Abstract
The M-Coffee server is a web server that makes it possible to compute multiple sequence alignments (MSAs) by running several MSA methods and combining their output into one single model. This allows the user to simultaneously run all his methods of choice without having to arbitrarily choose one of them. The MSA is delivered along with a local estimation of its consistency with the individual MSAs it was derived from. The computation of the consensus multiple alignment is carried out using a special mode of the T-Coffee package [Notredame, Higgins and Heringa (T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000; 302: 205–217); Wallace, O'Sullivan, Higgins and Notredame (M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006; 34: 1692–1699)] Given a set of sequences (DNA or proteins) in FASTA format, M-Coffee delivers a multiple alignment in the most common formats. M-Coffee is a freeware open source package distributed under a GPL license and it is available either as a standalone package or as a web service from www.tcoffee.org.
INTRODUCTION
The computation of an accurate multiple sequence alignment (MSA) is central to a large number of bioinformatics analyses, ranging from phylogeny, profile construction, structure prediction and more recently sequence/structure activity relationship. Despite its importance, the MSA problem has not yet met with a definitive answer and a wide variety of alternative methods are currently available (3,4). All these methods are meant to address the same problem in different ways. In recent years, many efforts have been undertaken to characterize their relative accuracy but the overall outcome suggests that there is no such thing as a perfect MSA method, with each individual method having specific strengths and weaknesses. In practice, evaluation is made using structure-based MSAs as a standard of truth and the expected accuracy of a method is deduced from its ability to produce a structurally correct sequence alignment while using sequence information only. At least five such collections of reference alignments (5–8) have been established, and although some methods give better average results than others, one cannot know in advance which method will outperform the others on a given dataset. As such, it is always possible for the worst method to outperform all the others on a specific dataset. For the biologist, this makes it impossible to use anything other than a weak statistical argument (i.e. best method on average) to choose one method among the others when computing an alignment.
The design of meta-methods (or jury-based methods) is one way of addressing such situations in biology. Meta-methods are meant to combine the output of several alternative methods into one final output. They are based on the empirical reasoning that errors produced by independent prediction systems should not be consistent, therefore suggesting agreement as an indication of correctness. Such an approach was successfully used in the field of gene predictions (9) or for secondary structure predictions (10). Combining alignments, however, is less simple than building consensus prediction and it is only in 1999 that an effective strategy was proposed by Bucka-Lassen (11). An alternative to the Bucka-Lassen strategy, using consistency, was later introduced in the T-Coffee (1) algorithm. Recently, this algorithm was further modified in order to address the problem of combining alternative MSAs into one (2). T-Coffee (1) is a progressive consistency-based algorithm that compiles an alignment on the basis of its consistency with a collection of pairwise constraints. In practice, the constraints correspond to pairs of residues that could end up aligned in the final alignment. These constraints, however, are not necessarily all compatible with one another and the goal of the algorithm is to fit as many as possible within the final alignment, while discarding those that were hopefully biologically less relevant. The term consistency refers to the notion that one tries to compute the alignment having the highest possible consistency with the constraint list. This notion was introduced by Gotoh (12) and later re-used in several algorithms (13,14). In 2000, Notredame et al. (1) described a variation of the progressive algorithm using consistency as a scoring scheme. This combination proved quite successful and is now at the core of several MSA packages (15–17). In its default mode, T-Coffee uses, as a list of constraints, all the pair-wise matches extracted from a compilation of all possible global pair-wise alignments and the 10 best local alignments from each pair of sequences. Yet, this is merely one of the possible recipes to assemble such a list of constraints, and alternatives are possible. For instance, ProbCons (16) uses suboptimal pairwise global alignments (as emitted by an HMM with posterior decoding); PCMA (15) uses pairwise profile comparisons and Expresso (18) uses a mixture of sequence and structure-based alignments. Following the same principle, it is also possible to generate alternative MSAs and compile them into a single list of constraints. This latest approach forms the basis of M-Coffee (2), where eight MSA methods are used to generate alternative MSAs. Extensive benchmarking showed that this combination results in a modest but consistent improvement over each individual method, with M-Coffee producing the best scoring alignment on two of three of the datasets contained in BaliBase (5), Prefab (6) and Homstrad (2).
Another interesting by-product of alignment combination is the possibility of estimating the local consistency between the final alignment and the individual alignments. This amounts to measuring, for every residue, the fraction of individual alignments that support its position in the final alignment. This measure is named the CORE index (Consistency of Overall Residue Evaluation) and was shown to be very informative with respect to the overall alignment accuracy (19). These initial reports recently gained further support thanks to some extensive analysis carried out by Sonhammer et al.(20) whose results indicate that the consistency between an MSA and a pre-computed collection of alternative alignments gives very reliable information with respect to the structural correctness of that alignment. As such, the local consistency measure appears to be one of the most reliable predictors of alignment accuracy available today.
The server we present here computes an alignment with eight of the most commonly used MSA packages. It then outputs a consensus alignment along with a CORE-based local evaluation that can either be color-coded or ASCII based. Two mirrors of these services currently run on separate clusters: one at the Swiss Institute of Bioinformatics on the Vital-IT framework, the other at the CNRS in Marseilles, France. Both mirrors can be accessed via the T-Coffee homepage: www.tcoffee.org and extra mirrors should be added in the close future.
METHODS
Primary library: computation of the initial MSAs
The principle of M-Coffee is to compute several alternative multiple alignments in order to combine them into one consensus alignment. By default, eight methods were chosen for this purpose: PCMA (Version 2.0) (15), POA (Version 2.0) (21), Dialign-t (Version 0.2.1) (22), MAFFT (Version 5.431, L-INS-i) (17), Muscle (Version 3.6), ProbCons (Version 1.2), ClustalW (23) and T-Coffee (1). Apart from MAFFT that is used in its most accurate mode (mafft- - localpair- -maxiterate 1000) all the methods are run on the initial dataset using the default parameters. This produces an MSA that is then turned into a T-Coffee primary library. All these libraries are then combined in order to generate an MSA.
M-Coffee alignment computation
In order to compute the final alignment, the server runs the following command:
t_coffee <seq> -method poa_msa, dialignt_msa, mafft_msa, clustalw_msa, muscle_msa, probcons_msa, t_coffee_msa, pcma_msa.
Using the M-Coffee server
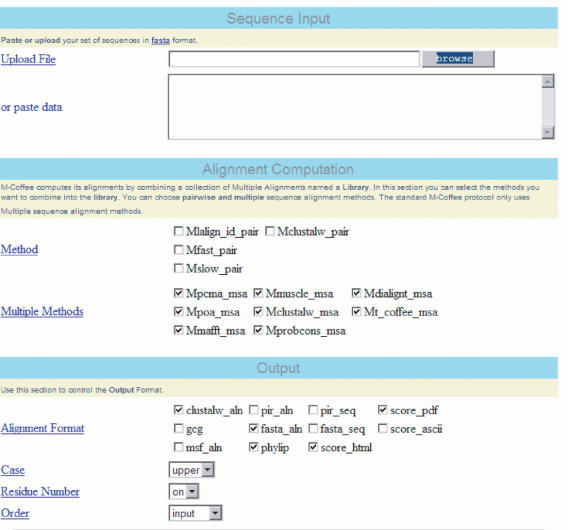
The server can be accessed at www.tcoffee.org. Following the M-Coffee link will either take the user to the regular or advanced mode. The regular mode merely requires the user to cut and paste a set of sequences in FASTA format. The advanced mode (Figure 1) offers more possibilities and guides the user with a series of bulleted points:
Cut and paste your sequences. Sequences should be in FASTA format. Duplicated names are now supported although not recommended.
Alignment computation. This section defines the way the primary library is computed. For instance, selecting only lalign_id_pair and slow_pair will lead to the computation of a regular T-Coffee MSA. The lower section (xxx_msa) displays the list of available MSA methods. Selecting only one of these methods will generate the corresponding alignment. Selecting several methods (or all of them, as in the regular mode displayed on Figure 1) will lead to a consensus T-Coffee MSA. If the MSA method one wants to combine is missing on this form, another server named ‘Combine’ should be used (accessible from www.tcoffee.org). The ‘Combine’ server works on the same principle as M-Coffee but does not compute the MSAs itself and requires the user to cut and paste pre-computed MSAs. At this point it should be used if one wants to incorporate specific constraints or structure-based sequence alignments.
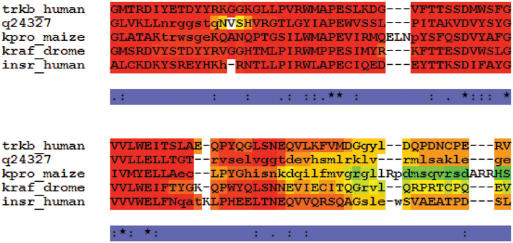
Output. The Output section makes it possible to control the output format. The most notable element is score_html that will cause the server to produce a colored version of the final alignment (Figure 2). In this output, residues are individually colored according to the consistency of their alignment with the T-Coffee library. Residues in red are in perfect agreement with every constituting multiple alignment while those in blue have the lowest agreement (i.e. the lowest support in the individual MSAs). Previous analysis indicates that 90% of the residues having a score of 7 or higher (dark yellow, orange and red) are correctly aligned (24). A text version of this output is available as score_ascii where each residue is replaced with its consistency estimation on a scale between 0 and 9 (9 corresponding to the red-brick residues in the color-output). These score_ascii files can be used to process multiple alignments (block extraction) using seq_reformat, one of the utilities distributed along with T-Coffee. For this purpose, users can download their alignment, the score_ascii file and use the command line version of T-Coffee with the following syntax:
Figure 1.
Method selection on the advanced M-Coffee server form. Each check box corresponds to either a pairwise (_pair) or a multiple sequence alignment method (_msa). Users should choose their methods of choice in order to combine them.
Figure 2.
Typical colored output. This output was obtained by using the kinase1_ref5 from BaliBase. Correctly aligned residues (as judged from the reference) are in upper case, non-correct ones are in lower case. In this colored output, each residue has a color that indicates the agreement of the individual MSAs with respect to the alignment of that specific residue. Dark red indicates residues aligned in a similar fashion among all the individual MSAs; blue indicates a very low agreement. Dark yellow, orange and red residues can be considered to be reliably aligned.
t_coffee -other_pg seq_reformat -in <aln> -struc_in <score_ascii> -struc_in_f number_aln -action +keep ‘[5-9]’
Where <aln> is the name of the alignment and <score_ascii> the name of the score_asccii file. This syntax will replace by a gap (‘-') every residue having an ascii_score lower than 5 (green and blue residues on the colored output).
CONCLUSION AND FUTURE DEVELOPMENTS
M-Coffee provides biologists with a useful alternative to the a priori choice of an MSA method. Although M-Coffee does not entirely solve the question of which method should be used, its local scoring scheme makes it easier to read the alignment and determine which portions are the most informative. Further developments will include making more methods available, as well as making it possible to combine sequences and structures, using the Expresso protocol.
ACKNOWLEDGEMENTS
The development of the server was supported by CNRS (Centre National de la Recherche Scientifique), by the Vital-IT frame work and by the European Union (ICGR-SIB FP6-026204). We thank Dr Pierre Pontarotti and Dr Vladimir Saudek for stimulating discussions. D.G.H. and I.M.W. are grateful to Science Foundation Ireland for funding. Funding to pay the Open Access publication charges for this article was provided by Départment des Science de la Vie, Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
REFERENCES
- 1.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 2.Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edgar RC, Batzoglou S. Multiple sequence alignment. Curr. Opin. Struct. Biol. 2006;16:368–373. doi: 10.1016/j.sbi.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Wallace IM, Blackshields G, Higgins DG. Multiple sequence alignments. Curr. Opin. Struct. Biol. 2005;15:261–266. doi: 10.1016/j.sbi.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Thompson JD, Koehl P, Ripp R, Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 6.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. Print 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuguchi K, Deane CM, Blundell TL, Overington JP. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghava GP, Searle SM, Audley PC, Barber JD, Barton GJ. OXBench: A benchmark for evaluation of protein multiple sequence alignment accuracy. BMC Bioinformatics. 2003;4:47. doi: 10.1186/1471-2105-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen JE, Salzberg SL. JIGSAW: integration of multiple sources of evidence for gene prediction. Bioinformatics. 2005;21:3596–3603. doi: 10.1093/bioinformatics/bti609. [DOI] [PubMed] [Google Scholar]
- 10.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 11.Bucka-Lassen K, Caprani O, Hein J. Combining many multiple alignments in one improved alignment. Bioinformatics. 1999;15:122–130. doi: 10.1093/bioinformatics/15.2.122. [DOI] [PubMed] [Google Scholar]
- 12.Gotoh O. Consistency of optimal sequence alignments. Bull. Math. Biol. 1990;52:509–525. doi: 10.1007/BF02462264. [DOI] [PubMed] [Google Scholar]
- 13.Vingron M, Argos P. Motif recognition and alignment for many sequences by comparison of dot-matrices. J. Mol. Biol. 1991;218:33–43. doi: 10.1016/0022-2836(91)90871-3. [DOI] [PubMed] [Google Scholar]
- 14.Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment [In Process Citation] Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 15.Pei J, Sadreyev R, Grishin NV. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 16.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notredame C, Abergel C. In: Bioinformatics and Genomes: Current Perspectives. Andrade M, editor. Horizon Scientific Press; 2003. pp. 30–50. [Google Scholar]
- 20.Lassmann T, Sonnhammer EL. Automatic assessment of alignment quality. Nucleic Acids Res. 2005;33:7120–7128. doi: 10.1093/nar/gki1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18:452–464. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian AR, Weyer-Menkhoff J, Kaufmann M, Morgenstern B. DIALIGN-T: An improved algorithm for segment-based multiple sequence alignment. BMC Bioinformatics. 2005;6:66. doi: 10.1186/1471-2105-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4690. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abergel C, Coutard B, Byrne D, Chenivesse S, Claude JB, Deregnaucourt C, Fricaux T, Gianesini-Boutreux C, Jeudy S, et al. Structural genomics of highly conserved microbial genes of unknown function in search of new antibacterial targets. J. Struct. Funct. Genomics. 2003;4:141–157. doi: 10.1023/a:1026177202925. [DOI] [PubMed] [Google Scholar]