C) Subgroup universal standard curve for group II strains (low-passage clinical bovine isolates) [log10 CFU inoculum size = 7.855 × e(−0.05052 × TTD) − 0.9633]. The blue line and red line show the best-fitting curve for the universal strains and specific strain tested, respectively. The dotted lines of the same color show the 95% PIs for the respective best-fit curves.
While the universal standard curve was accurate for all strains, by comparing cross-validations and pattern similarities of strain-specific curves, two subgroups within the universal standard curve became evident: subgroup I contained laboratory-adapted strains (Fig. 2B), and subgroup II was made up of recently isolated, low-passage clinical isolates (Fig. 2C). Strains ATCC 19698, K-10, JTC100, JTC114, and JTC303 belonged to subgroup I, and strains JTC102, B213, B236, B238, and B244 belonged to subgroup II. In subgroup I, none of the strains had identical strain-specific standard curves. The strain-specific standard curves for subgroup I were quite similar to the universal standard curve, but these strains tended to grow more slowly when inoculated at 103 to 107 CFU (Fig. 2B). In contrast, all subgroup II strains had identical strain-specific standard curves at 95% confidence intervals (CI) with CFU predictions similar to those of the strain-specific standard curves. Generally, subgroup II strains grew more rapidly than subgroup I strains (Fig. 2C). Data pooled by subgroup produced subgroup I and subgroup II standard curves. These two curves showed a better fit to the model, with R2 values of 0.97 and 0.99, respectively, and more accurately predicted CFU for the relevant strains than did the universal standard curve, whose prediction errors were ±0.65 and ±0.37 log10 CFU, respectively. Two human strains were excluded from the subgroup standard curves because their growth patterns did not fit either subgroup.
Reproducibility of the MGIT counting method.
MGIT CFU predictions, repeated in a second independent trial with all 12 strains, were compared using an unpaired t test with Welch's correction. No statistically significant difference was observed between the two experiments (P = 0.70). The correlation (r) between CFU estimations predicted by each experiment's universal standard curves was 0.99 when CFU estimations were compared at the same inoculum sizes (Fig. 3A). In addition, no significant difference was observed between CFU from plate counts and CFU estimations predicted by both universal standard curves (P > 0.05 by multiple comparison). Three M. paratuberculosis strains (ATCC 19698, JTC303, and JTC114) were selected for reproducibility analysis of strain-specific standard curves as well. Counting experiments for each strain were performed at least three times. The correlation between observed and predicted log10 CFU (using strain-specific models among repeated experiments for ATCC 19698, JTC303, and JTC114) was >0.99 (Fig. 3B).
FIG. 3.
MGIT counting model reproducibility. (A) Correlation between observed and predicted CFU using the universal standard curve for 12 M. paratuberculosis strains repeated twice. (B) Correlation between observed and predicted CFU using the M. paratuberculosis ATCC 19698 strain-specific model repeated three times. The dotted lines with the same color for each standard curve and green dotted lines show 95% PIs and 95% CIs, respectively.
Effect of inoculum volume.
The effect of inoculum volume at a uniform M. paratuberculosis concentration was tested over a wide range of dilutions. Tenfold serial dilutions of three M. paratuberculosis strains were prepared as described above. MGIT ParaTB tubes were inoculated with either 100 μl or 1,000 μl of M. paratuberculosis cell suspension. When predicted CFU counts for tubes with different inoculum volumes but the same expected number of M. paratuberculosis cells, e.g., 100 μl of 105 CFU/ml versus 1,000 μl of 104 CFU/ml, were compared, counts were highly correlated (r > 0.99) (Fig. 4).
FIG. 4.
TTD values correlated with CFU counts for MGIT ParaTB tubes inoculated with different volumes but the same CFU (100 μl of 105 CFU versus 1,000 μl of 104 CFU). Data for three M. paratuberculosis strains (ATCC 19698, JTC303, and JTC114) are shown. The dotted lines with the same color for each standard curve and green dotted lines show 95% PIs and 95% CIs, respectively.
Predicted log10 CFU for samples of unknown concentrations.
Triplicate CFU determinations for seven M. paratuberculosis strains tested over a wide range of cell concentrations were compared using the strain-specific, subgroup-specific, and universal models by multiple comparisons. When all results were combined, the CFU determinations based on the strain-specific, subgroup-specific, and universal models were highly correlated (r = 0.99, r = 0.98, and r = 0.97, respectively [95% CI]) (Fig. 5). The predicted numbers of cells for unknown samples tested in this experiment using the universal standard curve, subgroup-specific curves, and strain-specific curves erred by ±0.69, ±0.49, and ±0.35 log10 units, respectively (95% PI). These differences among the three models were not considered to be statistically significant (P = 0.77 by multiple comparison test).
FIG. 5.
Correlation between actual CFU and predicted CFU. (A) Predictive CFU based on the strain-specific standard curves. (B) Predictive CFU based on the subgroup-specific standard curves. (C) Predictive CFU based on the universal standard curve. The red dotted lines and green dotted lines show 95% PIs and 95% CIs, respectively.
Comparison of CFU prediction accuracy between the MGIT and BACTEC methods.
Using only M. paratuberculosis strain ATCC 19698, CFU predictions were compared using the BACTEC model and MGIT universal standard curve (triplicate determinations). While the predicted CFU counts were the same for both systems, for M. paratuberculosis at 103.5 to 106.5 CFU (P = 0.88 by multiple comparison test; r > 0.99), results were obtained in less than half the time with the MGIT method and over a greater dynamic range (6 logs versus 3 logs) compared to the BACTEC method (Table 1).
TABLE 1.
Comparison of predicted M. paratuberculosis counts by MGIT 960 and BACTEC 460 culture systems
| CFU by plate counting (log10) | MGIT 960
|
BACTEC 460
|
||
|---|---|---|---|---|
| CFU (mean ± SE) | Time to results (days) | CFU (mean ± SE) | Time to results (days) | |
| 6.49 | 6.74 ± 0.00 | 2 | 6.77 ± 0.12 | 10 |
| 5.49 | 5.76 ± 0.01 | 4 | 5.78 ± 0.10 | 15 |
| 4.49 | 4.73 ± 0.03 | 7 | 4.70 ± 0.12 | 19 |
| 3.49 | 3.19 ± 0.03 | 10 | 3.45 ± 0.11 | 28 |
| 2.49 | 2.50 ± 0.01 | 15 | UDa | >35 |
| 1.49 | 1.37 ± 0.03 | 20 | UD | |
| 0.49 | 0.34 ± 0.04 | 28 | UD | |
UD, undeterminable beyond 8 weeks of incubation.
Correlation of MGIT and spectrophotometric methods.
Suspensions of M. paratuberculosis cells from five different strains were adjusted to an OD600 of 1.00 for five different strains and then serially diluted 10-fold in PBS. Predicted log10 CFU concentrations for each dilution of each strain by both the universal and strain-specific standard curves matched plate counts between 10 and 10−5.0 dilutions (Table 2) (P > 0.05 by multiple comparison test). Significant differences at the lowest dilution (<10 CFU) and at the highest dilution (>10−6.5) were observed (i.e., TTD results varied across replicates) (P < 0.05 by multiple comparison test) (Table 2). Overall, however, CFU predictions at each dilution using the universal standard curve did not differ from plate counts (P = 0.43 by paired Wilcoxon signed-rank test). TTD intervals among dilutions were consistent and predictable regardless of the M. paratuberculosis strain used.
TABLE 2.
Correlation of M. paratuberculosis enumeration methods from a cell suspension adjusted to an OD600 of 1.00
| Dilution (cells/ml)a | Range (mean ± SE)
|
P value for comparison at each dilution between log10 CFUd | ||||
|---|---|---|---|---|---|---|
| TTD | Plate count (log10 CFU range) | Predicted CFU
|
TTD intervals between dilutions (days)b | |||
| Universal standard curve | Strain-specific standard curve | |||||
| 10−1 | 0.42-2.54 (1.17 ± 0.21) | 6.29-7.08 (6.59 ± 0.08) | 6.46-7.22 (6.94 ± 0.07) | 6.54-7.24 (6.78 ± 0.06) | 1.91-3.46 (2.54 ± 0.16) | 0.016* |
| 10−2 | 3.67-5.12 (4.55 ± 0.15) | 5.29-6.08 (5.59 ± 0.08) | 5.62-6.08 (5.80 ± 0.05) | 4.99-6.13 (5.62 ± 0.09) | 2.69-3.99 (3.09 ± 0.14) | 0.150 |
| 10−3 | 6.54-9.58 (7.79 ± 0.256) | 4.29-5.08 (4.59 ± 0.08) | 4.36-5.19 (4.84 ± 0.07) | 4.23-5.09 (4.66 ± 0.07) | 3.43-4.53 (3.63 ± 0.13) | 0.172 |
| 10−4 | 10.17-14.21 (11.76 ± 0.38) | 3.29-4.08 (3.59 ± 0.08) | 3.27-4.21 (3.83 ± 0.09) | 3.16-4.16 (3.66 ± 0.09) | 3.66-5.60 (4.34 ± 0.16) | 0.134 |
| 10−5 | 14.67-18.21 (16.18 ± 0.36) | 2.29-3.08 (2.59 ± 0.08) | 2.47-3.17 (2.87 ± 0.07) | 2.40-3.06 (2.74 ± 0.06) | 4.68-5.77 (5.40 ± 0.16) | 0.279 |
| 10−6 | 19.88-29.42 (24.04 ± 0.82) | 1.29-2.08 (1.59 ± 0.08) | 0.81-2.17 (1.54 ± 0.12) | 1.30-2.09 (1.54 ± 0.08) | 6.63-8.31 (7.30 ± 0.29) | 0.801 |
| 10−7 | 27.99-36.46 (33.35 ± 0.75) | 0.29-1.08 (0.59 ± 0.08) | 0.09-0.99 (0.40 ± 0.08) | 0.27-1.02 (0.59 ± 0.07) | NDc | 0.021* |
Counts reflect M. paratuberculosis CFU in 100 μl of this dilution.
Expected TTD interval ranges between dilutions. Values within this range at the defined dilutions reflect appropriate 10-fold serial dilutions.
ND, not determined.
P values were obtained by the Kruskal-Wallis test by multiple comparison between plate counts and predicted CFU from both standard curves at each dilution of all five strains. *, significant differences between groups (P < 0.05).
DISCUSSION
Standard plate counting methods for slowly growing mycobacteria are laborious, expensive, and time-consuming and frequently fail due to either contamination or medium dehydration during the required 30- to 50-day incubation (6, 10, 34). Spectrophotometric methods for the standardization of cell suspensions provide only rough estimations of bacterial cell concentrations, and replicated counts may vary by >1 log10 CFU/ml (7, 27).
The TTD data reported by the MGIT 960 system with the new ParaTB medium supplemented were highly correlated to colony counts determined by standard plate counting methods. Standard curves for all 12 strains approximated a linear relationship between CFU and TTD, but a one-phase exponential decay curve mode best fit the data. Pooling the data from 12 M. paratuberculosis strains created a universal standard growth curve with narrow prediction errors (<1 log10 CFU) that can be reliably used for quantification for a variety of experiments. While the greatest accuracy in predicting M. paratuberculosis CFU was obtained by the use of a strain-specific standard curve (±0.31 log10 CFU [95% PI]), this additional precision is not needed for most mycobacterial research trials. Note the differences in strain-specific standard curves for low-passage bovine strains of M. paratuberculosis (subgroup II) and laboratory-adapted strains (subgroup I). A characteristic of subgroup II strains was their identical strain-specific standard curves (95% CI). In contrast, subgroup I strains showed greater variability among strains; i.e., strain-specific standard curves varied more extensively, although the overall pattern of standard curves for these strains was similar. With the appropriate subgroup standard curve, CFU counts for recently isolated bovine organisms were as accurately predicted as with strain-specific standard curves (±0.37 versus ±0.31 log10 CFU [95% PI]).
Subgroup II strains grew faster than subgroup I strains, especially at higher inocula (>103 CFU). This growth rate difference between laboratory-adapted and low-passage clinical isolates could possibly be explained by the genetic differences between strains, by sample storage conditions, or by greater clumping at high inocula. Our group and others have shown the varied effect on growth rates and phenotypic characteristics caused by different types of media (11, 30). In this study, the laboratory-adapted strains had been subcultured in Watson-Reid medium, stored at −80°C for several months, and reconstituted in 7H9 broth, while the other strains were cultured in 7H9 broth since their initial isolation. This medium difference may contribute to growth rate variation. Interestingly, it has been reported that clumping increases the growth rate of mycobacteria both in vitro and in vivo (19, 21).
A reliable and accurate prediction of log10 M. paratuberculosis CFU was obtained between 101.5 and 106.5 CFU (dynamic range), with very low variation when the universal standard curve was applied for all study strains. However, both high concentrations of M. paratuberculosis cells (>106.5 CFU with a TTD of <3 days) and very low CFU (<101 CFU with a TTD of >30 days) cannot reliably be measured by these MGIT methods. The 95% prediction errors at these ranges were considerably wider than 1.0 log10 CFU. In addition, although the prediction errors were small in smaller inoculum sizes (inoculum of less than 1.0 log10 CFU), the coefficient variations were relatively high (i.e., 0.16% with an inoculum of 1 log10 CFU versus 0.09% with an inoculum of 6 log10 CFU). As a result, this counting method is not recommended for M. paratuberculosis cell concentrations of >106.5 or <101.0 per MGIT ParaTB tube. Another factor that may affect the quantification accuracy of this and other methods for counting of mycobacteria is the tendency of the organism to clump. Our protocols deliberately created homogenous single-cell suspensions to provide the most robust data for method validation, but in clinical samples, most of bacterial cells existed as clumps. Therefore, more work is needed to determine the utility of this method for clinical applications.
The one-phase exponential decay model with three parameters has been widely used to describe the kinetics of drug activity (14) and environmental factors with regard to bacterial growth (12). We found that this model was also effective for interpreting M. paratuberculosis growth data, producing standard curves of broad utility.
In comparison with the BACTEC 460 counting method, the MGIT 960 approach has better analytical sensitivity (101 CFU versus 103 CFU), determines its cell counts in less than half the time, is automated (no daily loading of vials into the BACTEC 460 instrument is required), and does not require radioisotopes. The BACTEC 460 method for M. paratuberculosis required at least 25 days to obtain the results for interpretation when inoculated with >103 CFU, while MGIT results can be obtained in less than 2 weeks. The internal coefficient of variation of CFU estimates between MGIT ParaTB tubes was exceedingly small (0.55% at a 95% CI): only two tubes were necessary for precise M. paratuberculosis CFU estimations.
Spectrophotometric (turbidimetry) measurements have long been used to estimate the number of bacteria in suspensions. Although neither very sensitive nor precise, it is a quick, inexpensive method for standardizing bacterial inocula prior to such studies as antibiotic susceptibility testing or experimental animal challenge. By combining spectrophotometry with MGIT, cells can be counted quickly and precisely. As a standard method, we propose that M. paratuberculosis cell suspensions be adjusted to a spectrophotometer OD600 of 1.00. This equates to roughly 108 CFU/ml of M. paratuberculosis. One hundred microliters of 10−1 to 10−3 dilutions of this suspension should then be inoculated into duplicate MGIT ParaTB tubes. With this method, accurate log10 M. paratuberculosis CFU can be obtained within 2 weeks. Dilution errors can quickly be detected by examining the TTD intervals between dilutions (Table 2). For example, if 106 CFU M. paratuberculosis were inoculated into supplemented MGIT ParaTB tubes, the TTD for a positive signal should be obtained in 4 days. The next serial dilution, 105 CFU, should signal positive between days 7 and 8 of incubation. Precision in M. paratuberculosis counting by the MGIT method can increase experimental precision and decrease the cost of studies of this emerging pathogen.
The universal, subgroup-specific, or strain-specific MGIT M. paratuberculosis counting curves can be used in any application requiring the quantification of viable M. paratuberculosis cell numbers, such as evaluations of physical factors such as radiation or pasteurization, chemical factors such as pH, salt, or disinfectants, and any in vitro growth promoter or suppressor. This system can also be used to measure whether antimycobacterial drugs are bacteriostatic or bactericidal by allowing the antimycobacterial agent-M. paratuberculosis interaction to take place outside the MGIT ParaTB medium and then transferring a small aliquot of the mixture into an MGIT tube for quantification.
Several studies demonstrated previously that M. paratuberculosis strains isolated from cattle are genetically conserved (23, 33). The similarity of strain-specific standard curves for subgroup II supports these findings. While the universal standard curve demonstrated in this study has broad utility, the subgroup II-specific standard curve might provide even more precise results for recently isolated, low-passage bovine isolates. Since some M. paratuberculosis strains isolated from sheep, bison, and nondomestic species were previously reported to display greater genetic and phenotypic diversity than bovine isolates (1, 13, 23), further work on quantification with this method with such isolates is needed.
Acknowledgments
The Board of Regents of the University of Wisconsin System gratefully acknowledges that the Awwa Research Foundation is the joint owner of the technical information upon which this publication is based. The Board of Regents of the University of Wisconsin System thanks the Foundation for its financial, technical, and administrative assistance in funding and managing the project through which this information was discovered. The comments and views detailed herein may not necessarily reflect the views of the Awwa Research Foundation, its officers, directors, affiliates or agents.
Partial funding for this work was provided by Cooperative Agreement 98-9613-0032-CA, Veterinary Services, Animal and Plant Health Inspection Service, USDA.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Amonsin, A., L. L. Li, Q. Zhang, J. P. Bannantine, A. S. Motiwala, S. Sreevatsan, and V. Kapur. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42:1694-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anklam, K. S., E. J. B. Manning, S. Sreevatsan, and M. T. Collin. 2005. Evaluation of a 3 PCR assay for identification of Mycobacterium avium subsp. paratuberculosis from liquid cultures, p. 549. In E. J. B. Manning and S. S. Nielsen (ed.), Proceedings of the 8th International Colloquium of Paratuberculosis. International Association for Paratuberculosis, Madison, WI.
- 3.Anklam, K. S., E. J. B. Manning, S. Sreevatsan, and M. T. Collins. 2004. Evaluation of a multiplex PCR assay for identification of Mycobacterium avium subsp. paratuberculosis from liquid cultures, p. 209. In J. T. Saliki (ed.), Proceedings of the 47th Annual Conference. American Association of Veterinary Laboratory Diagnosticians, Davis, CA.
- 4.Badak, F. Z., D. L. Kiska, S. Setterquist, C. Hartley, M. A. O'Connell, and R. L. Hopfer. 1996. Comparison of Mycobacteria Growth Indicator Tube with BACTEC 460 for detection and recovery of mycobacteria from clinical specimens. J. Clin. Microbiol. 34:2236-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiodini, R. J., and C. D. Buergelt. 1993. Susceptibility of Balb/c, C57/B6 and C57/B10 mice to infection with Mycobacterium paratuberculosis. J. Comp. Pathol. 109:309-319. [DOI] [PubMed] [Google Scholar]
- 7.Chiodini, R. J., and J. Hermon-Taylor. 1993. The thermal resistance of Mycobacterium paratuberculosis in raw milk under conditions simulating pasteurization. J. Vet. Diagn. Investig. 5:629-631. [DOI] [PubMed] [Google Scholar]
- 8.Collins, M. T., K. B. Kenefick, D. C. Sockett, R. S. Lambrecht, J. McDonald, and J. B. Jorgensen. 1990. Enhanced radiometric detection of Mycobacterium paratuberculosis by using filter-concentrated bovine fecal specimens. J. Clin. Microbiol. 28:2514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruciani, M., C. Scarparo, M. Malena, O. Bosco, G. Serpelloni, and C. Mengoli. 2004. Meta-analysis of BACTEC MGIT 960 and BACTEC 460 TB, with or without solid media, for detection of mycobacteria. J. Clin. Microbiol. 42:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damato, J. J., M. T. Collins, M. V. Rothlauf, and J. K. McClatchy. 1983. Detection of mycobacteria by radiometric and standard plate procedures. J. Clin. Microbiol. 17:1066-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Juan, L., J. Alvarez, B. Romero, J. Bezos, E. Castellanos, A. Aranaz, A. Mateos, and L. Dominguez. 2006. Comparison of four different culture media for isolation and growth of type II and type I/III Mycobacterium avium subsp. paratuberculosis strains isolated from cattle and goats. Appl. Environ. Microbiol. 72:5927-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson, J. S., G. R. Siragusa, and J. E. Wray, Jr. 1992. Predicting the growth of Salmonella typhimurium on beef by using the temperature function integration technique. Appl. Environ. Microbiol. 58:3482-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohmann, K., B. Strommenger, K. Stevenson, L. de Juan, J. Stratmann, V. Kapur, T. J. Bull, and G. F. Gerlach. 2003. Characterization of genetic differences between Mycobacterium avium subsp. paratuberculosis type I and type II isolates. J. Clin. Microbiol. 41:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edelstein, P. H., T. Shinzato, E. Doyle, and M. A. Edelstein. 2001. In vitro activity of gemifloxacin (SB-265805, LB20304a) against Legionella pneumophila and its pharmacokinetics in guinea pigs with L. pneumophila pneumonia. Antimicrob. Agents Chemother. 45:2204-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant, I. R., R. B. Kirk, E. Hitchings, and M. T. Rowe. 2003. Comparative evaluation of the MGIT and BACTEC culture systems for the recovery of Mycobacterium avium subsp. paratuberculosis from milk. J. Appl. Microbiol. 95:196-201. [DOI] [PubMed] [Google Scholar]
- 16.Grant, I. R., A. G. Williams, M. T. Rowe, and D. D. Muir. 2005. Efficacy of various pasteurization time-temperature conditions in combination with homogenization on inactivation of Mycobacterium avium subsp. paratuberculosis in milk. Appl. Environ. Microbiol. 71:2853-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumber, S., and R. J. Whittington. 2006. Comparison of BACTEC 460 and MGIT 960 systems for the culture of Mycobacterium avium subsp. paratuberculosis S strain and observations on the effect of inclusion of ampicillin in culture media to reduce contamination. Vet. Microbiol. 119:42-52. [DOI] [PubMed] [Google Scholar]
- 18.Hillemann, D., E. Richter, and S. Rusch-Gerdes. 2006. Use of the BACTEC Mycobacteria Growth Indicator Tube 960 automated system for recovery of mycobacteria from 9,558 extrapulmonary specimens, including urine samples. J. Clin. Microbiol. 44:4014-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoal-van Helden, E. G., D. Hon, L. A. Lewis, N. Beyers, and P. D. van Helden. 2001. Mycobacterial growth in human macrophages: variation according to donor, inoculum and bacterial strain. Cell Biol. Int. 25:71-81. [DOI] [PubMed] [Google Scholar]
- 20.Huang, T. S., C. S. Chen, S. S. Lee, W. K. Huang, and Y. C. Liu. 2001. Comparison of the BACTEC MGIT 960 and BACTEC 460TB systems for detection of mycobacteria in clinical specimens. Ann. Clin. Lab. Sci. 31:279-283. [PubMed] [Google Scholar]
- 21.Lambrecht, R. S., J. F. Carriere, and M. T. Collins. 1988. A model for analyzing growth kinetics of a slowly growing Mycobacterium sp. Appl. Environ. Microbiol. 54:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitritz, L., S. Schubert, B. Bucherl, A. Masch, J. Heesemann, and A. Roggenkamp. 2001. Evaluation of BACTEC MGIT 960 and BACTEC 460TB systems for recovery of mycobacteria from clinical specimens of a university hospital with low incidence of tuberculosis. J. Clin. Microbiol. 39:3764-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motiwala, A. S., M. Strother, A. Amonsin, B. Byrum, S. A. Naser, J. R. Stabel, W. P. Shulaw, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 41:2015-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motiwala, A. S., M. Strother, N. E. Theus, R. W. Stich, B. Byrum, W. P. Shulaw, V. Kapur, and S. Sreevatsan. 2005. Rapid detection and typing of strains of Mycobacterium avium subsp. paratuberculosis from broth cultures. J. Clin. Microbiol. 43:2111-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pardini, M., F. Varaine, M. Bonnet, G. Orefici, M. R. Oggioni, and L. Fattorini. 2006. Usefulness of the BACTEC MGIT 960 system for isolation of Mycobacterium tuberculosis from sputa subjected to long-term storage. J. Clin. Microbiol. 45:575-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddacliff, L. A., P. J. Nicholls, A. Vadali, and R. J. Whittington. 2003. Use of growth indices from radiometric culture for quantification of sheep strains of Mycobacterium avium subsp. paratuberculosis. Appl. Environ. Microbiol. 69:3510-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stabel, J. R., E. M. Steadham, and C. A. Bolin. 1997. Heat inactivation of Mycobacterium paratuberculosis in raw milk: are current pasteurization conditions effective? Appl. Environ. Microbiol. 63:4975-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung, N., and M. T. Collins. 1998. Thermal tolerance of Mycobacterium paratuberculosis. Appl. Environ. Microbiol. 64:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung, N., and M. T. Collins. 2000. Effect of three factors in cheese production (pH, salt, and heat) on Mycobacterium avium subsp. paratuberculosis viability. Appl. Environ. Microbiol. 66:1334-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung, N., and M. T. Collins. 2003. Variation in resistance of Mycobacterium paratuberculosis to acid environments as a function of culture medium. Appl. Environ. Microbiol. 69:6833-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Boxtel, R. M., R. S. Lambrecht, and M. T. Collins. 1990. Effects of colonial morphology and Tween 80 on antimicrobial susceptibility of Mycobacterium paratuberculosis. Antimicrob. Agents Chemother. 34:2300-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittington, R. J., I. Marsh, M. J. Turner, S. McAllister, E. Choy, G. J. Eamens, D. J. Marshall, and S. Ottaway. 1998. Rapid detection of Mycobacterium paratuberculosis in clinical samples from ruminants and in spiked environmental samples by modified BACTEC 12B radiometric culture and direct confirmation by IS900 PCR. J. Clin. Microbiol. 36:701-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, C. W., J. Glasner, M. Collins, S. Naser, and A. M. Talaat. 2006. Whole-genome plasticity among Mycobacterium avium subspecies: insights from comparative genomic hybridizations. J. Bacteriol. 188:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, B. Y., C. J. Czuprynski, and M. T. Collins. 1999. Intracellular fate of Mycobacterium avium subspecies paratuberculosis in monocytes from normal and infected, interferon-responsive cows as determined by a radiometric method. Can. J. Vet. Res. 63:56-61. [PMC free article] [PubMed] [Google Scholar]