Abstract
The Digene Hybrid Capture system cytomegalovirus (CMV) DNA (version 2.0), Roche CMV UL54 analyte-specific reagent, and QIAGEN RealArt CMV LightCycler PCR reagent tests were compared using whole-virus standards and plasma specimens collected from allogeneic-stem-cell transplant recipients. PCR assays showed better speed, sensitivity, and specificity.
The use of in-house-developed PCR-based assays for the quantification of cytomegalovirus (CMV) DNA in clinical specimens is hampered by variability in specimen type, nucleic acid purification, target sequence, and detection method (3). The CMV UL54 analyte-specific reagent (ASR) test (Roche Diagnostics, Indianapolis, IN) and RealArt CMV LightCycler PCR reagent test (QIAGEN, Germantown, MD) are two standardized real-time PCR tests that have yet to be clinically validated. Therefore, using whole-virus standards and clinical specimens, we compared these new ASR tests with our present method, the Hybrid Capture (HC) system CMV DNA test (version 2.0; Digene Corporation, Gaithersburg, MD).
The HC test was performed with 3.5 ml of whole blood according to the manufacturer's instructions (1). For PCR analyses, nucleic acid was purified with the MagNA Pure LightCycler total nucleic acid isolation kit (Roche Applied Science, Mannheim, Germany) on the MagNA Pure instrument (Roche Diagnostics). Two hundred microliters of plasma was concentrated into a 100-μl eluate, and sequential aliquots were used for both PCR assays on the same LightCycler 1.5 instrument. The Roche ASR test is designed to detect a 240-bp fragment of the CMV DNA polymerase gene (UL54). A 20-μl reaction volume (5 μl of eluate plus 15 μl of LightCycler FastStart DNA MasterPLUS mix) was used for PCR analysis. The lysis buffer was spiked with a recovery template from Roche before nucleic acid extraction to serve as a qualitative extraction and amplification control. The QIAGEN ASR test targeted a 105-bp fragment of the CMV immediate-early gene. The reaction volume was 25 μl (10 μl of eluate plus 15 μl of QIAGEN master mix with an internal control).
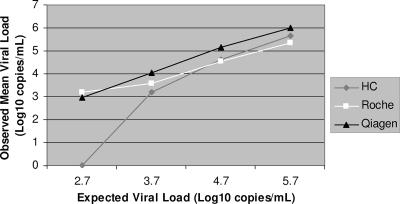
All three assays were tested using OptiQuant CMV DNA panels (AcroMetrix Corp.) that contained normal human plasma spiked with four concentrations of CMV strain AD169 (Fig. 1). Eight additional dilutions from 63 to 2,500 copies/ml were prepared from the panel with NAT dilution matrix (AcroMetrix) for analytic sensitivity analyses. After consent was obtained, whole blood was collected prospectively from adult and pediatric stem cell transplant recipients and stored in EDTA, and medical records were reviewed.
FIG. 1.
Digene HC system, Roche CMV UL54 ASR, and QIAGEN RealArt CMV LightCycler PCR reagent test comparisons using OptiQuant CMV DNA quantification panels. Expected OptiQuant viral load results: standard 1, log10 2.7 (500 copies/ml); standard 2, log10 3.7 (5,000 copies/ml); standard 3, log10 4.7 (50,000 copies/ml), and standard 4, log10 5.7 (500,000 copies/ml).
Probit analysis was used to determine the lower limit of detection, with a 95% confidence interval (CI), for each assay. Pearson correlation coefficients were applied for viral load comparisons; agreement between assays was assessed using the kappa statistic and bias plots. In three-way assay comparisons, the Friedman repeated-measures test was used for continuous variables and the Cochran-Mantel-Haenszel test was used for nominal variables. All P values and 95% CIs were two tailed.
The CMV DNA panels generated reproducible results (standard deviation range, log10 0.3 to log10 0.4 copies/ml). The PCR tests performed better at lower viral loads (Fig. 1), but only the results of the QIAGEN PCR test appeared to be log-linear across the range of CMV DNA panel concentrations. The lower limits of detection of each assay, in copies per milliliter, were as follows: HC system, 2,600 (95% CI, 1,756 to 5,869); Roche assay, 340 (95% CI, 178 to 3,005); and QIAGEN assay, 70 (the 95% CI was not computable).
Among 556 samples from 50 stem cell transplant recipients, CMV DNA was detected more often (P < 0.001) by the QIAGEN assay (79/556; 14.2%) than by the Roche (47/556; 8.5%) and HC (34/556; 6.1%) assays. The numbers of patients with detectable CMV DNA as determined by the three assays were similar (P = 0.13); however, 10 of 23 patients with discrepant results had single, positive results by HC only (likely false positive). Four patients had CMV detected by PCR only (the median viral load, in copies per milliliter, was 423 [range, 113 to 3,218] by the Roche PCR and 33 [range, 10 to 4,778] by the QIAGEN assay). None of these patients were treated with CMV-specific antiviral therapy, and none developed recognized sequelae of CMV infection.
There was moderate to substantial agreement among the three assays with clinical samples (Table 1). The Roche and QIAGEN assays had the strongest correlation (r = 0.79; P < 0.0001), followed by the HC and Roche assays (r = 0.48; P < 0.0001) and the HC and QIAGEN assays (r = 0.46; P < 0.0001). CMV viral loads detected with the QIAGEN assay tended to be lower than those detected with both the Roche (bias, log10 −0.4 [95% CI, log10 −0.6 to log10 −0.2]) and HC (bias, log10 −0.6 [95% CI, log10 −0.6 to log10 −0.3]) methods. Viral loads detected by the Roche assay were also lower than those detected by the HC system (bias, log10 −0.3 [95% CI, log10 0.0 to log10 −0.6]). PCR assays detected CMV DNA earlier posttransplant than the HC method (P = 0.001). The median time to the first positive result was 42.5 days (range, 18 to 171 days) with the HC system, 39.5 days (range, 12 to 171 days) with the Roche PCR, and 32 days (range, 12 to 171 days) with the QIAGEN assay. The median duration of CMV viremia upon the initiation of therapy was 7 days (range, 4 to 28 days) as determined by the HC system, 19 days (range, 4 to 28 days) as determined by the Roche assay, and 21 days (range, 11 to 35 days) as determined by the QIAGEN method. The CMV viral load in 4 (23.5%) of 17 patients as detected by PCR increased (2- to 25-fold) during the initial 1 to 2 weeks of treatment before declining to undetectable levels. This phenomenon was not observed with HC results. Increases in PCR-detected viral loads upon the initiation of therapy ranged from log10 0.2 to log10 1.3 and were not associated with treatment failure.
TABLE 1.
Concordance and kappa coefficients for the agreement between results of the Digene HC system, Roche CMV UL54 ASR, and QIAGEN RealArt CMV LightCycler PCR reagent tests for clinical specimens
| Test comparison | No. of identical results/total no. of results (% concordance) | Kappa coefficient |
|---|---|---|
| HC system versus Roche PCR | 513/556 (92.3) | 0.43 |
| HC system versus QIAGEN PCR | 487/556 (87.6) | 0.33 |
| Roche PCR versus QIAGEN PCR | 526/556 (94.6) | 0.64 |
Commercially available CMV reagents have the potential to promote standardization across laboratories and to enable better correlation with clinical trial results. The Roche and QIAGEN ASR tests were more sensitive than the HC assay, detected CMV DNA earlier after transplant, and remained positive longer once antiviral treatment was initiated. CMV viral loads detected by the HC system tended to be higher than those detected by PCR, in accordance with previous observations of higher viral loads in whole blood than in plasma (2). The QIAGEN assay was the more sensitive of the two PCRs (difference, log10 0.7) and performed better at the lower end of the dynamic range of standards, which may be related to the manufacturer's instructions to use 10 μl of purified DNA in the PCR compared with Roche's 5 μl.
Quantitative real-time PCR assays are likely to become the standard method for assessing CMV viral loads owing to speed, sensitivity, specificity, and improved work flow for the laboratory. Standardized CMV PCR tests should enable refined preemptive treatment strategies based on multicenter clinical trials.
Acknowledgments
This work was supported in part by NIH K12 award RR17630-03 (K.E.H.).
We thank AcroMetrix, Roche Diagnostics, and QIAGEN for the generous donation of reagents for assay comparisons and Kevin Anstrom and Christopher Woods for critical appraisal of the manuscript.
Footnotes
Published ahead of print on 21 February 2007.
REFERENCES
- 1.Mazzulli, T., L. W. Drew, B. Yen-Lieberman, D. Jekic-McMullen, D. J. Kohn, C. Isada, G. Moussa, R. Chua, and S. Walmsley. 1999. Multicenter comparison of the Digene Hybrid Capture CMV DNA assay (version 2.0), the pp65 antigenemia assay, and cell culture for detection of cytomegalovirus viremia. J. Clin. Microbiol. 37:958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razonable, R. R., R. A. Brown, J. Wilson, C. Groettum, W. Kremers, M. Espy, T. F. Smith, and C. V. Paya. 2002. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation 73:968-973. [DOI] [PubMed] [Google Scholar]
- 3.Von Muller, L., W. Hampl, J. Hinz, H. Meisel, A. Reip, E. Engelmann, R. Heilbronn, B. Gartner, O. Kramer, H. Einsele, H. Hebart, T. Ljubicic, J. Loffler, and T. Mertens. 2002. High variability between results of different in-house tests for cytomegalovirus (CMV) monitoring and a standardized quantitative plasma CMV PCR assay. J. Clin. Microbiol. 40:2285-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]