Abstract
A Legionella cluster was identified through retrospective PCR analysis of 240 throat swab samples from X-ray-confirmed pneumonia cases. These were identified among young and otherwise healthy U.S. military recruits during population-based surveillance for pneumonia pathogens. Results were confirmed by sequence analysis. Cases clustered tightly, suggesting a local environmental etiology.
CASE REPORT
Legionella pneumophila was identified by PCR in the oropharyngeal swabs of five male recruits aged 18 to 28 years who were diagnosed with, and treated for, X-ray-confirmed pneumonia at the Marine Corps Recruit Depot, San Diego, CA (MCRD). All five recruits were housed together in the same barracks as part of three platoons of the same training company, and initially all reported ill with pneumonia between 3 November and 8 November 2004. Positive samples were collected on the 3rd, 5th, and 8th of November, along with five concurrent but otherwise identical negative samples taken from recruits with pneumonia from other companies at MCRD with use of the same batch of sampling reagents. Identical reagents were also used to collect 24 more nonconcurrent negative samples from pneumonia patients at the same site. During the week in which the positive samples were collected there were nine active companies at the training center. Clustering of all five cases in one company was highly significant (P = 0.001, Fisher's exact test, weighted for company size). There was no significant clustering in platoons. Testing extended to 240 samples collected during the years 2004 and 2005, all collected by the same methods and all stored and processed together, and the cluster appeared to be tightly limited in time. No other samples were identified as L. pneumophila positive.
Prior to recruitment, one of the five recruits was a regular smoker, one was a light smoker, and three had never smoked. None of the five reported a history of chronic cough, cough at night, asthma, or shortness of breath. One of the five reported a history of hay fever; the others reported no such history. All five reported to basic training on the same day from different states and reported to sick call 22 to 27 days later. Since the period of L. pneumophila incubation is 2 to 10 days, the timing strongly suggests that the affected recruits were infected after arrival at the training base. Electronic record-keeping systems indicated that none of the recruits were hospitalized overnight.
Records showed that all patients had been prophylactically treated with bicillin (benzathine penicillin G) approximately 1 month before onset of symptoms. After identification of pneumonia, all were treated with various antibiotic regimens including penicillin only (n = 1), quinolones only (n = 1), and both quinolones and cephalosporins (n = 3), some intravenously. While none of the patients were hospitalized, all were taken out of training and reassigned to a medical rehabilitation platoon for periods ranging from 9 to 18 days, with an average of 12 days. This range was not significantly different from what was reported for other (non-Legionella-associated) pneumonias. Throat swabs were taken when the patients initially reported with pneumonia symptoms, and upon positive chest X-ray, patients were enrolled in the Naval Health Research Center's population-based pneumonia surveillance study. Oropharyngeal swabs (sterile polyester-tipped applicators; Puritan, Guilford, ME) were placed in 1 ml TE buffer (0.01 M Tris, 0.001 M EDTA [both from Sigma, St. Louis, MO] in diethyl pyrocarbonate-treated molecular-biology-grade water [Quality Biologicals]) and transported on dry ice to the testing facility, where the samples were stored at −80°C.
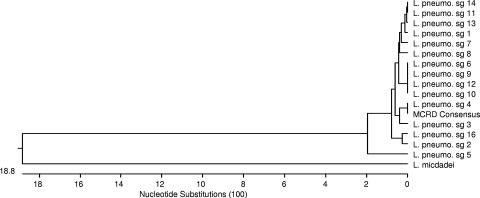
Retrospective identifications were made using a newly developed multiplex PCR test for the atypical pneumonia pathogens L. pneumophila, Mycoplasma pneumoniae, Chlamydophila pneumoniae, and Bordetella pertussis (8). This test was applied retrospectively to 240 samples collected at four military recruit training facilities as part of ongoing pneumonia surveillance of recruits. Two-hundred-microliter sample aliquots were extracted for PCR using the QIAGEN blood kit (QIAGEN, Valencia, CA) following the manufacturer's instructions. Five microliters of the resulting 200-μl extracts was used for PCR. Negative controls included a blank swab in TE buffer, and all controls yielded negative results. L. pneumophila-positive results were confirmed by replication of initial PCR and sequencing (in both directions) of the band-purified amplicons using an ABI 3130 capillary sequencer and BigDye reagents (both from Applied Biosystems, Foster City, CA) per the manufacturer's instructions. All sequences were identical (MCRD consensus, Fig. 1), and all were an exact match to L. pneumophila serogroup 4 in the region analyzed. The serogroup 4 sequence is not shared by other L. pneumophila serogroups. Phylogenic analysis of the MCRD consensus sequence is based on GenBank sequences (http://www.ncbi.nlm.nih.gov/) homologous to bases 82 to 509 of the L. pneumophila serogroup 4 macrophage infectivity potentiator protein gene (mip) (GenBank accession no. AF022318) (10). This sequence and others used for comparison were identified using the Basic Local Alignment Search Tool (1). Virus transport medium samples from the same cases were tested by PCR and culture for other common bacterial and viral pathogens associated with respiratory disease including Streptococcus pyogenes, respiratory syncytial virus, coronavirus, adenovirus, influenza virus, and rhinovirus. Three of the five were positive by at least one PCR test for rhinovirus—a rate not significantly different from that seen in general among both healthy and sick individuals in this population (Naval Health Research Center surveillance using published methods [11]; data not shown), and four of the five were positive for adenovirus by both culture and PCR, a much higher rate than that seen in healthy recruits but not significantly different from the rate seen among individuals reporting with febrile respiratory illness or other pneumonias (with or without apparent typical or atypical bacterial pneumonia agent diagnoses). The adenovirus tests (both culture and PCR) were performed under College of American Pathologists diagnostic accreditation, using previously described protocols (9).
FIG. 1.
Phylogenic analysis of the training center strain (represented by the MCRD consensus sequence) is based on GenBank sequences homologous to bases 939529 to 939956 of the L. pneumophila strain Lens complete genome (GenBank sequence accession no. NC_006369). sg, serogroup.
Identification of L. pneumophila could not be confirmed by culture, most likely due to the unpreserved nature of samples, suboptimal collection media, and the length of storage. Serologic antibody testing for L. pneumophila was ambiguous and essentially irrelevant, as the convalescent-phase serum was collected 3 weeks after the acute-phase serum, and the serological kit used suggests that conversion may require up to 9 weeks for L. pneumophila.
To further verify the results, all five positive samples as well as five temporally concurrent negative samples from pneumonia patients in different companies at the same base were retested with the primary PCR (8). All of these were originally extracted using the same batch of QIAGEN reagents. The original extracts were also tested using a published real-time primer and probe set for L. pneumophila (7) and a published 16S ribosomal sequencing method (4), using the manufacturers’ instructions with a few cycling parameter adjustments for local optimization. The results strongly supported the original results, with four of the five positive results being verified by the real-time PCR test and three of the five being verified by the 16S sequencing method, while all five coextracted samples from other companies were negative by all methods. The original samples were not available for reextraction, and hence these five negative samples served as the control for the extraction reagents.
Nineteen samples were collected during November 2004 at the MCRD. Of these, six were from B company, five of these were positive, and all five were collected in the same week. All affected patients were in the same company and hence shared much of their living and training quarters. However, this company occupied multiple buildings. Records were not available to determine the exact living quarters of the recruits. Therefore, patients were contacted at the time of writing and asked to recall their quarters when they became ill. Of the three who responded, two shared quarters in one barracks, while the third resided in another building.
Given the distribution of recruits at the training center during the time of the cluster, it is statistically unlikely that these would be clustered in the same company by chance. This suggests a common source in the recruits’ shared environment, probably a shared training facility outside the barracks. L. pneumophila is known to cause building-specific outbreaks (5), and infection by person-to-person contact has not been reported in the literature (12). Exposure to Legionella comes from environmental sources, such as fresh water and water systems for heating and cooling (12).
L. pneumophila-associated pneumonia is not generally thought to affect young adults, but rather the elderly, the immunocompromised, or those with underlying respiratory ailments (3). Cases affecting generally healthy young adults have rarely been reported, and those cases appear to have been isolated (6, 13). Given the age and general health of the recruits, identification of this cluster may suggest a previously unrecognized susceptibility of military recruit populations. Recruits in training are known to be highly susceptible to a variety of respiratory diseases.
The fact that four out of five of the patients also harbored adenovirus suggests that the very high rate and continuous occurrence of adenovirus-associated febrile respiratory illness in this population (2) may act as a predisposing factor for L. pneumophila colonization. It is impossible to say whether the L. pneumophila identified in these patients was the primary factor in their pneumonias, as adenovirus is strongly associated with pneumonia in recruit populations. We do not claim to demonstrate a causal link but feel that it is important to note the observed association in order to encourage continued surveillance for this potentially severe pathogen in recruits reporting with pneumonia. If similar future cases were recognized more quickly, greater efforts could be made to identify environmental sources, to utilize the proper serological and culture measures for legionellosis case definition, and to better define the impact of this pathogen on the affected recruits’ health.
Unique medical factors related to recruit respiratory health include crowding, stress, sleep deprivation, a large number of people brought together from different geographic environments, prophylactic antibiotic treatment, and multiple vaccinations, the last two of which are specifically directed at preventing the spread of respiratory disease and pneumonia in this highly susceptible population.
The 240 tested pneumonia samples were collected between September 2004 and November 2005. No samples other than those discussed were positive for Legionella. Given the tightness of the temporal clustering of the Legionella-positive samples, the cluster appears to have been self-limiting. Such incidents may be sporadic and thereby undetectable in the absence of active, ongoing surveillance.
(Portions of the work presented here have been previously presented at the Force Health Protection Conference [2006], the Navy Environmental Health Center Conference [2006], and the International Conference on Emerging Infectious Diseases [2006].)
Acknowledgments
This paper represents report 06-29, supported by the Department of Defense, under work unit 60501.
The views expressed in this article are those of the authors and do not reflect the official policy or position of the Department of the Navy, Department of the Army, Department of Defense, or the U.S. Government.
This research has been conducted in compliance with all applicable federal regulations governing the protection of human subjects in research (protocol NHRC.2003.0024).
Footnotes
Published ahead of print on 25 April 2007.
REFERENCES
- 1.Altschul, S. F., W. Fish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Blasiole, D. A., D. Metzgar, L. T. Daum, M. A. K. Ryan, J. Wu, C. Wills, C. T. Le, N. E. Freed, C. J. Hansen, G. C. Gray, and K. L. Russell. 2004. Molecular analysis of adenovirus isolates from vaccinated and unvaccinated young adults. J. Clin. Microbiol. 42:1686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiman, R. F., and J. C. Butler. 1998. Legionnaires’ disease: clinical, epidemiological, and public health perspectives. Semin. Respir. Infect. 13:84-89. [PubMed] [Google Scholar]
- 4.Cloud, J. L., K. C. Carroll, P. Pixton, M. Erali, and D. R. Hillyard. 2000. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J. Clin. Microbiol. 38:1709-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepard, and P. S. Brachman. 1977. Legionnaires’ disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 6.Karim, A., S. Ahmed, and L. J. Rossoff. 2002. Legionnaire's disease associated with acute encephalitis and arrhythmia. Crit. Care Med. 30:1028-1029. [DOI] [PubMed] [Google Scholar]
- 7.Khanna, M., J. Fan, K. Pehler-Harrington, C. Waters, P. Douglass, J. Stallock, S. Kehl, and K. J. Henrickson. 2005. The Pneumoplex assays, a multiplex PCR-enzyme hybridization assay that allows simultaneous detection of five organisms, Mycoplasma pneumoniae, Chlamydia (Chlamydophila) pneumoniae, Legionella pneumophila, Legionella micdadei, and Bordetella pertussis, and its real-time counterpart. J. Clin. Microbiol. 43:565-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonough, E. A., C. P. Barrozo, K. L. Russell, and D. Metzgar. 2005. A multiplex PCR for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and Bordetella pertussis, in clinical specimens. Mol. Cell. Probes 19:314-322. [DOI] [PubMed] [Google Scholar]
- 9.Metzgar, D., M. Osuna, S. Yingst, M. Rakha, K. Earhart, D. Elyan, H. Esmat, M. A. Darwish, A. Kajon, J. Wu, G. C. Gray, M. A. K. Ryan, and K. L. Russell. 2005. PCR analysis of Egyptian respiratory adenovirus isolates, including identification of species, serotypes, and coinfections. J. Clin. Microbiol. 43:5743-5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratcliff, R. M., J. A. Lanser, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36:1560-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santti, J., T. Hyypia, and P. Halonen. 1997. Comparison of PCR primer pairs in the detection of human rhinoviruses in nasopharyngeal aspirates. J. Virol. Methods 66:139-147. [DOI] [PubMed] [Google Scholar]
- 12.Thibodeau, K. P., and A. J. Viera. 2004. Atypical pathogens and challenges in community-acquired pneumonia. Am. Fam. Physician 69:1700-1706. [PubMed] [Google Scholar]
- 13.Tsai, H. C., S. S. Lee, W. R. Lin, C. K. Huang, Y. S. Chen, S. R. Wann, H. H. Lin, M. Y. Yen, and Y. C. Liu. 2001. Legionnaires’ disease in an immunocompetent young adult. Kaohsiung J. Med. Sci. 17:331-335. [PubMed] [Google Scholar]