Abstract
Anaplasma phagocytophilum is a widely distributed tick-borne pathogen of humans, livestock, and companion animals. We used in silico methods to identify 10 variable-number tandem-repeat (VNTR) loci within the genome sequence of the A. phagocytophilum HZ strain and used these data to develop a multilocus VNTR-based typing scheme for the species. Having confirmed the stability of four of the loci in replicates of the A. phagocytophilum strain that had been subjected to different numbers of passages through cell cocultures in vitro, we then used this typing scheme to discriminate between 20 A. phagocytophilum strains of diverse geographical and host provenances. Extensive diversity was found at each of the four loci studied, with total allele numbers ranging from 13 to 18 and Hunter-Gaston discriminatory index values ranging from 0.93 to 0.99. Only 2 of the 20 strains examined shared alleles at all four loci. The discriminatory power of VNTR analysis was found to be greater than that of either partial msp4 or 16S rRNA gene sequence comparison. The extremely high sensitivity of this novel approach to the genetic fingerprinting of A. phagocytophilum strains should serve well in molecular epidemiological studies of infection transmission, particularly when fine-scale strain delineation is required.
Anaplasma phagocytophilum has long been recognized as a pathogen of veterinary importance, primarily causing tick-borne fever in sheep and cattle but also being associated with infections in other domesticated animals including cats, dogs, and horses. However, more recently it has emerged as a zoonotic agent, causing human granulocytic anaplasmosis (HGA). Following its first description in the United States just over a decade ago, HGA has been reported across Europe and North and South America, and in the United States at least, HGA is now recognized as being among the most medically important tick-borne diseases (8).
A. phagocytophilum is transmitted by ticks of the genus Ixodes and is thought to exploit a wide range of mammals as reservoir hosts. In Europe, Ixodes ricinus, which has a broad host range, is generally considered the most important vector for A. phagocytophilum; surveys of questing ticks belonging to this species collected across the continent have, in some locales, revealed the presence of the bacterium at a prevalence of over 20% (12, 20). However, maintenance of A. phagocytophilum in places where I. ricinus is absent indicates that other Ixodes species are also likely competent vectors (2). The ability of different Ixodes species to transmit A. phagocytophilum is also well recognized in North America, where Ixodes scapularis and Ixodes pacificus are important vectors on the eastern and western sides of the continent, respectively (30, 34). In eastern Asia, A. phagocytophilum has been detected in questing Ixodes persulcatus and Ixodes ovatus ticks (3, 16, 25, 29). In addition to an ability to exploit different Ixodes species as vectors, A. phagocytophilum is thought to be capable of exploiting a wide range of wildlife species as reservoir hosts. Apparently asymptomatic infections have been detected in various mammal species, most frequently cervids and rodents, in Asia, Europe, and North America (2, 3, 16, 21, 22, 26, 27, 34).
Given the broad geographical and biological diversity of vectors and hosts exploited by A. phagocytophilum, it is clear that the species can be maintained in a number of different enzootic cycles in different ecosystems. One of the consequences of these geographical and ecological differences should be the existence of marked diversification among A. phagocytophilum strains, but as yet, only relatively limited measurable genetic diversity within the species has been encountered. However, this observation may be as much a consequence of methodological insensitivity as of a biological foundation. To date, genetic fingerprinting of A. phagocytophilum has almost exclusively relied on determination and comparison of nucleotide sequences, and analyses of various loci, including the 16S rRNA-encoding gene, the groESL operon (encoding a chaperonin/heat shock protein), gltA (encoding citrate synthase), msp2 (p44) and msp4 (both of which encode surface proteins), and ankA (encoding a putatively translocated protein), have been reported elsewhere (4, 6, 15, 18, 23, 32).
Among these loci, it is comparative 16S rRNA-encoding gene sequence analysis that has been used most often. Although only a few alleles have been encountered, in the United States at least, there is evidence that different alleles could be indicators of ecological differences among strains (21, 24). However, in contrast, in Europe, alleles considered indicative of different strain ecologies in North America have been encountered in the same species of host and even in sheep belonging to the same flock (31). Comparative sequence analysis of partial msp4 has recently been described and is potentially a useful new tool for A. phagocytophilum strain differentiation (6). Survey of partial msp4 sequences belonging to about 50 A. phagocytophilum strains from diverse sources worldwide demonstrated the existence of 10 alleles, the sequences of which differed by as much as 9%. Some correlation between msp4 allele type and strain provenance was observed, although half of the strains studied possessed the same msp4 sequence (6). To date, the greatest heterogeneity among A. phagocytophilum strains has been revealed by comparison of ankA sequences (23, 36). In one survey of strains infecting I. ricinus ticks in Germany, as much as 25% sequence dissimilarity was detected among complete ank open reading frames (ORFs), with 20 alleles being encountered among the 24 strains examined (36). However, given the large size (almost 4,000 bp) of ankA ORFs and the likely requirement for degenerate primer sets to amplify ankA fragments from different A. phagocytophilum strains, widespread adoption of this approach would clearly entail considerable resources and expenses. In this study we assessed the feasibility of using multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA) as a novel approach to the genetic fingerprinting of A. phagocytophilum. MLVA has previously been shown to offer very good discriminatory capacity for other species of bacteria, such as Borrelia burgdorferi, Francisella tularensis, and the obligate intracellular pathogen Coxiella burnetii (9, 10, 33). Having identified 10 loci within the A. phagocytophilum HZ strain genome sequence that possessed VNTRs, we examined the extent of heterogeneity at four of these loci among a panel of European A. phagocytophilum strains, thereby obtaining preliminary evidence for the usefulness of MLVA as a sensitive tool for discrimination of the species.
MATERIALS AND METHODS
A. phagocytophilum samples.
DNA extracts were prepared from either A. phagocytophilum-infected ISE6 cell lines or naturally infected animal blood using either a previously described alkaline lysis method (17) or the QIAamp DNA minikit (QIAGEN, Ltd., West Sussex, United Kingdom). Samples originated from Denmark, Slovenia, and the United Kingdom and were derived from symptomatic and asymptomatically infected animals. Samples were available from a variety of mammalian hosts including humans, companion animals, livestock, and wild-living ruminants (Table 1). For all the samples from naturally infected hosts used in this study, the presence of A. phagocytophilum infections was initially determined using a previously described real-time PCR assay targeting the msp2 hypervariable region flanking sequence (5).
TABLE 1.
Details of the provenances of samples used in this study and of VNTR, partial msp4, and 16S rRNA gene data obtained from them
| Strain | Country of origin | Host species | Repeat motif copy no.b
|
Sequence type
|
||||
|---|---|---|---|---|---|---|---|---|
| VNTR1 | VNTR4 | VNTR5 | VNTR8 | msp4 | 16S rRNA gene | |||
| HZ | United States | Human | 52 | 37 | 17 | 19 | i | Ac |
| SL0474a | Slovenia | Roe deer | 32 | 1 | 16 | 23 | ii | B |
| SL2514a | Slovenia | Dog | 94 | 36 | 29 | 33 | iii | C |
| SL3537a | Slovenia | Human | 35 | 39 | 21 | 59 | iii | A |
| SL3777a | Slovenia | Boar | 14 | 49 | 36 | 33 | iii | A |
| SL3883a | Slovenia | Boar | 50 | 40 | 31 | 41 | iii | A |
| AC804a | England | Dog | 6 | 1 | 30 | 1 | iv | A |
| Cairn | Scotland | Sheep | 44 | 21 | 14 | 38 | v | D |
| Feral goat | Scotland | Goat | 46 | 19 | 14 | 38 | vi | E |
| Harris | Scotland | Sheep | 46 | 19 | 14 | 38 | vi | B |
| Old Sourhope | Scotland | Sheep | 49 | 33 | 15 | 18 | vi | D |
| Perth | Scotland | Sheep | 44 | 44 | 20 | 38 | vii | D |
| R153 | Scotland | Sheep | 79 | 22 | 22 | 18 | vi | D |
| ZW122 | Wales | Sheep | 22 | 29 | 3 | 39 | viii | D |
| ZW129 | England | Sheep | 51 | 50 | 42 | 33 | ix | D |
| ZW144 | England | Cow | 32 | 39 | 34 | 37 | x | D |
| D016a | Denmark | Dog | 30 | 43 | 14 | 45 | iii | C |
| D233a | Denmark | Dog | 1 | 49 | 17 | 30 | iii | C |
| D309a | Denmark | Dog | 25 | 50 | 30 | 65 | iii | C |
| D399a | Denmark | Dog | 58 | 13 | 17 | 38 | xi | A |
These strains were not isolated; rather, DNA extracts were prepared from infected-blood samples.
Repeat motif copy numbers in bold were determined by sequencing, those in normal text were determined by fragment analysis, and those in underlined bold were determined by both approaches.
The partial 16S rRNA gene sequences have all been previously encountered, and examples of each exist in GenBank with the following accession numbers: A, AF507941; B, AY176589; C, AY281796; D, AY176587; E, AY176588.
Genomic analysis.
In May 2004, all 16 contigs available from the ongoing A. phagocytophilum HZ strain genomic sequencing project were downloaded from the TIGR webpage (www.tigr.org) and used to detect potential VNTR loci. The sequence was screened for the presence of tandem-repeat motifs with Tandem Repeats Finder software (1).
Amplification of VNTR loci and determination of sequences and sizes of amplicons.
PCR primer sets were designed around each potential VNTR locus, and the abilities of assays incorporating these primers (obtained from MWG-Biotech AG, Ebersberg, Germany) to amplify each locus among a small number of isolates were determined. Loci that repeatedly yielded amplification products and which appeared polymorphic were chosen for further evaluation. For each of these chosen loci, amplification products were sequenced and the data obtained were used to inform improved primer design. PCR assays incorporating new primer pairs were optimized in terms of thermal cycle and reaction mix formulation and then used to survey all samples included in the study. The success of each assay was assessed by UV illumination of ethidium bromide-stained 1% agarose gels on which 5-μl aliquots of amplification products had been electrophoretically resolved. When fragment analysis was employed to determine the size of amplification products, the PCRs used to generate these products incorporated a relevant 6-carboxyfluorescein-labeled forward primer. All thermal cycles were carried out on a Bio-Rad DNA Engine (Bio-Rad, Hertfordshire, United Kingdom), and all PCR reagents were obtained from ABgene (Surrey, United Kingdom).
Amplification products were processed for either sequence determination, fragment analysis, or both. For sequencing, amplification products were purified using a QIAquick PCR purification kit (QIAGEN), and the nucleotide base sequence of both strands of each product was determined using standard methods, incorporating the same primers as used in the PCRs described above, by a commercial sequencing service (Advanced Biotechnology Centre, Imperial College, London, United Kingdom). For each amplification product, sequence data derived from each primer were compared, verified, and combined using AlignPlus 4 (Scientific and Educational Software, NC). For fragment analysis, amplification products were analyzed directly at various dilutions (undiluted to 1/100 depending on the amount of product) using capillary electrophoresis on a PRISM 3730 automated DNA sequencer (Applied Biosystems Ltd., Cheshire, United Kingdom) together with X-rhodamine-labeled MapMarker1000 (Bioventures Inc., TN). Output data were processed using Genescan software (Applied Biosystems).
Amplification and sequence determination of msp4 fragments.
Nucleotide sequence data available for A. phagocytophilum msp4, as previously described (6), were obtained from GenBank and aligned with one another using AlignPlus 4, and this alignment was used to inform the design of primers for use in a nested PCR assay. The resulting assay consisted of two identical thermal cycles, comprising 94°C for 5 min; then 40 cycles of 94°C for 10 s, 58°C for 10 s, and 72°C for 50 s; and then 72°C for 5 min. First-round reaction mixtures (25 μl) contained 12.5 μl of 2× PCR Mastermix (ABgene), 1 μl of a 10-pmol μl−1 solution of primers MSP4AP5 and MSP4AP3 (6), 9.5 μl of water, and 1 μl of DNA extract. Second-round reaction mixtures (25 μl) contained 12.5 μl of 2× Mastermix (ABgene), 1 μl of a 10-pmol μl−1 solution of primers msp4f (5′ CTA TTG GYG GNG CYA GAGT) and msp4r (5′ GTT CAT CGA AAA TTC CGT GGT A), 9.5 μl of water, and 1 μl of the first-round postamplification mix. The success of amplification reactions was verified as described above, and amplification products were purified and then sequenced as described above using assays incorporating the same primers as used in the second round of the msp4 PCR. For each amplification product, sequence data derived from each primer were compared, verified, and combined using AlignPlus 4 and then primer sequences at both extremities were removed. The resulting 301-bp sequences were then compared with one another and with previously determined A. phagocytophilum msp4 sequences using AlignPlus 4. For phylogenetic studies, partial msp4 sequences were aligned with one another using Clustal X (35) and inferences were drawn from this alignment using programs within the PHYLIP suite (11).
Amplification and sequence determination of 16S rRNA gene fragments.
16S rRNA gene fragments were amplified from nucleic acid extracts prepared from A. phagocytophilum strains, and then amplification products were sequenced as previously described (2). Sequences were handled using AlignPlus 4, as described above.
Determination of the discriminatory ability of VNTR.
The abilities of individual or grouped VNTRs to discriminate among the A. phagocytophilum strains studied were assessed using the Hunter-Gaston discriminatory index (HGDI) (14). This index is based on the probability that two unrelated strains will be placed into different typing groups and was described particularly for use on representative (nonlocal) collections of distinct strains. An acceptable level for discrimination clearly depends on a number of factors, but the authors suggest that an index of greater than 0.90 is desirable if typing results are to be interpreted with confidence (14).
Nucleotide sequence accession numbers.
The novel partial msp4 sequences reported in this study have been deposited in GenBank with the following accession numbers: SL0474 (type ii) msp4, EF442003; SL2514 (type iii) msp4, EF442004; AC804 (type iv) msp4, EF442005; Cairn (type v) msp4, EF442006; Feral goat (type vi) msp4, EF442007; Perth (type vii) msp4, EF442008, ZW122 (type viii) msp4, EF442009; ZW129 (type ix) msp4, EF442010; ZW144 (type x) msp4, EF442011; D399 (type xi) msp4, EF442012.
RESULTS
Identification of VNTR loci in the A. phagocytophilum HZ strain genome sequence and characterization of loci selected for MLVA-based genetic fingerprinting.
Our interrogation of the genome sequence of A. phagocytophilum HZ strain revealed 10 potential VNTR loci (Table 2). All but one of these lay outside ORFs or pseudogenes, as subsequently described in the annotated genome (13). The exception, VNTR7, lay within a 273-bp putative ORF with no known homologs. This locus was excluded from further analysis. The VNTR5 and VNTR6 loci lay close to one another (Table 2), approximately 600 bp apart on the genome; hence, only one (VNTR5) was used for further analysis. The sizes of the VNTR motifs were similar to those commonly reported for other bacteria (ranging from 7 to 15 bp), but the number of repeats encountered at some loci was high (up to 80) (Table 2). Attempted amplification of the eight remaining loci, using primers designed by reference to the HZ genome sequence, from a small panel of A. phagocytophilum DNA extracts, yielded mixed results: some loci yielded a single amplification product for all the extracts tested, but others either failed to yield a detectable amplification product with all or some of the extracts or yielded multiple amplification products of various sizes from the same DNA extract. Following optimization attempts, which incorporated various modifications of the amplification procedure (temperature gradient PCRs and nested PCRs) and repeated extraction of A. phagocytophilum DNA, four loci were considered unsuitable for our needs. The remaining four loci (VNTR1, VNTR4, VNTR5, and VNTR8) were explored in 20 A. phagocytophilum strains (Table 1), using PCRs incorporating locus-specific primers (Table 3). A total of 51 amplification products were analyzed by sequencing. However, many contained VNTRs that were too long (>600 bp) to reliably permit complete coverage using this approach; thus, the VNTR number could be accurately estimated only using fragment analysis. Once fragment analysis had been introduced and evaluated, it was used in place of sequencing to estimate repeat motif number in amplification products generated later in our study.
TABLE 2.
Details of the 10 VNTR loci identified in the A. phagocytophilum HZ strain genome sequence
| VNTR locus | Genome coordinate | Repeat motif sequence | No. of repeats |
|---|---|---|---|
| VNTR1 | 1026286 | CTCTGGTCT | 52 |
| VNTR2 | 937744 | TGGGCTAT | 32 |
| VNTR3 | 308635 | GTCAGGAA | 80 |
| VNTR4 | 293445 | TTGCTCA | 37 |
| VNTR5 | 214596 | TGAAAGGTATCGCGG | 17 |
| VNTR6 | 215448 | ATGTGCT | 49 |
| VNTR7 | 1444211 | GAAGGCA | 12 |
| VNTR8 | 1168712 | GGAGACGTACT | 19 |
| VNTR9 | 645705 | TGACTGGG | 35 |
| VNTR10 | 657819 | TGTTCC | 12 |
TABLE 3.
Details of PCR primers used for amplification of A. phagocytophilum VNTR loci
| VNTR locus | Forward primer | Reverse primer | Size range of PCR products (bp) | Range of VNTR repeat units | No. of allelesa | HGDI |
|---|---|---|---|---|---|---|
| VNTR1 | TGTGCAGGTTTAGAGGCAA | CATAGATCTATGTGATCTCT | 164-1001 | 1-94 | 18 | 0.989 |
| VNTR4 | GAAAGAGATGTTCTCAGCTT | GCAAAGGGTTTAACCTATGA | 410-753 | 1-50 | 15 | 0.974 |
| VNTR5 | GACCATAAGTTGAAGACAGGGA | AATCCGGCTTATCTCCACCTG | 192-777 | 3-42 | 15 | 0.947 |
| VNTR8 | TGCATGCATATTCTCGAGGATT | GTCCGTGGTCTATCTTATACA | 202-906 | 1-65 | 13 | 0.926 |
Number of alleles observed among the 20 A. phagocytophilum strains examined.
For each locus, the numbers of repeat motifs in amplification products from at least four A. phagocytophilum strains were estimated using both sequencing and fragment analysis. For one locus (VNTR8) these estimates matched, but for the remaining three, fragment analysis either underestimated (VNTR4) or overestimated (VNTR1 and VNTR5) by two repeats. The stability of each VNTR locus was examined using the Old Sourhope strain of A. phagocytophilum. DNA extracts were prepared from isolates of this strain recovered following 4th and 20th passages in coculture with ISE6 cells. The numbers of repeats present at all four VNTR loci were the same in the two extracts.
The range in the number of repeats found among the 20 strains examined at the four loci was broad (Table 1). At one extreme, for three loci, at least one strain possessed only a single copy of the VNTR motif. At the other extreme, one strain possessed 94 repeat motifs at the VNTR1 locus. At each VNTR locus, sequence analysis of amplification products indicated high conservation of repeat motifs among the 20 A. phagocytophilum strains examined. Among the eight VNTR1 loci sequenced, all possessed complete and identical repeat motifs. Among 15 VNTR4 loci, all possessed complete repeat motifs but that of the HZ strain differed from that of the other 14 strains by a single substitution. Among the 16 VNTR5 loci examined, all possessed complete repeat motifs, but three variants were observed (Table 4), differing by substitutions and insertions/deletions. Among the 12 VNTR8 loci examined, the repeat motifs were complete and identical in 11 strains, but one (ZW129), although complete, possessed a single substitution.
TABLE 4.
Variation in VNTR5 repeat motifs observed among the 16 A. phagocytophilum strains for which sequence data were available
| A. phagocytophilum strain designation(s) | VNTR5 repeat motif |
|---|---|
| HZ, R153 | TGAA-AGGTATCGCGG |
| SL2514, FG, H, OS, PR, D016, D233, D309 | TGTAGAGGTATCGCGG |
| SL0474, SL3537, SL3777, SL3883, AC804, D399 | TATAGAGGTATCGCGG |
Application of VNTR analysis to the genetic fingerprinting of A. phagocytophilum and comparison with partial msp4 and 16S rRNA gene sequence analysis.
The HGDI was used to assess the abilities of the four VNTR loci to differentiate among the 20 studied A. phagocytophilum strains. Between 13 and 18 alleles were encountered at each locus, with VNTR1 demonstrating the greatest discrimination (HGDI = 0.989) and VNTR8 the lowest (HGDI = 0.926) (Table 3). Eighteen of the 20 A. phagocytophilum strains examined shared at least one allele with at least one other strain, and these common alleles were shared by up to five different strains (Table 1). Apart from the two strains that shared alleles at all four loci, no clear pattern of relatedness could be discerned among the strains examined (Table 1), and given the opportunistic manner in which strains were selected for this study, further exploration of possible correlation between the MLVA fingerprint of a strain and its host species or country of origin was not carried out.
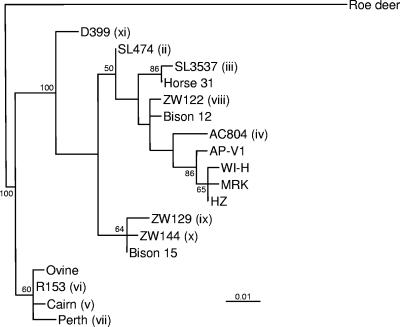
Comparison of partial msp4 sequences identified 11 different sequence types among the 20 A. phagocytophilum strains (Table 1), yielding an HGDI of 0.858. Two of the sequence types were shared by more than one strain, with type iii being shared by most of the strains of Slovenian or Danish origin and type vi being shared by four strains recovered from United Kingdom sheep, including the two which possessed identical VNTR alleles (Table 1). Comparison of the 10 previously unreported msp4 sequences used in our study with the nine different partial (301-bp) sequences previously found among a wide diversity of North American and European A. phagocytophilum isolates (6) revealed that all were similar (generally <5% dissimilarity) but new. Phylogenetic inference based on parsimony analysis of an alignment of these 19 sequences indicated that the strains examined in our study shared a close and specific evolutionary relationship with those previously examined (Fig. 1). The topology of this reconstruction was very similar to those of reconstructions inferred using maximum-likelihood and distance matrix-based methods (data not shown) and to that described in previous studies (6). Furthermore, strong bootstrap support was obtained for some of the proposed branching orders (Fig. 1). Perhaps of most significance was the clustering of ruminant (mainly sheep) isolates from Norway and the United Kingdom on a deeply diverging lineage; however, United Kingdom sheep-associated isolates were also encountered elsewhere in the tree, and all the 10 new sequence types were far less divergent from the majority of previously characterized strains than the “roe deer” sequence type associated with isolates obtained from a German roe deer (6).
FIG. 1.
Phylogenetic dendrogram inferred using parsimony approach from alignment of 301-bp sequences for the representatives of the 10 novel partial msp4 sequence types encountered in this study (ii to xi) and representatives of nine partial msp4 sequence types described previously (6). Numbers on the branches indicate the percent support for each of the branching orders proposed, as determined by bootstrap analysis of 1,000 replicates.
Comparison of partial 16S rRNA gene sequences identified five sequence types among the 20 strains examined, yielding an HGDI of 0.774. All sequence types but one were possessed by more than one strain, and one sequence type was shared by seven strains (Table 1). Sequence analysis revealed that the five sequence types were derived from only four point substitutions within a 497-bp alignment (<1% dissimilarity). All five sequence types had been encountered in previous studies.
There did not appear to be any clear correlation between the three approaches to genetic fingerprinting used in this study. The distribution of 16S rRNA gene sequence types among the strains studied did not bear any apparent congruence to that of msp4 sequence types, and neither data set demonstrated an obvious relationship with the MLVA. Remarkably, the “Feral goat” and “Harris” strains, which were indistinguishable by VNTR and msp4-based analysis, belong to different 16S rRNA gene sequence types (Table 1). Thus, combination of the approaches to fingerprinting used in this study resulted in all 20 strains studied being distinguished from one another.
DISCUSSION
The recent release of the A. phagocytophilum genome sequence has provided a resource of exceptional importance for improving our understanding of the biology of this enigmatic organism and the epidemiology of the infections that it causes (13). One of the most immediate benefits from this release has been to provide a shortcut for identifying loci around the genome that have the potential for hypervariability. Exploitation of such loci in novel approaches to the detection of intraspecies genetic delineation has been reported for numerous other bacterial pathogens (19), and in this study, we add A. phagocytophilum to this list.
MLVA of A. phagocytophilum appears to be a practical and potentially very useful addition to the repertoire of molecular methods currently used to explore the genetic diversity of the species. We found all four VNTR loci to be stable with time/passage and began to explore the usefulness of MLVA for genetic fingerprinting of the species. In our hands, MLVA proved to be a markedly more sensitive approach to delineating among A. phagocytophilum strains than either msp4 or 16S rRNA gene-based analyses, which are two of the most widely applied approaches currently in use (6, 22). The MLVA scheme described herein distinguished between 19 of the 20 strains examined, yielding an HGDI of 0.995, whereas msp4 and 16S rRNA gene analyses yielded HGDIs of 0.858 and 0.774, respectively.
Although we have demonstrated the usefulness of MLVA for the delineation of A. phagocytophilum strains, our study has not provided a clear indication that this approach is also useful for comparative assessment of genetic relatedness among strains. The number of shared alleles encountered at each VNTR locus was remarkably low compared to MLVA schemes described for other bacteria (19). Although this heterogeneity is useful in that each locus has strong discriminatory power, among the strains that we studied it was just too extensive to be practical for comparative purposes. This observation does not, however, exclude the application of our MLVA scheme to this end; it may well be useful for comparing strains that may be linked epidemiologically, for example, those circulating in a particular flock of sheep or among the inhabitants of a specific woodland. There is, perhaps, some support for this scenario in our results, with strains infecting Scottish sheep sharing markedly more alleles with one another than with strains not infecting Scottish sheep.
The use of specific PCRs for MLVA permits the analysis of A. phagocytophilum strains in infected tissues, thereby circumventing the need to culture these highly fastidious organisms. Although methods for the recovery of A. phagocytophilum from infected material are well established and relatively reliable, they require tissue culture facilities that may not be available in routine bacteriology laboratories. Thus, the development of genetic fingerprinting tools for which isolation of A. phagocytophilum is not a prerequisite, such as the MLVA scheme described herein, has clear practical advantages. Indeed, although culture-reliant methods such as pulsed-field gel electrophoresis have been applied to the study of A. phagocytophilum genetic diversity (7), PCR-based methods have been used in an overwhelming proportion of the studies reported.
Our characterization of nine novel msp4 sequence types among the 19 previously uncharacterized A. phagocytophilum strains that we examined almost doubles the number currently reported. That none of the msp4 sequence types that we described has previously been encountered is surprising. The paper in which the development and application of comparative analysis of msp4 sequences for delineation of A. phagocytophilum were first reported (6) described 10 sequence types among 50 strains recovered from eight different host species (human, dog, horse, white-tailed deer, roe deer, donkey, bison, and sheep) in five different European countries (Switzerland, Italy, Germany, Poland, and Norway) and the United States. Our encounter with an entirely novel set of sequence types in a smaller group of strains of a less varied provenance suggests that the vast majority of msp4 sequence types remain to be discovered. Interestingly, although our findings point to serious shortcomings in the current knowledge of msp4 heterogeneity and thus, perhaps, the folly of attempting epidemiological inferences from the limited data set that we have, in our sample, human and ruminant-associated strains were distinct, as previously reported (6). Clearly, more rigorous comparisons are necessary before any significance can be put on this apparent delineation.
Our finding of incongruence between the approaches to A. phagocytophilum genetic fingerprinting is not entirely unexpected. von Loewenich et al. (36) made similar observations when comparing intraspecies delineations derived from comparison of ankA, 16S rRNA gene, and groEL sequence data. Our findings are also consistent with the conclusion drawn by these authors (36) that the sequence variability at one genetic locus is not sufficient to determine the genetic diversity of a certain strain. Furthermore, given that, in general, no clear association between genotype and any epidemiological parameter has emerged, we echo previous workers in suggesting that a framework for the discovery of biologically meaningful genetic variation among A. phagocytophilum strains can be constructed only with a better understanding of ecological differences within the species. Specifically we need to further assess the degree to which strains have adapted to exploit differing transmission pathways comprising different host and vector species, particularly as the limited experimental work in this area has provided support for the existence of such adaptation (4, 24, 28).
Acknowledgments
This work was funded by project grant 070675/Z/03/Z from The Wellcome Trust.
Footnotes
Published ahead of print on 18 April 2007.
REFERENCES
- 1.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bown, K. J., M. Begon, M. Bennett, Z. Woldehiwet, and N. H. Ogden. 2003. Seasonal dynamics of Anaplasma phagocytophila in a rodent-tick (Ixodes trianguliceps) system, United Kingdom. Emerg. Infect. Dis. 9:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, W. C., L. Zhan, J. He, J. E. Foley, S. J. De Vlas, X. M. Wu, H. Yang, J. H. Richardus, and J. D. Habbema. 2006. Natural Anaplasma phagocytophilum infection of ticks and rodents from a forest area of Jilin Province, China. Am. J. Trop. Med. Hyg. 75:664-668. [PubMed] [Google Scholar]
- 4.Casey, A. N., R. J. Birtles, A. D. Radford, K. J. Bown, N. P. French, Z. Woldehiwet, and N. H. Ogden. 2004. Groupings of highly similar major surface protein (p44)-encoding paralogues: a potential index of genetic diversity amongst isolates of Anaplasma phagocytophilum. Microbiology 150:727-734. [DOI] [PubMed] [Google Scholar]
- 5.Courtney, J. W., L. M. Kostelnik, N. S. Zeidner, and R. F. Massung. 2004. Multiplex real-time PCR for detection of Anaplasma phagocytophilum and Borrelia burgdorferi. J. Clin. Microbiol. 42:3164-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de la Fuente, J., R. F. Massung, S. J. Wong, F. K. Chu, H. Lutz, M. Meli, F. D. von Loewenich, A. Grzeszczuk, A. Torina, S. Caracappa, A. J. Mangold, V. Naranjo, S. Stuen, and K. M. Kocan. 2005. Sequence analysis of the msp4 gene of Anaplasma phagocytophilum strains. J. Clin. Microbiol. 43:1309-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumler, J. S., K. M. Asanovich, and J. S. Bakken. 2003. Analysis of genetic identity of North American Anaplasma phagocytophilum strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:3392-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumler, J. S., K. S. Choi, J. C. Garcia-Garcia, N. S. Barat, D. G. Scorpio, J. W. Garyu, D. J. Grab, and J. S. Bakken. 2005. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg. Infect. Dis. 11:1828-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farlow, J., D. Postic, K. L. Smith, Z. Jay, G. Baranton, and P. Keim. 2002. Strain typing of Borrelia burgdorferi, Borrelia afzelii, and Borrelia garinii by using multiple-locus variable-number tandem repeat analysis. J. Clin. Microbiol. 40:4612-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Grzeszczuk, A., and J. Stanczak. 2006. Highly variable year-to-year prevalence of Anaplasma phagocytophilum in Ixodes ricinus ticks in northeastern Poland: a 4-year follow-up. Ann. N. Y. Acad. Sci. 1078:309-311. [DOI] [PubMed] [Google Scholar]
- 13.Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inokuma, H., P. Brouqui, M. Drancourt, and D. Raoult. 2001. Citrate synthase gene sequence: a new tool for phylogenetic analysis and identification of Ehrlichia. J. Clin. Microbiol. 39:3031-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, C. M., Y. H. Yi, D. H. Yu, M. J. Lee, M. R. Cho, A. R. Desai, S. Shringi, T. A. Klein, H. C. Kim, J. W. Song, L. J. Baek, S. T. Chong, M. L. O'Guinn, J. S. Lee, I. Y. Lee, J. H. Park, J. Foley, and J. S. Chae. 2006. Tick-borne rickettsial pathogens in ticks and small mammals in Korea. Appl. Environ. Microbiol. 72:5766-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurtenbach, K., M. Peacey, S. G. Rijpkema, A. N. Hoodless, P. A. Nuttall, and S. E. Randolph. 1998. Differential transmission of the genospecies of Borrelia burgdorferi sensu lato by game birds and small rodents in England. Appl. Environ. Microbiol. 64:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, Q., Y. Rikihisa, R. F. Massung, Z. Woldehiwet, and R. C. Falco. 2004. Polymorphism and transcription at the p44-1/p44-18 genomic locus in Anaplasma phagocytophilum strains from diverse geographic regions. Infect. Immun. 72:5574-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindstedt, B. A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 26:2567-2582. [DOI] [PubMed] [Google Scholar]
- 20.Mantelli, B., E. Pecchioli, H. C. Hauffe, R. Rosa, and A. Rizzoli. 2006. Prevalence of Borrelia burgdorferi s.l. and Anaplasma phagocytophilum in the wood tick Ixodes ricinus in the Province of Trento, Italy. Eur. J. Clin. Microbiol. Infect. Dis. 25:737-739. [DOI] [PubMed] [Google Scholar]
- 21.Massung, R. F., J. W. Courtney, S. L. Hiratzka, V. E. Pitzer, G. Smith, and R. L. Dryden. 2005. Anaplasma phagocytophilum in white-tailed deer. Emerg. Infect. Dis. 11:1604-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massung, R. F., M. J. Mauel, J. H. Owens, N. Allan, J. W. Courtney, K. C. Stafford, III, and T. N. Mather. 2002. Genetic variants of Ehrlichia phagocytophila, Rhode Island and Connecticut. Emerg. Infect. Dis. 8:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massung, R. F., J. H. Owens, D. Ross, K. D. Reed, M. Petrovec, A. Bjoersdorff, R. T. Coughlin, G. A. Beltz, and C. I. Murphy. 2000. Sequence analysis of the ank gene of granulocytic ehrlichiae. J. Clin. Microbiol. 38:2917-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massung, R. F., R. A. Priestley, N. J. Miller, T. N. Mather, and M. L. Levin. 2003. Inability of a variant strain of Anaplasma phagocytophilum to infect mice. J. Infect. Dis. 188:1757-1763. [DOI] [PubMed] [Google Scholar]
- 25.Ohashi, N., M. Inayoshi, K. Kitamura, F. Kawamori, D. Kawaguchi, Y. Nishimura, H. Naitou, M. Hiroi, and T. Masuzawa. 2005. Anaplasma phagocytophilum-infected ticks, Japan. Emerg. Infect. Dis. 11:1780-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovec, M., A. Bidovec, J. W. Sumner, W. L. Nicholson, J. E. Childs, and T. Avsic-Zupanc. 2002. Infection with Anaplasma phagocytophila in cervids from Slovenia: evidence of two genotypic lineages. Wien. Klin. Wochenschr. 114:641-647. [PubMed] [Google Scholar]
- 27.Polin, H., P. Hufnagl, R. Haunschmid, F. Gruber, and G. Ladurner. 2004. Molecular evidence of Anaplasma phagocytophilum in Ixodes ricinus ticks and wild animals in Austria. J. Clin. Microbiol. 42:2285-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pusterla, N., J. B. Pusterla, U. Braun, and H. Lutz. 1999. Experimental cross-infections with Ehrlichia phagocytophila and human granulocytic ehrlichia-like agent in cows and horses. Vet. Rec. 145:311-314. [DOI] [PubMed] [Google Scholar]
- 29.Rar, V. A., N. V. Fomenko, A. K. Dobrotvorsky, N. N. Livanova, S. A. Rudakova, E. G. Fedorov, V. B. Astanin, and O. V. Morozova. 2005. Tickborne pathogen detection, Western Siberia, Russia. Emerg. Infect. Dis. 11:1708-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richter, P. J., Jr., R. B. Kimsey, J. E. Madigan, J. E. Barlough, J. S. Dumler, and D. L. Brooks. 1996. Ixodes pacificus (Acari: Ixodidae) as a vector of Ehrlichia equi (Rickettsiales: Ehrlichieae). J. Med. Entomol. 33:1-5. [DOI] [PubMed] [Google Scholar]
- 31.Stuen, S., I. Van De Pol, K. Bergstrom, and L. M. Schouls. 2002. Identification of Anaplasma phagocytophila (formerly Ehrlichia phagocytophila) variants in blood from sheep in Norway. J. Clin. Microbiol. 40:3192-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumner, J. W., W. L. Nicholson, and R. F. Massung. 1997. PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J. Clin. Microbiol. 35:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svraka, S., R. Toman, L. Skultety, K. Slaba, and W. L. Homan. 2006. Establishment of a genotyping scheme for Coxiella burnetii. FEMS Microbiol. Lett. 254:268-274. [DOI] [PubMed] [Google Scholar]
- 34.Telford, S. R., III, J. E. Dawson, P. Katavolos, C. K. Warner, C. P. Kolbert, and D. H. Persing. 1996. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc. Natl. Acad. Sci. USA 93:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Loewenich, F. D., B. U. Baumgarten, K. Schroppel, W. Geissdorfer, M. Rollinghoff, and C. Bogdan. 2003. High diversity of ankA sequences of Anaplasma phagocytophilum among Ixodes ricinus ticks in Germany. J. Clin. Microbiol. 41:5033-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]