Abstract
We examined failures of commercial human immunodeficiency virus type 1 (HIV-1) viral load assays of 1,195 plasma samples from Brazilian patients. Assay failure was assumed for samples in which the virus was undetectable by commercial assay but which tested positive by real-time reverse transcription-PCR of the HIV-1 long terminal repeat (LTR) region or if the viral load differed by >2 log10 from that determined by LTR assay. Failure rates for Bayer Versant bDNA 3.0, Roche Amplicor Monitor v1.5, and bioMerieux NucliSens QT were 0.68, 0.47, and 4.33%, respectively. NucliSens may be inadequate for use in Brazil.
Worldwide, over 38 million people are estimated to be infected with human immunodeficiency virus type 1 (HIV-1) (15). About 630,000 of them live in Brazil. In Brazil, HIV-infected individuals receive treatment free of charge, which creates one of the largest populations on antiretroviral therapy worldwide (10). Determination of viral load is an integral part of therapy, necessary to recognize treatment failure and to prevent transmission of resistant strains. However, commercial viral load assays were originally designed to detect subtype B viruses, which are most prevalent in North America and Western Europe (14). It is likely that such assays will be less appropriate for other virus populations, and several earlier studies focused on this problem. Unfortunately, most studies used small numbers of clinical samples, in the range of 100 specimens (1, 4, 12, 13). No study has prospectively evaluated a routine testing program, including all major viral load assays, and none used a uniform gold standard method for comparison.
In view of the large numbers of patients under monitored therapy in Brazil, the significance of wrong viral load quantification was determined in this specific setting. We analyzed 1,195 samples from three different regions of Brazil. In a recent multicenter study on HIV-1 quantification (6), viral loads in these samples were tested with either the Bayer Versant bDNA 3.0, bioMerieux NucliSens HIV-1 QT, or Roche Amplicor Monitor v1.5 assay. Viral loads in all samples were also determined with a new real-time reverse transcription-PCR test for the highly conserved 5′ long terminal repeat (LTR) region. This assay detected non-B subtypes with improved efficiency (6). It served as our gold standard.
We selected all samples that tested negative in any of the three commercial assays and showed viral loads above 5,000 copies/ml by real-time LTR reverse transcription-PCR. In addition, samples that were underquantified by at least a factor of 100 in the commercial assays compared to results with LTR PCR were also included.
To make sure that underlying data were correct, all LTR viral loads originally determined in the previous study were confirmed by retesting the original plasma samples with the LTR assay. Viral loads determined by commercial assays were not retested, since these were issued during ongoing treatment by certified Brazilian viral load laboratories, which are subjected to strict quality control standards by the Brazilian Ministry of Health.
As shown in Table 1, the Bayer Versant bDNA 3.0 and Roche Monitor v1.5 showed significant discrepancies in 0.68% and 0.47% of samples, respectively. The NucliSens assay, however, gave discrepant results for 4.33% of the tested samples. This rate was significantly higher than those with the other commercial methods (x2 = 20.36, P < 0.0001). To exclude a systematic technical error in the gold standard method, 10 of the 14 samples with discrepant results by NucliSens were retested with Roche Amplicor (no material was left from the remaining four samples). Indeed, the virus was detected by Amplicor in 7 out of 10 samples. The median discrepancy between LTR and Amplicor results was 0.95 log10 for the positive samples. For comparison, the median difference between LTR and NucliSens was 4.44 log10.
TABLE 1.
Comparison of undetected or underquantified viral load as determined by the Bayer Versant bDNA 3.0, Roche Amplicor Monitor v1.5, and Biomerieux NucliSens QT assays
| Assay | Subtypea (p24/pol) | No. of samples | Log deviationc (range) | No. of deviant samples/samples tested |
|---|---|---|---|---|
| Roche Amplicor Monitor v1.5 | F/F | 1 | 4.68 | 2/430 |
| FB/BF | 1 | 4.23 | ||
| Bayer Versant bDNA v3.0 | B/B | 1 | 5.58 | 3/442 |
| C/C | 1 | 2.40 | ||
| F/C | 1 | 4.49 | ||
| Biomerieux NucliSens HIV-1 QT | B/B | 11 | 4.54 (2.3-5.7) | 14/323 |
| F/recb | 3 | 3.99 (1.7-5.5) |
Phylogenetic analysis comprised HXB2 genome residues 1345 to 1839 and HXB2 2266 to 3257. The primer sequences used for amplification and sequencing were HXB2 886 to 908, 1307 to 1328, 1344 to 1363, 1344 to 1369, 2155 to 2174, 2220 to 2242, 2600 to 2620 (sense) and 2176 to 2152, 1843 to 1822, 1736 to 1760, 1654 to1675, 3334 to 3311, 3308 to 3288, and 3298 to 3276 (antisense). Typing of the circulating recombinant form was based on SimPlot analysis of gag (p24)/pol genes.
Includes one F/B, one F/BF, and one F/FB gag (p24)/pol recombinant sample.
The median expressed as log10(LTR assay − commercial assay).
The reasons for failure were determined by sequencing the gag and pol genes directly from the samples. Phylogenetic analysis was performed using TreeCon 1.3. Phylogenetic typing was confirmed with Simplot 3.5.1 (8), using the subtype reference alignment of the Los Alamos HIV database supplemented by extra Brazilian B-, C-, and F-subtype sequences (three per subtype).
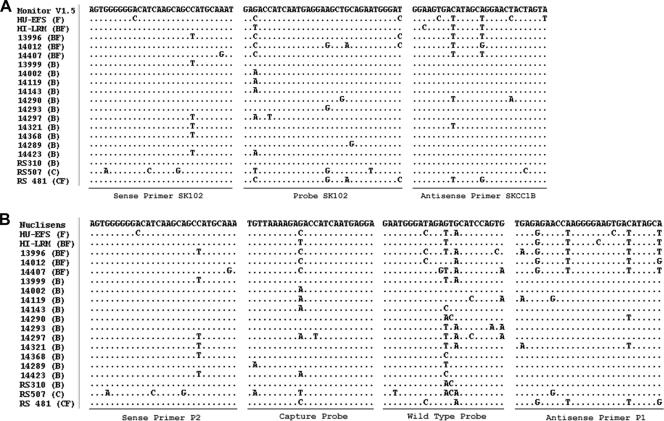
The numbers of mismatches for both the Monitor v1.5 and the NucliSens assays were highest for F, BF, C, and CF samples (Fig. 1). Both samples in which the virus was not detected by Monitor v1.5 had subtype F signatures in their amplicon target regions (gag) and revealed a common mismatch pattern in the hybridization domain of the reverse primer. To our surprise, the majority of samples with results discrepant by NucliSens contained pure subtype-B viruses (11 of 14) (Table 1); the remainder were also F recombinants. Oligonucleotide mismatches are shown in Fig. 1 for all BF recombinants and four subtype-B samples. For the remaining subtype-B samples, such mismatches could not be confirmed. Interestingly, there are other recent reports of such observations, suggesting that oligonucleotide mismatches may not be the only reason for assay failure with the NuciSens system (1, 12).
FIG. 1.
Nucleotide mismatches of Amplicor Monitor v1.5 (A) and NucliSens HIV-1 QT (B) assays at oligonucleotide binding sites in the gag p24 gene. The viral loads in samples HU-EFS and HI-LRM were underquantified by Monitor v1.5. The viral loads in samples 13996 to 14423 were underquantified by NucliSens. Samples RS310 to RS481 were underquantified by bDNA v3.0. Primer and probe sequences were obtained from N. L. Michael et al. (9) and C. Christopherson et al. (5) for Monitor v1.5 and B. van Gemen et al. (16) and I. von Truchsess et al. (18) for NucliSens, respectively.
For the bDNA assay, it was not possible to determine the contribution of virus variability to misquantification because its 98 probes in the pol gene are unpublished. Its design should be relatively stable against nucleotide mismatches (7, 13). Interestingly, for two of the three samples misquantified with this assay, the alignment in Fig. 1 suggests that they would predictably test negative with the NucliSens and Monitor assays (no retesting was possible due to lack of plasma). These viruses were C subtypes or C recombinants.
This is the first study to permit a quantitative estimation of HIV-1 viral load assay failure within Brazil, one of the largest HIV-1 treatment settings worldwide. Approximately 83% of Brazilian HIV-1 strains are subtype B, 14% are F, and 3% are C. Subtype C is predominant, occurring at a rate of up to 50%, in the south of Brazil, whereas subtype F and B/F recombinants are predominant in the north, occurring at rates of up to 50% (2, 3, 11, 17). The conformity of our samples with this spectrum of HIV-1 subtypes and the large number of samples analyzed justify extrapolation of our study to the Brazilian HIV-1 population. Currently, NucliSens is being applied for HIV-1 viral load determination in 32 of 73 Brazilian Ministry of Health reference laboratories, many of which are situated in the regions with the highest prevalence of subtype-F and F-recombinant viruses. One hundred eighty thousand individuals are on antiretroviral therapy in Brazil, receiving up to four viral load determinations per year. According to these numbers, the observed rate of 4.3% samples for which the virus was undetectable or seriously underquantified with the Biomerieux NucliSens assay can be extrapolated to at least 2,580 patients annually. Whether this assay is appropriate for Brazil at all should be reexamined.
Nucleotide sequence accession numbers.
All nucleotide sequences determined in this study are available from GenBank, accession numbers EF075946 to EF075983.
Acknowledgments
This study was partially funded by Brazilian Ministry of Health grants CFA 273/04 and CFA 238/05. The Bernhard Nocht Institute receives funding from the German Ministry of Health for operating the National Reference Centre for Tropical Infectious Diseases. L. K. de Souza Luna is receiving a scholarship from the Brazilian National Council of Technological and Scientific Development CNPQ. The authors declare that they have no conflicts of interest.
We are grateful to Daniel J. Morgan for revision of the manuscript and Carla Baroni of the Centro Biomédico da UFES—Núcleo de Doenças Infecciosas, Vitória/Espírito Santo, and Mara Liane Rieck of the Laboratório do Hospital Nossa Senhora da Conceição, Porto Alegre/Rio Grande do Sul, for kindly contributing samples.
Footnotes
Published ahead of print on 28 March 2007.
REFERENCES
- 1.Antunes, R., S. Figueiredo, I. Bartolo, M. Pinheiro, L. Rosado, I. Soares, H. Lourenco, and N. Taveira. 2003. Evaluation of the clinical sensitivities of three viral load assays with plasma samples from a pediatric population predominantly infected with human immunodeficiency virus type 1 subtype G and BG recombinant forms. J. Clin. Microbiol. 41:3361-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bongertz, V., D. C. Bou-Habib, L. F. Brigido, M. Caseiro, P. J. Chequer, J. C. Couto-Fernandez, P. C. Ferreira, B. Galvao-Castro, D. Greco, M. L. Guimaraes, M. I. Linhares de Carvalho, M. G. Morgado, C. A. Oliveira, S. Osmanov, C. A. Ramos, M. Rossini, E. Sabino, A. Tanuri, and M. Ueda, et al. 2000. HIV-1 diversity in Brazil: genetic, biologic, and immunologic characterization of HIV-1 strains in three potential HIV vaccine evaluation sites. J. Acquir. Immune Defic. Syndr. 23:184-193. [DOI] [PubMed] [Google Scholar]
- 3.Brindeiro, R. M., R. S. Diaz, E. C. Sabino, M. G. Morgado, I. L. Pires, L. Brigido, M. C. Dantas, D. Barreira, P. R. Teixeira, and A. Tanuri. 2003. Brazilian Network for HIV Drug Resistance Surveillance (HIV-BResNet): a survey of chronically infected individuals. AIDS (London) 17:1063-1069. [DOI] [PubMed] [Google Scholar]
- 4.Burgisser, P., P. Vernazza, M. Flepp, J. Boni, Z. Tomasik, U. Hummel, G. Pantaleo, and J. Schupbach. 2000. Performance of five different assays for the quantification of viral load in persons infected with various subtypes of HIV-1. J. Acquir. Immune Defic. Syndr. 23:138-144. [DOI] [PubMed] [Google Scholar]
- 5.Christopherson, C., J. Sninsky, and S. Kwok. 1997. The effects of internal primer-template mismatches on RT-PCR: HIV-1 model studies. Nucleic Acids Res. 25:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten, C., M. Panning, J. F. Drexler, F. Hansel, C. Pedroso, J. Yeats, L. K. de Souza Luna, M. Samuel, B. Liedigk, U. Lippert, M. Sturmer, H. W. Doerr, C. Brites, and W. Preiser. 2006. Ultrasensitive monitoring of HIV-1 viral load by a low-cost real-time reverse transcription-PCR assay with internal control for the 5′ long terminal repeat domain. Clin. Chem. 52:1258-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagodzinski, L. L., D. L. Wiggins, J. L. McManis, S. Emery, J. Overbaugh, M. Robb, S. Bodrug, and N. L. Michael. 2000. Use of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J. Clin. Microbiol. 38:1247-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michael, N. L., S. A. Herman, S. Kwok, K. Dreyer, J. Wang, C. Christopherson, J. P. Spadoro, K. K. Young, V. Polonis, F. E. McCutchan, J. Carr, J. R. Mascola, L. L. Jagodzinski, and M. L. Robb. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 37:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ministério da Saúde/Coordenação Nacional de DST e Aids. 2002. A experiência do programa Brasileiro de Aids. Ministério da Saúde/Coordenação Nacional de DST e Aids, Brasília, Brazil.
- 11.Soares, M. A., T. De Oliveira, R. M. Brindeiro, R. S. Diaz, E. C. Sabino, L. Brigido, I. L. Pires, M. G. Morgado, M. C. Dantas, D. Barreira, P. R. Teixeira, S. Cassol, and A. Tanuri. 2003. A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS 17:11-21. [DOI] [PubMed] [Google Scholar]
- 12.Swanson, P., C. de Mendoza, Y. Joshi, A. Golden, R. L. Hodinka, V. Soriano, S. G. Devare, and J. Hackett, Jr. 2005. Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity on performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J. Clin. Microbiol. 43:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson, P., V. Soriano, S. G. Devare, and J. Hackett, Jr. 2001. Comparative performance of three viral load assays on human immunodeficiency virus type 1 (HIV-1) isolates representing group M (subtypes A to G) and group O: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR version 1.5, and Quantiplex HIV-1 RNA version 3.0. J. Clin. Microbiol. 39:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takebe, Y., S. Kusagawa, and K. Motomura. 2004. Molecular epidemiology of HIV: tracking AIDS pandemic. Pediatr. Int. 46:236-244. [DOI] [PubMed] [Google Scholar]
- 15.UNAIDS. 2006. Report on the global AIDS epidemic. Joint United Nations Programme on HIV/AIDS/World Health Organization, Geneva, Switzerland.
- 16.van Gemen, B., R. van Beuningen, A. Nabbe, D. van Strijp, S. Jurriaans, P. Lens, and T. Kievits. 1994. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J. Virol. Methods 49:157-167. [DOI] [PubMed] [Google Scholar]
- 17.Vicente, A. C., K. Otsuki, N. B. Silva, M. C. Castilho, F. S. Barros, D. Pieniazek, D. Hu, M. A. Rayfield, G. Bretas, and A. Tanuri. 2000. The HIV epidemic in the Amazon Basin is driven by prototypic and recombinant HIV-1 subtypes B and F. J. Acquir. Immune Defic. Syndr. 23:327-331. [DOI] [PubMed] [Google Scholar]
- 18.von Truchsess, I., B. Harris, H. M. Schatzl, and J. Hackett, Jr. 2006. The first B/G intersubtype recombinant form of human immunodeficiency virus type 1 (HIV-1) identified in Germany was undetected or underquantitated by some commercial viral load assays. J. Med. Virol. 78:311-317. [DOI] [PubMed] [Google Scholar]