Abstract
Mycobacterium massiliense is a rapidly growing mycobacterium that is indistinguishable from Mycobacterium chelonae/M. abscessus by partial 16S rRNA gene sequencing. We sequenced rpoB, sodA, and hsp65 genes from isolates previously identified as being M. chelonae/M. abscessus and identified M. massiliense from isolates from two patients with invasive disease representing the first reported cases in the United States.
Rapidly growing mycobacterium infections are increasing in the United States (7) and are difficult to speciate by conventional methods. Partial 16S rRNA gene sequencing is the most widely used method for the identification of nontuberculous mycobacteria (6, 8, 12), but this gene target is often limited by the lack of sequence divergence among closely related Mycobacterium species (15). Mycobacterium chelonae and M. abscessus are two species that share the same 16S rRNA gene sequence, and since distinguishing these two species is clinically relevant, assays targeting base pair differences within the 16S-23S rRNA internal transcribed spacer (ITS) region have been developed (5). The ITS assay has proven to be valuable but cannot differentiate M. abscessus from M. massiliense and M. bolletii, two new species of mycobacteria that share the same 16S rRNA gene sequence with M. chelonae/M. abscessus (1, 4). Although M. massiliense and M. bolletii have not been described in the United States, these two species may have been misclassified by previous assays and may remain undetected as emerging pathogens.
We sequenced portions of the rpoB, sodA, and hsp65 genes to gain a better understanding of the frequency of detection of M. massiliense or M. bolletii among clinical isolates identified as being M. chelonae/M. abscessus by 16S and ITS assays. From this analysis, we found four isolates from two patients with identifications consistent with the novel species M. massiliense and report their clinical case histories. To our knowledge, these are the first reported cases of invasive infections from M. massiliense in the United States.
Case reports. (i) Patient 1.
A 43-year-old female from Nevada with multiple sclerosis and pacemaker placement 11 months previously presented with pacemaker pocket infection. An erythematous “lump” developed at her pacemaker site that required local incision and drainage (no culture). She was treated empirically with vancomycin but developed fever and increasing pain at the site. Intraoperative cultures from surgical debridement grew colonies of acid-fast bacilli, which were identified as being M. abscessus. She remained on vancomycin. The fever persisted, and 2 weeks later, all components of the pacemaker were removed surgically, with intraoperative cultures again being positive for M. abscessus. The isolate was susceptible only to clarithromycin (MIC < 0.12 μg/ml) and amikacin (MIC < 16 μg/ml). She was discharged and received 6 months of clarithromycin treatment.
(ii) Patient 2.
A 29-year-old female from Florida with chronic myelogenous leukemia received an allogeneic hematopoietic stem cell transplant that was complicated by chronic graft-versus-host disease. Five months following the hematopoietic stem cell transplant, she developed fever and cough. Cultures from bronchoalveolar lavage and multiple blood cultures recovered rapidly growing mycobacteria identified as being M. abscessus. The isolate was susceptible to linezolid (MIC < 8 mg/ml), clarithromycin (MIC < 0.12 μg/ml), and amikacin (MIC < 16 μg/ml). She received 6 weeks of treatment with intravenous tigecycline, oral moxifloxacin, and oral azithromycin.
Over a 7-month period, 63 clinical isolates, representing 58 patients, that were previously identified as harboring M. chelonae (n = 8) and M. abscessus (n = 55) by 16S and ITS sequence analyses (5) were retrieved retrospectively. DNA extractions, PCR, and sequencing reactions were performed as previously described (13) with amplification and sequencing primers targeting the rpoB (2), sodA (3), and hsp65 (14) genes. Neighbor-joining trees were constructed by using MEGA v3.1 (9). Isolates were identified by using the rpoB sequence criteria described previously by Adekambi et al. (2). Susceptibility testing was performed by broth microdilution according to CLSI (formerly NCCLS) standard M24-A (11). Doxycycline susceptibility testing was performed by Etest (AB Biodisk, Solna, Sweden).
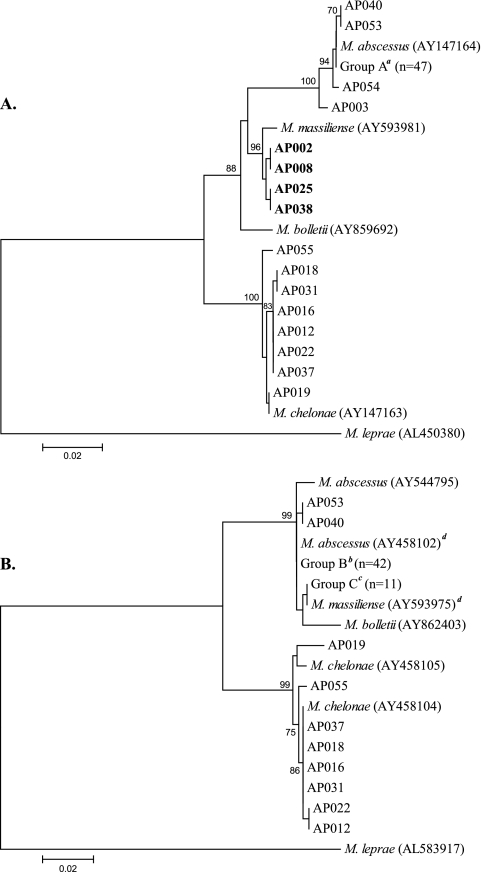
A comparison of rpoB, sodA, and hsp65 sequences is shown in Table 1. Four isolates identified as being M. abscessus by ITS assay were identified as being M. massiliense by rpoB sequencing (Fig. 1). The rpoB sequencing results agreed with results for the ITS assay for the remaining 59 isolates. With sodA sequencing, only M. chelonae was differentiated from M. massiliense and M. abscessus (Figure). A total of 11 isolates had 100% identity with the previously published sodA sequence for M. massiliense. However, the rpoB sequence identified these 11 isolates as being M. massiliense (n = 4) or M. abscessus (n = 7). A comparison of the hsp65 sequences showed results that were similar to those of sodA comparisons. The 11 isolates that shared hsp65 identity with M. massiliense correlated with the 11 isolates that shared 100% sodA identity with M. massiliense. No isolates were identified as being M. bolletii. Susceptibility patterns for all 63 isolates are shown in Table 2. All M. massiliense isolates were resistant to doxycycline.
TABLE 1.
Comparison of gene sequences from clinical isolates and reference strains of M. abscessus, M. chelonae, and M. massiliense
| Organism | No. of isolates found by ITS assay |
rpoBa
|
sodAb
|
hsp65c
|
|||
|---|---|---|---|---|---|---|---|
| No. of isolates | % Identity | No. of isolates | % Identity | No. of isolates | % Identity | ||
| M. abscessus | 55 | 47 | 100 | 42 | 100 | 44 | 100 |
| 2 | 99.9 | 2 | 99.8 | ||||
| 1 | 99.6 | ||||||
| 1 | 99.3 | ||||||
| M. chelonae | 8 | 1 | 100 | 4 | 100 | 6 | 100 |
| 4 | 99.7 | 2 | 99.8 | 1 | 99.8 | ||
| 3 | 99.6 | 1 | 99.5 | 1 | 98.8 | ||
| 1 | 98.4 | ||||||
| M. massiliense | NAd | 4 | 99.3 | 11 | 100 | 11 | 100 |
rpoB reference sequences under GenBank accession numbers AY147164 for M. abscessus, AY147163 for M. chelonae, and AY593981 M. massiliense.
sodA reference sequences under GenBank accession numbers AY458102 for M. abscessus, AY458104 for M. chelonae, and AY593975 for M. massiliense.
hsp65 reference sequences under GenBank accession numbers AY498743 for M. abscessus, AY458082 for M. chelonae, and AY596465 for M. massiliense.
NA, not applicable.
FIG. 1.
Phylogenetic trees of rpoB (A) and sodA (B) gene sequences. Bootstrapping values over 70% are recorded at the tree nodes. Mycobacterium leprae was used as an outgroup. Study isolates are designated AP001 to AP063. Isolates identified as being M. massiliense by rpoB sequencing are in boldface type. Isolates AP002 and AP008 represent patient 1, and isolates AP025 and AP038 represent patient 2. aIncludes isolates AP001, AP004 to AP007, AP009 to AP011, AP013 to AP015, AP017, AP020, AP021, AP023, AP024, AP026 to AP030, AP032 to AP036, AP039, AP041 to AP052, and AP056 to AP063. bIncludes isolates AP004 to AP007, AP009 to AP011, AP013 to AP015, AP017, AP020, AP021, AP023, AP024, AP026 to AP028, AP030, AP032 to AP036, AP039, AP041, AP042, AP044 to AP052, AP056, AP059, and AP061 to AP063. cIncludes isolates AP001, AP002, AP003, AP008, AP025, AP029, AP038, AP043, AP057, AP058, and AP060. dPublished sodA sequences for M. massiliense (GenBank accession number AY593975) and M. abscessus (accession number AY458102) differed by only 2 bp.
TABLE 2.
Antimicrobial susceptibilities of isolates by broth microdilution method
| Identification based on rpoB sequence (no. of isolates) | MIC (μg/ml) (no. of isolates)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Linezolid | Clarithromycin | Amikacin | Cefoxitin | Imipenem | Tobramycin | Ciprofloxacin | Gatifloxacin | Minocycline | Trimethoprim- sulfamethoxazole | |
| M. abscessusa (51) | 2 (1) | 0.12 (20) | 8 (12) | 16 (9) | 1 (2) | 4 (2) | 0.25 (1) | 0.5 (1) | 16 (2) | 4/76 (2) |
| 4 (4) | 0.25 (8) | 16 (23) | 32 (31) | 4 (7) | 8 (12) | 4 (6) | 4 (9) | 64 (47) | 8/152 (13) | |
| 8 (15) | 0.5 (8) | 32 (14) | 64 (9) | 8 (27) | 16 (30) | 8 (8) | 8 (10) | 16/304 (34) | ||
| 16 (22) | 1 (10) | 16 (10) | 32 (5) | 16 (20) | 16 (29) | |||||
| 32 (7) | 2 (5) | 32 (3) | 32 (14) | |||||||
| M. massiliense (4) | 32 (4) | 0.12 (4) | 16 (4) | 32 (4) | 8 (4) | 16 (3) | 16 (3) | 16 (4) | 8 (2) | 8/152 (2) |
| 32 (1) | 32 (1) | 64 (2) | 16/304 (2) | |||||||
| M. chelonae (8) | 2 (1) | 0.12 (5) | 8 (1) | 256 (1) | 4 (1) | 1 (4) | 0.5 (1) | 1 (1) | 32 (3) | 4/76 (2) |
| 4 (4) | 0.25 (3) | 16 (3) | 512 (7) | 8 (3) | 2 (4) | 1 (1) | 2 (3) | 64 (5) | 8/152 (4) | |
| 8 (3) | 32 (4) | 16 (4) | 2 (2) | 4 (2) | 16/304 (2) | |||||
| 4 (2) | 8 (2) | |||||||||
| 8 (1) | ||||||||||
| 16 (1) | ||||||||||
Two M. abscessus isolates had susceptibility testing performed for clarithromycin only.
With rpoB sequencing, 51 isolates originally identified as M. abscessus isolates by ITS assay were confirmed to be M. abscessus isolates, with 4 isolates recharacterized as harboring M. massiliense. To our knowledge, this is the first report to describe the isolation and identification of M. massiliense isolates collected in the United States associated with invasive infections, and it suggests that M. massiliense may be more commonly encountered but may be potentially misclassified as M. abscessus.
Unlike partial 16S rRNA gene sequencing, where interspecies similarity is high, with ≤0.4% sequence differences, the rpoB gene sequence is more variable, with 2 to 3% sequence differences correctly classifying most nontuberculous mycobacteria (9). In this study, we found partial rpoB gene sequencing to be a more discriminating gene target than sodA and hsp65 for M. massiliense, a unique observation that conflicts with data from previous reports (4, 10). Additionally, other investigators previously proposed that susceptibility to doxycycline may serve as a surrogate marker to differentiate M. abscessus from M. massiliense (4), but we could not confirm this finding.
Distinguishing M. chelonae from M. abscessus is clinically important because of their unique susceptibility patterns and disease manifestations. Our understanding of M. massiliense representing a distinct clinical and taxonomical entity from M. abscessus is still evolving. For our two patients, no histories of unusual exposure to animals or environmental sources were present, and their clinical presentations and susceptibility patterns were similar to those of infections caused by M. abscessus.
Overall, rpoB gene sequencing is emerging as the preferred tool to identify mycobacteria taxa (1-4) and enabled us to identify M. massiliense from two patients with invasive infection. Although the clinical significance of routinely identifying certain mycobacteria remains unclear, the 16S rRNA gene and ITS regions are often inadequate to completely capture the microbial diversity of mycobacteria, and alternative gene targets such as the rpoB gene should be considered to recognize emerging mycobacterial pathogens with the potential for invasive disease.
Acknowledgments
This work was supported by the Associated Regional and University Pathologists Institute for Clinical and Experimental Pathology, an enterprise of the University of Utah and its Department of Pathology.
We have no conflict of interest regarding the software, products, or concepts used in this study.
Footnotes
Published ahead of print on 4 April 2007.
REFERENCES
- 1.Adekambi, T., P. Berger, D. Raoult, and M. Drancourt. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133-143. [DOI] [PubMed] [Google Scholar]
- 2.Adekambi, T., P. Colson, and M. Drancourt. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adekambi, T., and M. Drancourt. 2004. Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54:2095-2105. [DOI] [PubMed] [Google Scholar]
- 4.Adékambi, T., M. Reynaud-Gaubert, G. Greub, M.-J. Gevaudan, B. La Scola, D. Raoult, and M. Drancourt. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J. Clin. Microbiol. 42:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloud, J. L., K. Hoggan, E. Belousov, S. Cohen, B. Brown-Elliot, L. Mann, R. Wilson, W. Aldous, R. J. Wallace, and G. L. Woods. 2005. Use of the MGB Eclipse system and SmartCycler PCR for differentiation of Mycobacterium chelonae and M. abscessus. J. Clin. Microbiol. 43:4205-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloud, J. L., H. Neal, R. Rosenberry, C. Y. Turenne, M. Jama, D. R. Hillyard, and K. C. Carroll. 2002. Identification of Mycobacterium spp. by using a commercial 16S ribosomal DNA sequencing kit and additional sequencing libraries. J. Clin. Microbiol. 40:400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Groote, M. A., and G. Huitt. 2006. Infections due to rapidly growing mycobacteria. Clin. Infect. Dis. 42:1756-1763. [DOI] [PubMed] [Google Scholar]
- 8.Hall, L., K. A. Doerr, S. L. Wohlfiel, and G. D. Roberts. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA 3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 10.McNabb, A., K. Adie, M. Rodrigues, W. A. Black, and J. Isaac-Renton. 2006. Direct identification of mycobacteria in primary liquid detection media by partial sequencing of the 65-kilodalton heat shock protein gene. J. Clin. Microbiol. 44:60-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NCCLS. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes: approved standard. NCCLS document M24-A. NCCLS, Wayne, PA. [PubMed]
- 12.Patel, J. B., D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmon, K. E., A. C. Croft, and C. A. Petti. 2006. Application of SmartGene IDNS software for partial 16S rRNA gene sequences for a diverse group of bacteria in a clinical laboratory. J. Clin. Microbiol. 44:4400-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tortoli, E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319-354. [DOI] [PMC free article] [PubMed] [Google Scholar]